An Overview of the Relevance of Human Gut and Skin Microbiome in Disease: The Influence on Atopic Dermatitis
Abstract
:1. Introduction
2. Human Microbiome
2.1. Microbiome Types
2.1.1. Oral Cavity
2.1.2. Nasopharynx
2.1.3. Skin
2.1.4. Gastrointestinal Tract
2.1.5. Urogenital Tract
3. Microbiome Evolution throughout Life
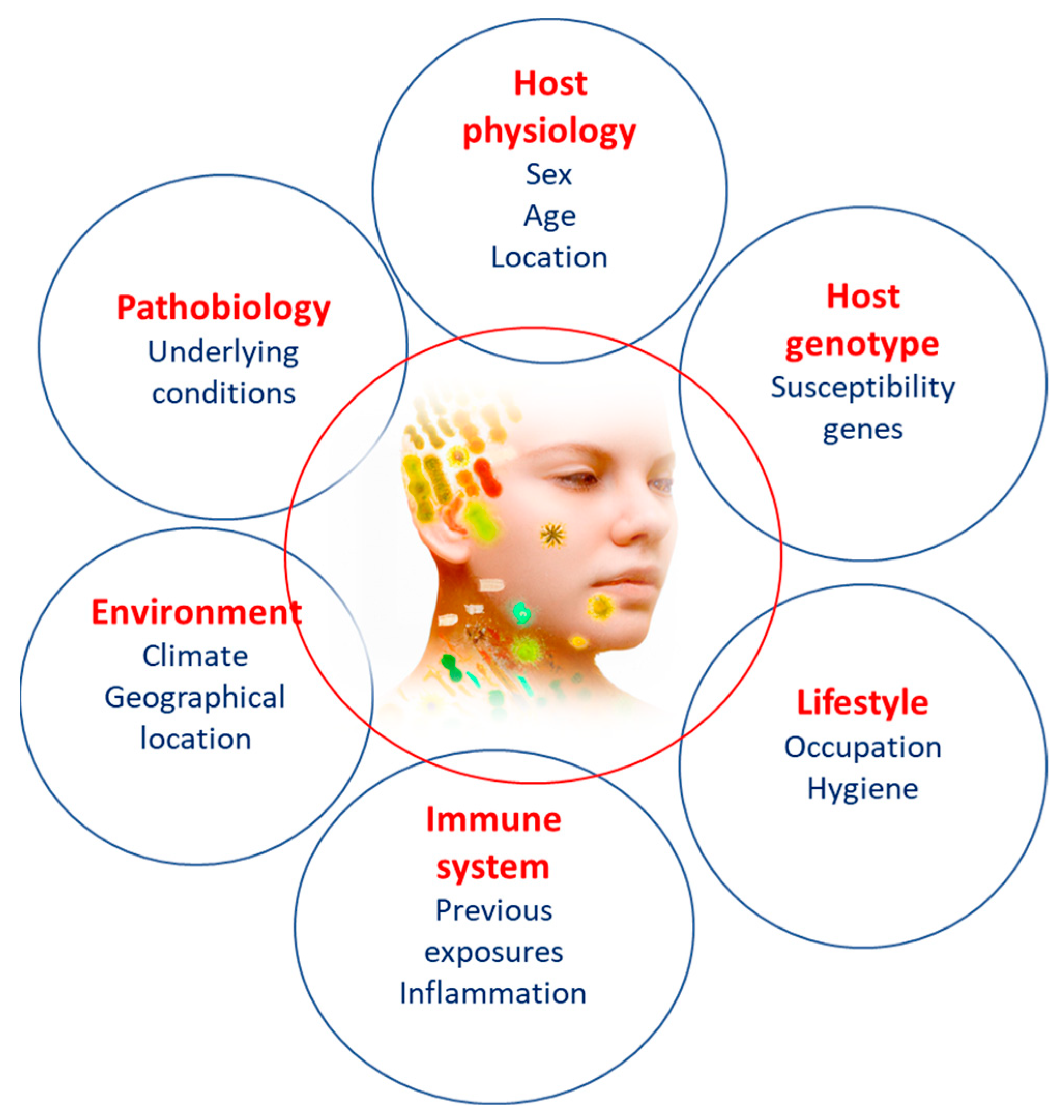
4. Factors Influencing Microbiome
4.1. The Effects of Diet
4.2. Antibiotics and Drugs
4.3. Oxidative Stress
4.4. Socioeconomic Status
5. Microbiome Influence on Atopic Dermatitis
5.1. Atopic Dermatitis
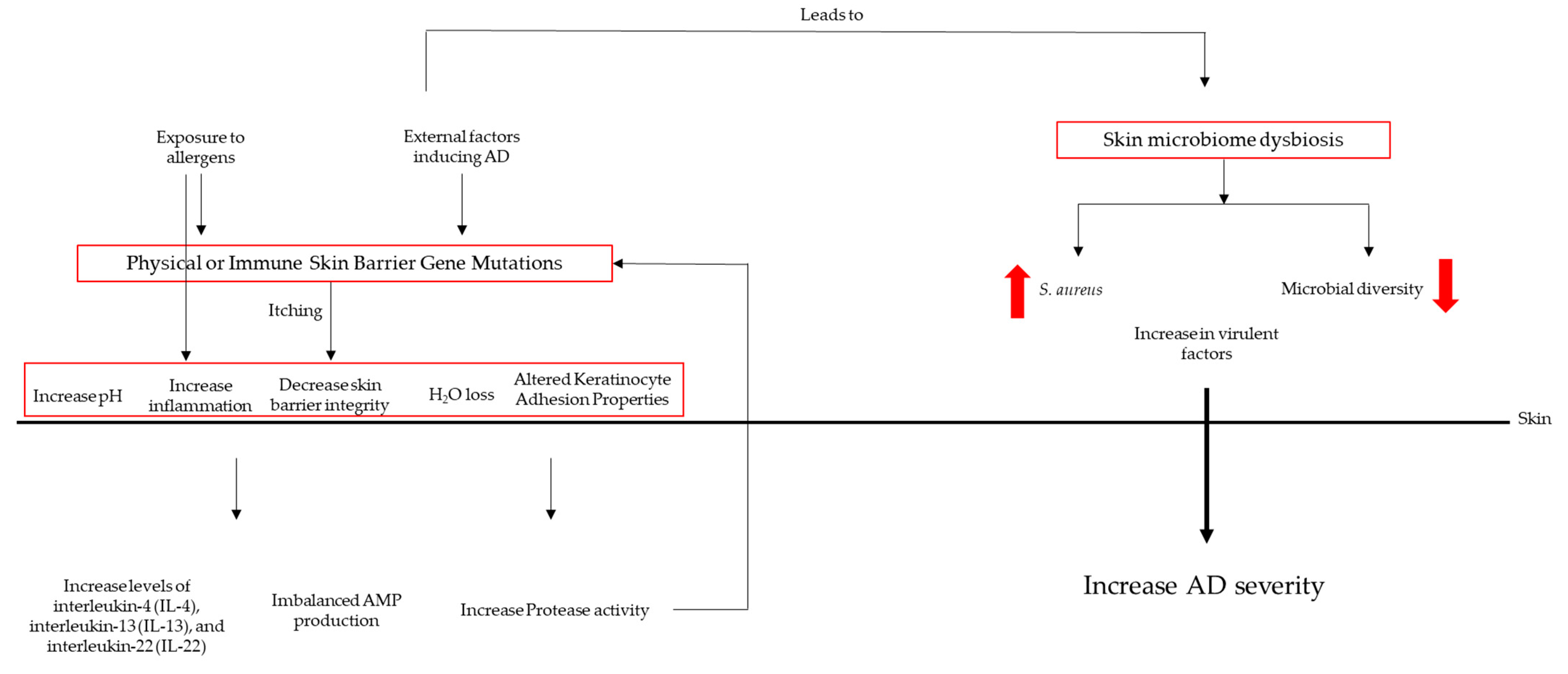
5.2. Dysbiosis in AD Patients
5.3. Microbial Colonization
5.4. The Role of S. aureus in Atopic Dermatitis
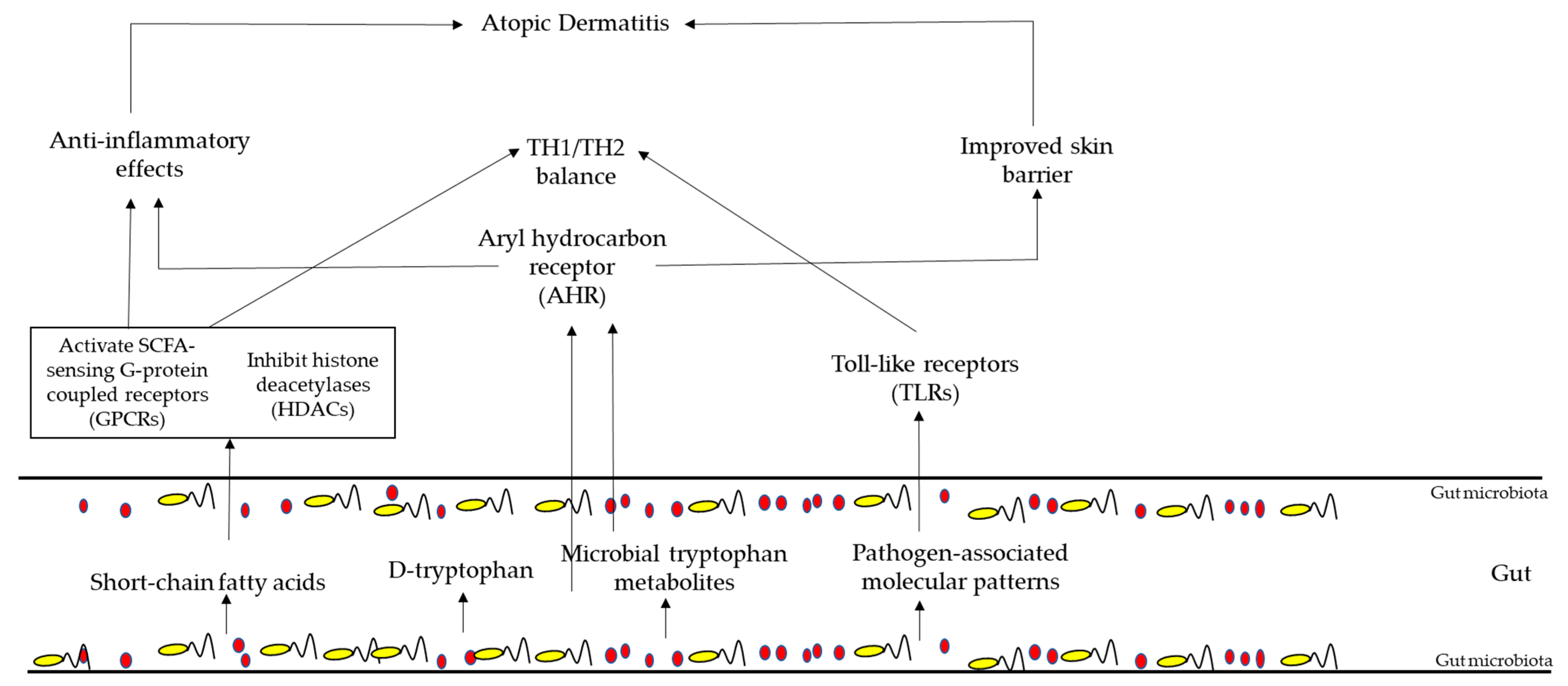
5.5. Gut Microbiota in AD Patient
6. Therapies Targeting Gut Microbiota’s Composition
Therapies Targeting Microbiota Composition in Atopic Dermatitis
7. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ursell, L.K.; Metcalf, J.L.; Parfrey, L.W.; Knight, R. Defining the human microbiome. Nutr. Rev. 2012, 70 (Suppl. S1), S38–S44. [Google Scholar] [CrossRef]
- Khanna, S.; Tosh, P.K. A clinician’s primer on the role of the microbiome in human health and disease. Mayo Clin. Proc. 2014, 89, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Vijay, A.; Valdes, A.M. Role of the gut microbiome in chronic diseases: A narrative review. Eur. J. Clin. Nutr. 2022, 76, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Xia, Y.L. Microbiome in Colonic Carcinogenesis. Compr. Physiol. 2023, 13, 4685–4708. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Birmingham, A.; Knight, R. Context and the human microbiome. Microbiome 2015, 3, 52. [Google Scholar] [CrossRef]
- Gupta, V.K.; Paul, S.; Dutta, C. Geography, Ethnicity or Subsistence-Specific Variations in Human Microbiome Composition and Diversity. Front. Microbiol. 2017, 8, 1162. [Google Scholar] [CrossRef]
- Pasolli, E.; Asnicar, F.; Manara, S.; Zolfo, M.; Karcher, N.; Armanini, F.; Beghini, F.; Manghi, P.; Tett, A.; Ghensi, P.; et al. Extensive Unexplored Human Microbiome Diversity Revealed by Over 150,000 Genomes from Metagenomes Spanning Age, Geography, and Lifestyle. Cell 2019, 176, 649–662.e20. [Google Scholar] [CrossRef]
- Honda, K.; Littman, D.R. The microbiota in adaptive immune homeostasis and disease. Nature 2016, 535, 75–84. [Google Scholar] [CrossRef]
- Chong-Nguyen, C.; Duboc, H.; Sokol, H. The gut microbiota, a new cardiovascular risk factor? Presse Med. 2017, 46, 708–713. [Google Scholar] [CrossRef]
- Griffin, J.L.; Wang, X.; Stanley, E. Does our gut microbiome predict cardiovascular risk? A review of the evidence from metabolomics. Circ. Cardiovasc. Genet. 2015, 8, 187–191. [Google Scholar] [CrossRef]
- Griffiths, K.; Aggarwal, B.B.; Singh, R.B.; Buttar, H.S.; Wilson, D.; De Meester, F. Food Antioxidants and Their Anti-Inflammatory Properties: A Potential Role in Cardiovascular Diseases and Cancer Prevention. Diseases 2016, 4, 28. [Google Scholar] [CrossRef] [PubMed]
- Hrestak, D.; Matijasic, M.; Cipcic Paljetak, H.; Ledic Drvar, D.; Ljubojevic Hadzavdic, S.; Peric, M. Skin Microbiota in Atopic Dermatitis. Int. J. Mol. Sci. 2022, 23, 3503. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Zhang, L.; Chen, Y.; Wang, H.; Xie, J. Gut microbiota and atopic dermatitis: A two-sample Mendelian randomization study. Front. Med. 2023, 10, 1174331. [Google Scholar] [CrossRef] [PubMed]
- Pothmann, A.; Illing, T.; Wiegand, C.; Hartmann, A.A.; Elsner, P. The Microbiome and Atopic Dermatitis: A Review. Am. J. Clin. Dermatol. 2019, 20, 749–761. [Google Scholar] [CrossRef]
- Demessant-Flavigny, A.L.; Connetable, S.; Kerob, D.; Moreau, M.; Aguilar, L.; Wollenberg, A. Skin microbiome dysbiosis and the role of Staphylococcus aureus in atopic dermatitis in adults and children: A narrative review. J. Eur. Acad. Dermatol. Venereol. 2023, 37 (Suppl. S5), 3–17. [Google Scholar] [CrossRef]
- Mazur, M.; Tomczak, H.; Lodyga, M.; Plagens-Rotman, K.; Merks, P.; Czarnecka-Operacz, M. The Intestinal and Skin Microbiome in Patients with Atopic Dermatitis and Their Influence on the Course of the Disease: A Literature Review. Healthcare 2023, 11, 766. [Google Scholar] [CrossRef]
- Walker, R.L.; Vlamakis, H.; Lee, J.W.J.; Besse, L.A.; Xanthakis, V.; Vasan, R.S.; Shaw, S.Y.; Xavier, R.J. Population study of the gut microbiome: Associations with diet, lifestyle, and cardiometabolic disease. Genome. Med. 2021, 13, 188. [Google Scholar] [CrossRef]
- Aggarwal, N.; Kitano, S.; Puah, G.R.Y.; Kittelmann, S.; Hwang, I.Y.; Chang, M.W. Microbiome and Human Health: Current Understanding, Engineering, and Enabling Technologies. Chem. Rev. 2023, 123, 31–72. [Google Scholar] [CrossRef]
- Morgan, X.C.; Segata, N.; Huttenhower, C. Biodiversity and functional genomics in the human microbiome. Trends. Genet. 2013, 29, 51–58. [Google Scholar] [CrossRef]
- Peroni, D.G.; Nuzzi, G.; Trambusti, I.; Di Cicco, M.E.; Comberiati, P. Microbiome Composition and Its Impact on the Development of Allergic Diseases. Front. Immunol. 2020, 11, 700. [Google Scholar] [CrossRef]
- Hou, K.; Wu, Z.X.; Chen, X.Y.; Wang, J.Q.; Zhang, D.; Xiao, C.; Zhu, D.; Koya, J.B.; Wei, L.; Li, J.; et al. Microbiota in health and diseases. Signal. Transduct. Target. Ther. 2022, 7, 135. [Google Scholar] [CrossRef] [PubMed]
- Willis, J.R.; Gabaldon, T. The Human Oral Microbiome in Health and Disease: From Sequences to Ecosystems. Microorganisms 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Avila, M.; Ojcius, D.M.; Yilmaz, O. The oral microbiota: Living with a permanent guest. DNA Cell Biol. 2009, 28, 405–411. [Google Scholar] [CrossRef] [PubMed]
- Jenkinson, H.F.; Lamont, R.J. Oral microbial communities in sickness and in health. Trends Microbiol. 2005, 13, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; He, J.; Xue, J.; Wang, Y.; Li, K.; Zhang, K.; Guo, Q.; Liu, X.; Zhou, Y.; Cheng, L.; et al. Oral cavity contains distinct niches with dynamic microbial communities. Environ. Microbiol. 2015, 17, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Struzycka, I. The oral microbiome in dental caries. Pol. J. Microbiol. 2014, 63, 127–135. [Google Scholar] [CrossRef]
- Zhang, J.S.; Chu, C.H.; Yu, O.Y. Oral Microbiome and Dental Caries Development. Dent. J. 2022, 10, 184. [Google Scholar] [CrossRef]
- Zheng, L.; Cao, T.; Xiong, P.; Ma, Y.; Wei, L.; Wang, J. Characterization of the oral microbiome and gut microbiome of dental caries and extrinsic black stain in preschool children. Front. Microbiol. 2023, 14, 1081629. [Google Scholar] [CrossRef]
- Jhajharia, K.; Parolia, A.; Shetty, K.V.; Mehta, L.K. Biofilm in endodontics: A review. J. Int. Soc. Prev. Community Dent. 2015, 5, 1–12. [Google Scholar] [CrossRef]
- Hao, Y.; Zeng, Z.; Peng, X.; Ai, P.; Han, Q.; Ren, B.; Li, M.; Wang, H.; Zhou, X.; Zhou, X.; et al. The human oral-nasopharynx microbiome as a risk screening tool for nasopharyngeal carcinoma. Front. Cell. Infect. Microbiol. 2022, 12, 1013920. [Google Scholar] [CrossRef]
- Wang, J.; Feng, J.; Zhu, Y.; Li, D.; Wang, J.; Chi, W. Diversity and Biogeography of Human Oral Saliva Microbial Communities Revealed by the Earth Microbiome Project. Front. Microbiol. 2022, 13, 931065. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.G.; Sarkar, S.; Umar, S.; Lee, S.T.M.; Thomas, S.M. The Contribution of the Human Oral Microbiome to Oral Disease: A Review. Microorganisms 2023, 11, 318. [Google Scholar] [CrossRef] [PubMed]
- Dewhirst, F.E.; Chen, T.; Izard, J.; Paster, B.J.; Tanner, A.C.; Yu, W.H.; Lakshmanan, A.; Wade, W.G. The human oral microbiome. J. Bacteriol. 2010, 192, 5002–5017. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ganly, I. The oral microbiome and oral cancer. Clin. Lab. Med. 2014, 34, 711–719. [Google Scholar] [CrossRef]
- Nearing, J.T.; DeClercq, V.; Langille, M.G.I. Investigating the oral microbiome in retrospective and prospective cases of prostate, colon, and breast cancer. NPJ Biofilms Microbiomes 2023, 9, 23. [Google Scholar] [CrossRef]
- Smedra, A.; Berent, J. The Influence of the Oral Microbiome on Oral Cancer: A Literature Review and a New Approach. Biomolecules 2023, 13, 815. [Google Scholar] [CrossRef]
- Thu, M.S.; Chotirosniramit, K.; Nopsopon, T.; Hirankarn, N.; Pongpirul, K. Human gut, breast, and oral microbiome in breast cancer: A systematic review and meta-analysis. Front. Oncol. 2023, 13, 1144021. [Google Scholar] [CrossRef]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef]
- Bryan, N.S.; Tribble, G.; Angelov, N. Oral Microbiome and Nitric Oxide: The Missing Link in the Management of Blood Pressure. Curr. Hypertens Rep. 2017, 19, 33. [Google Scholar] [CrossRef]
- Bui, F.Q.; Almeida-da-Silva, C.L.C.; Huynh, B.; Trinh, A.; Liu, J.; Woodward, J.; Asadi, H.; Ojcius, D.M. Association between periodontal pathogens and systemic disease. Biomed. J. 2019, 42, 27–35. [Google Scholar] [CrossRef]
- Sanz, M.; Marco Del Castillo, A.; Jepsen, S.; Gonzalez-Juanatey, J.R.; D’Aiuto, F.; Bouchard, P.; Chapple, I.; Dietrich, T.; Gotsman, I.; Graziani, F.; et al. Periodontitis and cardiovascular diseases: Consensus report. J. Clin. Periodontol. 2020, 47, 268–288. [Google Scholar] [CrossRef] [PubMed]
- Jourdain, S.; Smeesters, P.R.; Denis, O.; Dramaix, M.; Sputael, V.; Malaviolle, X.; Van Melderen, L.; Vergison, A. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin. Microbiol. Infect. 2011, 17, 907–914. [Google Scholar] [CrossRef] [PubMed]
- Jourdain, S.; Dreze, P.A.; Vandeven, J.; Verhaegen, J.; Van Melderen, L.; Smeesters, P.R. Sequential multiplex PCR assay for determining capsular serotypes of colonizing S. pneumoniae. BMC Infect. Dis. 2011, 11, 100. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Kang, J.H. Effects of nasopharyngeal microbiota in respiratory infections and allergies. Clin. Exp. Pediatr. 2021, 64, 543–551. [Google Scholar] [CrossRef]
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef]
- Dickson, R.P.; Erb-Downward, J.R.; Martinez, F.J.; Huffnagle, G.B. The Microbiome and the Respiratory Tract. Annu. Rev. Physiol. 2016, 78, 481–504. [Google Scholar] [CrossRef]
- Rosenthal, M.; Goldberg, D.; Aiello, A.; Larson, E.; Foxman, B. Skin microbiota: Microbial community structure and its potential association with health and disease. Infect Genet. Evol. 2011, 11, 839–848. [Google Scholar] [CrossRef]
- Grice, E.A.; Segre, J.A. The skin microbiome. Nat. Rev. Microbiol. 2011, 9, 244–253. [Google Scholar] [CrossRef]
- Santiago-Rodriguez, T.M.; Le Francois, B.; Macklaim, J.M.; Doukhanine, E.; Hollister, E.B. The Skin Microbiome: Current Techniques, Challenges, and Future Directions. Microorganisms 2023, 11, 1222. [Google Scholar] [CrossRef]
- Oh, J.; Byrd, A.L.; Park, M.; Program, N.C.S.; Kong, H.H.; Segre, J.A. Temporal Stability of the Human Skin Microbiome. Cell 2016, 165, 854–866. [Google Scholar] [CrossRef]
- Skowron, K.; Bauza-Kaszewska, J.; Kraszewska, Z.; Wiktorczyk-Kapischke, N.; Grudlewska-Buda, K.; Kwiecinska-Pirog, J.; Walecka-Zacharska, E.; Radtke, L.; Gospodarek-Komkowska, E. Human Skin Microbiome: Impact of Intrinsic and Extrinsic Factors on Skin Microbiota. Microorganisms 2021, 9, 543. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A. The intersection of microbiome and host at the skin interface: Genomic- and metagenomic-based insights. Genome Res. 2015, 25, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Grice, E.A.; Kong, H.H.; Conlan, S.; Deming, C.B.; Davis, J.; Young, A.C.; Program, N.C.S.; Bouffard, G.G.; Blakesley, R.W.; Murray, P.R.; et al. Topographical and temporal diversity of the human skin microbiome. Science 2009, 324, 1190–1192. [Google Scholar] [CrossRef] [PubMed]
- Capone, K.A.; Dowd, S.E.; Stamatas, G.N.; Nikolovski, J. Diversity of the human skin microbiome early in life. J. Invest Dermatol. 2011, 131, 2026–2032. [Google Scholar] [CrossRef]
- Oh, J.; Conlan, S.; Polley, E.C.; Segre, J.A.; Kong, H.H. Shifts in human skin and nares microbiota of healthy children and adults. Genome. Med. 2012, 4, 77. [Google Scholar] [CrossRef]
- Holland, K.T.; Bojar, R.A. Cosmetics: What is their influence on the skin microflora? Am. J. Clin. Dermatol. 2002, 3, 445–449. [Google Scholar] [CrossRef]
- Costello, E.K.; Lauber, C.L.; Hamady, M.; Fierer, N.; Gordon, J.I.; Knight, R. Bacterial community variation in human body habitats across space and time. Science 2009, 326, 1694–1697. [Google Scholar] [CrossRef]
- Byrd, A.L.; Belkaid, Y.; Segre, J.A. The human skin microbiome. Nat. Rev. Microbiol. 2018, 16, 143–155. [Google Scholar] [CrossRef]
- Schommer, N.N.; Gallo, R.L. Structure and function of the human skin microbiome. Trends Microbiol. 2013, 21, 660–668. [Google Scholar] [CrossRef]
- Kim, G.; Kim, M.; Kim, M.; Park, C.; Yoon, Y.; Lim, D.H.; Yeo, H.; Kang, S.; Lee, Y.G.; Beak, N.I.; et al. Spermidine-induced recovery of human dermal structure and barrier function by skin microbiome. Commun. Biol. 2021, 4, 231. [Google Scholar] [CrossRef]
- Gao, Z.; Tseng, C.H.; Pei, Z.; Blaser, M.J. Molecular analysis of human forearm superficial skin bacterial biota. Proc. Natl. Acad. Sci. USA 2007, 104, 2927–2932. [Google Scholar] [CrossRef] [PubMed]
- Schneider, A.M.; Nelson, A.M. Skin microbiota: Friend or foe in pediatric skin health and skin disease. Pediatr. Dermatol. 2019, 36, 815–822. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4578–4585. [Google Scholar] [CrossRef]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
- Sidebottom, A.M. A Brief History of Microbial Study and Techniques for Exploring the Gastrointestinal Microbiome. Clin. Colon. Rectal. Surg. 2023, 36, 98–104. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, S.; Zou, H.; Han, Z.; Xie, T.; Zhang, B.; Dai, D.; Yin, X.; Liang, Y.; Kou, Y.; et al. Correlation of the gut microbiome and immune-related adverse events in gastrointestinal cancer patients treated with immune checkpoint inhibitors. Front. Cell. Infect. Microbiol. 2023, 13, 1099063. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Blaser, M.J.; Ley, R.E.; Knight, R. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology 2011, 140, 1713–1719. [Google Scholar] [CrossRef]
- Yamashita, T.; Emoto, T.; Sasaki, N.; Hirata, K.I. Gut Microbiota and Coronary Artery Disease. Int. Heart J. 2016, 57, 663–671. [Google Scholar] [CrossRef]
- Dong, C.; Yang, Y.; Wang, Y.; Hu, X.; Wang, Q.; Gao, F.; Sun, S.; Liu, Q.; Li, L.; Liu, J.; et al. Gut microbiota combined with metabolites reveals unique features of acute myocardial infarction patients different from stable coronary artery disease. J. Adv. Res. 2023, 46, 101–112. [Google Scholar] [CrossRef]
- Arora, T.; Sharma, R. Fermentation potential of the gut microbiome: Implications for energy homeostasis and weight management. Nutr. Rev. 2011, 69, 99–106. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Louis, P.; Duncan, S.H. The role of the gut microbiota in nutrition and health. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef] [PubMed]
- Wan, Z.; Zheng, J.; Zhu, Z.; Sang, L.; Zhu, J.; Luo, S.; Zhao, Y.; Wang, R.; Zhang, Y.; Hao, K.; et al. Intermediate role of gut microbiota in vitamin B nutrition and its influences on human health. Front. Nutr. 2022, 9, 1031502. [Google Scholar] [CrossRef] [PubMed]
- Prakash, S.; Rodes, L.; Coussa-Charley, M.; Tomaro-Duchesneau, C. Gut microbiota: Next frontier in understanding human health and development of biotherapeutics. Biologics 2011, 5, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, M.C. What is the Healthy Gut Microbiota Composition? A Changing Ecosystem across Age, Environment, Diet, and Diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Tsukumo, D.M.; Carvalho, B.M.; Carvalho Filho, M.A.; Saad, M.J. Translational research into gut microbiota: New horizons on obesity treatment: Updated 2014. Arch. Endocrinol. Metab. 2015, 59, 154–160. [Google Scholar] [CrossRef]
- Sartor, R.B. Microbial influences in inflammatory bowel diseases. Gastroenterology 2008, 134, 577–594. [Google Scholar] [CrossRef]
- Delgado, S.; Cabrera-Rubio, R.; Mira, A.; Suarez, A.; Mayo, B. Microbiological survey of the human gastric ecosystem using culturing and pyrosequencing methods. Microb. Ecol. 2013, 65, 763–772. [Google Scholar] [CrossRef]
- Walker, A.W.; Duncan, S.H.; Louis, P.; Flint, H.J. Phylogeny, culturing, and metagenomics of the human gut microbiota. Trends Microbiol. 2014, 22, 267–274. [Google Scholar] [CrossRef]
- Turroni, F.; Ozcan, E.; Milani, C.; Mancabelli, L.; Viappiani, A.; van Sinderen, D.; Sela, D.A.; Ventura, M. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 2015, 6, 1030. [Google Scholar] [CrossRef]
- Riviere, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and Butyrate-Producing Colon Bacteria: Importance and Strategies for Their Stimulation in the Human Gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Fakharian, F.; Thirugnanam, S.; Welsh, D.A.; Kim, W.K.; Rappaport, J.; Bittinger, K.; Rout, N. The Role of Gut Dysbiosis in the Loss of Intestinal Immune Cell Functions and Viral Pathogenesis. Microorganisms 2023, 11, 1849. [Google Scholar] [CrossRef]
- Parfrey, L.W.; Knight, R. Spatial and temporal variability of the human microbiota. Clin. Microbiol. Infect. 2012, 18 (Suppl. S4), 8–11. [Google Scholar] [CrossRef]
- Whiteside, S.A.; Razvi, H.; Dave, S.; Reid, G.; Burton, J.P. The microbiome of the urinary tract—A role beyond infection. Nat. Rev. Urol. 2015, 12, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.S.; Lee, J.W. Urinary Tract Infection and Microbiome. Diagnostics 2023, 13, 1921. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.; Kasper, D.L. Microbiota-stimulated immune mechanisms to maintain gut homeostasis. Curr. Opin. Immunol. 2010, 22, 455–460. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.M.; Hilt, E.E.; Rosenfeld, A.B.; Zilliox, M.J.; Thomas-White, K.; Fok, C.; Kliethermes, S.; Schreckenberger, P.C.; Brubaker, L.; Gai, X.; et al. The female urinary microbiome: A comparison of women with and without urgency urinary incontinence. mBio 2014, 5, e01283-01214. [Google Scholar] [CrossRef]
- Karstens, L.; Asquith, M.; Davin, S.; Stauffer, P.; Fair, D.; Gregory, W.T.; Rosenbaum, J.T.; McWeeney, S.K.; Nardos, R. Does the Urinary Microbiome Play a Role in Urgency Urinary Incontinence and Its Severity? Front. Cell. Infect. Microbiol. 2016, 6, 78. [Google Scholar] [CrossRef]
- Mestrovic, T.; Matijasic, M.; Peric, M.; Cipcic Paljetak, H.; Baresic, A.; Verbanac, D. The Role of Gut, Vaginal, and Urinary Microbiome in Urinary Tract Infections: From Bench to Bedside. Diagnostics 2020, 11, 7. [Google Scholar] [CrossRef]
- Ling, Z.; Kong, J.; Liu, F.; Zhu, H.; Chen, X.; Wang, Y.; Li, L.; Nelson, K.E.; Xia, Y.; Xiang, C. Molecular analysis of the diversity of vaginal microbiota associated with bacterial vaginosis. BMC Genom. 2010, 11, 488. [Google Scholar] [CrossRef] [PubMed]
- Walther-Antonio, M.R.; Jeraldo, P.; Berg Miller, M.E.; Yeoman, C.J.; Nelson, K.E.; Wilson, B.A.; White, B.A.; Chia, N.; Creedon, D.J. Pregnancy’s stronghold on the vaginal microbiome. PLoS ONE 2014, 9, e98514. [Google Scholar] [CrossRef] [PubMed]
- Walker, R.W.; Clemente, J.C.; Peter, I.; Loos, R.J.F. The prenatal gut microbiome: Are we colonized with bacteria in utero? Pediatr. Obes. 2017, 12 (Suppl. S1), 3–17. [Google Scholar] [CrossRef] [PubMed]
- Senn, V.; Bassler, D.; Choudhury, R.; Scholkmann, F.; Righini-Grunder, F.; Vuille-Dit-Bile, R.N.; Restin, T. Microbial Colonization From the Fetus to Early Childhood-A Comprehensive Review. Front. Cell. Infect. Microbiol. 2020, 10, 573735. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.; Corwin, E.J.; Brennan, P.A.; Jordan, S.; Murphy, J.R.; Dunlop, A. The Infant Microbiome: Implications for Infant Health and Neurocognitive Development. Nurs. Res. 2016, 65, 76–88. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Florova, V.; Romero, R.; Borisov, A.B.; Winters, A.D.; Galaz, J.; Gomez-Lopez, N. Sneathia: An emerging pathogen in female reproductive disease and adverse perinatal outcomes. Crit. Rev. Microbiol. 2021, 47, 517–542. [Google Scholar] [CrossRef]
- Walsh, C.; Lane, J.A.; van Sinderen, D.; Hickey, R.M. Human milk oligosaccharides: Shaping the infant gut microbiota and supporting health. J. Funct. Foods 2020, 72, 104074. [Google Scholar] [CrossRef]
- Rodriguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Health Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Fiocchi, A.; Burks, W.; Bahna, S.L.; Bielory, L.; Boyle, R.J.; Cocco, R.; Dreborg, S.; Goodman, R.; Kuitunen, M.; Haahtela, T.; et al. Clinical Use of Probiotics in Pediatric Allergy (CUPPA): A World Allergy Organization Position Paper. World Allergy Organ. J. 2012, 5, 148–167. [Google Scholar] [CrossRef]
- Smythe, P.; Wilkinson, H.N. The Skin Microbiome: Current Landscape and Future Opportunities. Int. J. Mol. Sci. 2023, 24, 3950. [Google Scholar] [CrossRef]
- Bajinka, O.; Tan, Y.; Abdelhalim, K.A.; Ozdemir, G.; Qiu, X. Extrinsic factors influencing gut microbes, the immediate consequences and restoring eubiosis. AMB Express 2020, 10, 130. [Google Scholar] [CrossRef] [PubMed]
- Zoetendal, E.G.; Raes, J.; van den Bogert, B.; Arumugam, M.; Booijink, C.C.G.M.; Troost, F.J.; Bork, P.; Wels, M.; de Vos, W.M.; Kleerebezem, M. The human small intestinal microbiota is driven by rapid uptake and conversion of simple carbohydrates. ISME J. 2012, 6, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.J.; Ye, Q.N.; Sun, M.X.; Wang, L.L.; Tan, Y.R.; Wu, G.J. Saturated hydrogen improves lipid metabolism disorders and dysbacteriosis induced by a high-fat diet. Exp. Biol. Med. 2020, 245, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Silva, F.M.D.E.; de Oliveira, E.E.; Ambrosio, M.G.E.; Ayupe, M.C.; de Souza, V.P.; Gameiro, J.; Reis, D.R.D.; Machado, M.A.; Macedo, G.C.; Mattes, J.; et al. High-fat diet-induced obesity worsens TH2 immune response and immunopathologic characteristics in murine model of eosinophilic oesophagitis. Clin. Exp. Allergy 2020, 50, 244–255. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, M.; Pang, X.; Zhao, Y.; Wang, L.; Zhao, L. Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 2012, 6, 1848–1857. [Google Scholar] [CrossRef]
- Nettleton, J.E.; Reimer, R.A.; Shearer, J. Reshaping the gut microbiota: Impact of low calorie sweeteners and the link to insulin resistance? Physiol. Behav. 2016, 164, 488–493. [Google Scholar] [CrossRef]
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef]
- Rogers, M.A.M.; Aronoff, D.M. The influence of non-steroidal anti-inflammatory drugs on the gut microbiome. Clin. Microbiol. Infect. 2016, 22, 178.e171–178.e179. [Google Scholar] [CrossRef]
- Freedberg, D.E.; Toussaint, N.C.; Chen, S.P.; Ratner, A.J.; Whittier, S.; Wang, T.C.; Wang, H.H.; Abrams, J.A. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology 2015, 149, 883–885.e9. [Google Scholar] [CrossRef]
- Vich Vila, A.; Collij, V.; Sanna, S.; Sinha, T.; Imhann, F.; Bourgonje, A.R.; Mujagic, Z.; Jonkers, D.; Masclee, A.A.M.; Fu, J.; et al. Impact of commonly used drugs on the composition and metabolic function of the gut microbiota. Nat. Commun. 2020, 11, 362. [Google Scholar] [CrossRef]
- Ni, Q.; Zhang, P.; Li, Q.; Han, Z. Oxidative Stress and Gut Microbiome in Inflammatory Skin Diseases. Front. Cell. Dev. Biol. 2022, 10, 849985. [Google Scholar] [CrossRef] [PubMed]
- Bellavia, M.; Tomasello, G.; Romeo, M.; Damiani, P.; Lo Monte, A.I.; Lozio, L.; Campanella, C.; Marino Gammazza, A.; Rappa, F.; Zummo, G.; et al. Gut microbiota imbalance and chaperoning system malfunction are central to ulcerative colitis pathogenesis and can be counteracted with specifically designed probiotics: A working hypothesis. Med. Microbiol. Immunol. 2013, 202, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Li, S.; Yang, L.; Huang, P.; Li, W.; Wang, S.; Zhao, G.; Zhang, M.; Pang, X.; Yan, Z.; et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 2013, 4, 2163. [Google Scholar] [CrossRef] [PubMed]
- Reijnders, D.; Goossens, G.H.; Hermes, G.D.; Neis, E.P.; van der Beek, C.M.; Most, J.; Holst, J.J.; Lenaerts, K.; Kootte, R.S.; Nieuwdorp, M.; et al. Effects of Gut Microbiota Manipulation by Antibiotics on Host Metabolism in Obese Humans: A Randomized Double-Blind Placebo-Controlled Trial. Cell. Metab. 2016, 24, 341. [Google Scholar] [CrossRef]
- Williams, M.R.; Gallo, R.L. The role of the skin microbiome in atopic dermatitis. Curr. Allergy Asthma Rep. 2015, 15, 65. [Google Scholar] [CrossRef]
- Ziehfreund, S.; Tizek, L.; Hangel, N.; Fritzsche, M.C.; Weidinger, S.; Smith, C.; Bryce, P.J.; Greco, D.; van den Bogaard, E.H.; Flohr, C.; et al. Requirements and expectations of high-quality biomarkers for atopic dermatitis and psoriasis in 2021-a two-round Delphi survey among international experts. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1467–1476. [Google Scholar] [CrossRef]
- Harder, I.; Stolzl, D.; Sander, N.; Hartmann, J.; Rodriguez, E.; Mazur, C.; Kerzel, S.; Kabesch, M.; Kuster, D.; Schmitt, J.; et al. Effects of Early Emollient Use in Children at High Risk of Atopic Dermatitis: A German Pilot Study. Acta Derm. Venereol. 2023, 103, adv5671. [Google Scholar] [CrossRef]
- Paller, A.S.; Weidinger, S.; Capozza, K.; Pink, A.E.; Tang, M.; Guillaume, X.; Praestgaard, A.; Leclerc, M.; Chuang, C.C.; Thomas, R.B.; et al. Similarities and Differences in the Perception of Atopic Dermatitis Burden Between Patients, Caregivers, and Independent Physicians (AD-GAP Survey). Dermatol. Ther. 2023, 13, 961–980. [Google Scholar] [CrossRef]
- Kapur, S.; Watson, W.; Carr, S. Atopic dermatitis. Allergy Asthma Clin. Immunol. 2018, 14, 52. [Google Scholar] [CrossRef]
- Weidinger, S.; Novak, N. Atopic dermatitis. Lancet 2016, 387, 1109–1122. [Google Scholar] [CrossRef]
- Tokura, Y.; Hayano, S. Subtypes of atopic dermatitis: From phenotype to endotype. Allergol. Int. 2022, 71, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Blicharz, L.; Rudnicka, L.; Czuwara, J.; Waskiel-Burnat, A.; Goldust, M.; Olszewska, M.; Samochocki, Z. The Influence of Microbiome Dysbiosis and Bacterial Biofilms on Epidermal Barrier Function in Atopic Dermatitis-An Update. Int. J. Mol. Sci. 2021, 22, 8403. [Google Scholar] [CrossRef] [PubMed]
- Lunjani, N.; Ahearn-Ford, S.; Dube, F.S.; Hlela, C.; O’Mahony, L. Mechanisms of microbe-immune system dialogue within the skin. Genes. Immun. 2021, 22, 276–288. [Google Scholar] [CrossRef]
- Kim, K.; Kim, H.; Sung, G.Y. An Interleukin-4 and Interleukin-13 Induced Atopic Dermatitis Human Skin Equivalent Model by a Skin-On-A-Chip. Int. J. Mol. Sci. 2022, 23, 2116. [Google Scholar] [CrossRef]
- Yamasaki, K.; Gallo, R.L. Antimicrobial peptides in human skin disease. Eur. J. Dermatol. 2008, 18, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Villarreal, M.; Stewart, S.; Choi, J.; Ganguli-Indra, G.; Babineau, D.C.; Philpot, C.; David, G.; Yoshida, T.; Boguniewicz, M.; et al. Altered composition of epidermal lipids correlates with Staphylococcus aureus colonization status in atopic dermatitis. Br. J. Dermatol. 2017, 177, e125–e127. [Google Scholar] [CrossRef]
- Tamai, M.; Yamazaki, Y.; Ito, T.; Nakagawa, S.; Nakamura, Y. Pathogenic role of the staphylococcal accessory gene regulator quorum sensing system in atopic dermatitis. Front. Cell Infect. Mi 2023, 13, 435. [Google Scholar] [CrossRef]
- Ring, J.; Ring, J.; Przybilla, B.; Ruzicka, T. Handbook of Atopic Eczema, 2nd ed.; Springer: Berlin/Heidelberg, Germany; New York, NY, USA, 2006. [Google Scholar]
- Paller, A.S.; Kong, H.H.; Seed, P.; Naik, S.; Scharschmidt, T.C.; Gallo, R.L.; Luger, T.; Irvine, A.D. The microbiome in patients with atopic dermatitis. J. Allergy Clin. Immunol. 2019, 143, 26–35. [Google Scholar] [CrossRef]
- Fyhrquist, N.; Muirhead, G.; Prast-Nielsen, S.; Jeanmougin, M.; Olah, P.; Skoog, T.; Jules-Clement, G.; Feld, M.; Barrientos-Somarribas, M.; Sinkko, H.; et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat. Commun. 2019, 10, 4703. [Google Scholar] [CrossRef]
- D’Auria, E.; Banderali, G.; Barberi, S.; Gualandri, L.; Pietra, B.; Riva, E.; Cerri, A. Atopic dermatitis: Recent insight on pathogenesis and novel therapeutic target. Asian Pac. J. Allergy Immunol. 2016, 34, 98–108. [Google Scholar] [CrossRef]
- Park, Y.M.; Lee, S.Y.; Kang, M.J.; Kim, B.S.; Lee, M.J.; Jung, S.S.; Yoon, J.S.; Cho, H.J.; Lee, E.; Yang, S.I.; et al. Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant. Allergy Asthma Immunol. Res. 2020, 12, 322–337. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.J.; Kang, M.J.; Lee, S.Y.; Lee, E.; Kim, K.; Won, S.; Suh, D.I.; Kim, K.W.; Sheen, Y.H.; Ahn, K.; et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J. Allergy Clin. Immunol. 2018, 141, 1310–1319. [Google Scholar] [CrossRef] [PubMed]
- Yap, G.C.; Loo, E.X.; Aw, M.; Lu, Q.; Shek, L.P.; Lee, B.W. Molecular analysis of infant fecal microbiota in an Asian at-risk cohort-correlates with infant and childhood eczema. BMC Res. Notes 2014, 7, 166. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, C.; Tan, J.; Macia, L.; Mackay, C.R. The nutrition-gut microbiome-physiology axis and allergic diseases. Immunol. Rev. 2017, 278, 277–295. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.L.; Martoni, C.J.; Prakash, S. Cholesterol lowering and inhibition of sterol absorption by Lactobacillus reuteri NCIMB 30242: A randomized controlled trial. Eur. J. Clin. Nutr. 2012, 66, 1234–1241. [Google Scholar] [CrossRef]
- Fuentes, M.C.; Lajo, T.; Carrion, J.M.; Cune, J. Cholesterol-lowering efficacy of Lactobacillus plantarum CECT 7527, 7528 and 7529 in hypercholesterolaemic adults. Br. J. Nutr. 2013, 109, 1866–1872. [Google Scholar] [CrossRef]
- Lau, K.; Srivatsav, V.; Rizwan, A.; Nashed, A.; Liu, R.; Shen, R.; Akhtar, M. Bridging the Gap between Gut Microbial Dysbiosis and Cardiovascular Diseases. Nutrients 2017, 9, 859. [Google Scholar] [CrossRef]
- Shinohara, K.; Ohashi, Y.; Kawasumi, K.; Terada, A.; Fujisawa, T. Effect of apple intake on fecal microbiota and metabolites in humans. Anaerobe 2010, 16, 510–515. [Google Scholar] [CrossRef]
- Femia, A.P.; Luceri, C.; Bianchini, F.; Salvadori, M.; Salvianti, F.; Pinzani, P.; Dolara, P.; Calorini, L.; Caderni, G. Marie Menard apples with high polyphenol content and a low-fat diet reduce 1,2-dimethylhydrazine-induced colon carcinogenesis in rats: Effects on inflammation and apoptosis. Mol. Nutr. Food Res. 2012, 56, 1353–1357. [Google Scholar] [CrossRef]
- Dewulf, E.M.; Cani, P.D.; Claus, S.P.; Fuentes, S.; Puylaert, P.G.; Neyrinck, A.M.; Bindels, L.B.; de Vos, W.M.; Gibson, G.R.; Thissen, J.P.; et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut 2013, 62, 1112–1121. [Google Scholar] [CrossRef]
- Scott, K.P.; Martin, J.C.; Duncan, S.H.; Flint, H.J. Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria, in vitro. FEMS Microbiol. Ecol. 2014, 87, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Koutsos, A.; Tuohy, K.M.; Lovegrove, J.A. Apples and cardiovascular health—Is the gut microbiota a core consideration? Nutrients 2015, 7, 3959–3998. [Google Scholar] [CrossRef] [PubMed]
- Selak, M.; Riviere, A.; Moens, F.; Van den Abbeele, P.; Geirnaert, A.; Rogelj, I.; Leroy, F.; De Vuyst, L. Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl. Microbiol. Biotechnol. 2016, 100, 4097–4107. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.H.; Diven, M.A.; Huff, L.W.; Paulos, C.M. Harnessing the Microbiome to Enhance Cancer Immunotherapy. J. Immunol. Res. 2015, 2015, 368736. [Google Scholar] [CrossRef]
- Borgo, F.; Verduci, E.; Riva, A.; Lassandro, C.; Riva, E.; Morace, G.; Borghi, E. Relative Abundance in Bacterial and Fungal Gut Microbes in Obese Children: A Case Control Study. Child Obes. 2017, 13, 78–84. [Google Scholar] [CrossRef]
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73. [Google Scholar] [CrossRef]
- Hu, X.F.; Zhang, W.Y.; Wen, Q.; Chen, W.J.; Wang, Z.M.; Chen, J.; Zhu, F.; Liu, K.; Cheng, L.X.; Yang, J.; et al. Fecal microbiota transplantation alleviates myocardial damage in myocarditis by restoring the microbiota composition. Pharmacol. Res. 2019, 139, 412–421. [Google Scholar] [CrossRef]
- Leshem, A.; Horesh, N.; Elinav, E. Fecal Microbial Transplantation and Its Potential Application in Cardiometabolic Syndrome. Front. Immunol. 2019, 10, 1341. [Google Scholar] [CrossRef]
- Kim, E.S.; Yoon, B.H.; Lee, S.M.; Choi, M.; Kim, E.H.; Lee, B.W.; Kim, S.Y.; Pack, C.G.; Sung, Y.H.; Baek, I.J.; et al. Fecal microbiota transplantation ameliorates atherosclerosis in mice with C1q/TNF-related protein 9 genetic deficiency. Exp. Mol. Med. 2022, 54, 103–114. [Google Scholar] [CrossRef]
- Toh, Z.Q.; Anzela, A.; Tang, M.L.; Licciardi, P.V. Probiotic therapy as a novel approach for allergic disease. Front. Pharmacol. 2012, 3, 171. [Google Scholar] [CrossRef]
- Rusu, E.; Enache, G.; Cursaru, R.; Alexescu, A.; Radu, R.; Onila, O.; Cavallioti, T.; Rusu, F.; Posea, M.; Jinga, M.; et al. Prebiotics and probiotics in atopic dermatitis. Exp. Ther. Med. 2019, 18, 926–931. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghazzewi, F.H.; Tester, R.F. Impact of prebiotics and probiotics on skin health. Benef. Microbes 2014, 5, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.S.; Trivedi, M.K.; Jha, A.; Lin, Y.F.; Dimaano, L.; Garcia-Romero, M.T. Synbiotics for Prevention and Treatment of Atopic Dermatitis: A Meta-analysis of Randomized Clinical Trials. JAMA Pediatr. 2016, 170, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.L.; Lodge, C.J. Examining the Evidence for Using Synbiotics to Treat or Prevent Atopic Dermatitis. JAMA Pediatr. 2016, 170, 201–203. [Google Scholar] [CrossRef] [PubMed]
- Dissanayake, E.; Tani, Y.; Nagai, K.; Sahara, M.; Mitsuishi, C.; Togawa, Y.; Suzuki, Y.; Nakano, T.; Yamaide, F.; Ohno, H.; et al. Skin Care and Synbiotics for Prevention of Atopic Dermatitis or Food Allergy in Newborn Infants: A 2 × 2 Factorial, Randomized, Non-Treatment Controlled Trial. Int. Arch. Allergy Immunol. 2019, 180, 202–211. [Google Scholar] [CrossRef] [PubMed]
- Caballero-Franco, C.; Keller, K.; De Simone, C.; Chadee, K. The VSL#3 probiotic formula induces mucin gene expression and secretion in colonic epithelial cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G315–G322. [Google Scholar] [CrossRef] [PubMed]
- Malik, K.; Heitmiller, K.D.; Czarnowicki, T. An Update on the Pathophysiology of Atopic Dermatitis. Dermatol. Clin. 2017, 35, 317–326. [Google Scholar] [CrossRef]
- Li, L.; Han, Z.; Niu, X.; Zhang, G.; Jia, Y.; Zhang, S.; He, C. Probiotic Supplementation for Prevention of Atopic Dermatitis in Infants and Children: A Systematic Review and Meta-analysis. Am. J. Clin. Dermatol. 2019, 20, 367–377. [Google Scholar] [CrossRef]
- Alam, M.J.; Xie, L.; Yap, Y.A.; Marques, F.Z.; Robert, R. Manipulating Microbiota to Treat Atopic Dermatitis: Functions and Therapies. Pathogens 2022, 11, 642. [Google Scholar] [CrossRef]
- Shimamori, Y.; Mitsunaka, S.; Yamashita, H.; Suzuki, T.; Kitao, T.; Kubori, T.; Nagai, H.; Takeda, S.; Ando, H. Staphylococcal Phage in Combination with Staphylococcus Epidermidis as a Potential Treatment for Staphylococcus Aureus-Associated Atopic Dermatitis and Suppressor of Phage-Resistant Mutants. Viruses 2020, 13, 7. [Google Scholar] [CrossRef]
- Totte, J.E.E.; van Doorn, M.B.; Pasmans, S. Successful Treatment of Chronic Staphylococcus aureus-Related Dermatoses with the Topical Endolysin Staphefekt SA.100: A Report of 3 Cases. Case Rep. Dermatol. 2017, 9, 19–25. [Google Scholar] [CrossRef]
- de Wit, J.; Totte, J.E.E.; van Mierlo, M.M.F.; van Veldhuizen, J.; van Doorn, M.B.A.; Schuren, F.H.J.; Willemsen, S.P.; Pardo, L.M.; Pasmans, S. Endolysin treatment against Staphylococcus aureus in adults with atopic dermatitis: A randomized controlled trial. J. Allergy Clin. Immunol. 2019, 144, 860–863. [Google Scholar] [CrossRef]
- Ahn, K. The Effect of Prebiotics on Atopic Dermatitis. Allergy Asthma Immun. 2023, 15, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.Z.; Tang, L.; Nie, T.T.; Fang, M.Y.; Cao, X.Q. Fructo-oligofructose ameliorates 2,4-dinitrofluorobenzene-induced atopic dermatitis-like skin lesions and psychiatric comorbidities in mice. J. Sci. Food Agr. 2023, 103, 5004–5018. [Google Scholar] [CrossRef] [PubMed]
- Husein-ElAhmed, H.; Steinhoff, M. Meta-analysis on preventive and therapeutic effects of probiotic supplementation in infant atopic dermatitis. J. Dtsch. Dermatol. Ges. 2023, 21, 833–843. [Google Scholar] [CrossRef] [PubMed]
- Kawano, K.; Iyori, K.; Kondo, N.; Yamakawa, S.; Fujii, T.; Funasaka, K.; Hirooka, Y.; Tochio, T. Clinical effects of combined Lactobacillus paracasei and kestose on canine atopic dermatitis. Pol. J. Vet. Sci. 2023, 26, 131–136. [Google Scholar] [CrossRef]
- Kim, S.; Han, S.Y.; Lee, J.Y.; Kim, N.R.; Lee, B.R.; Kim, H.; Kwon, M.; Ahn, K.; Noh, Y.; Kim, S.J.; et al. Bifidobacterium longum and Galactooligosaccharide Improve Skin Barrier Dysfunction and Atopic Dermatitis-like Skin. Allergy Asthma Immun. 2022, 14, 549–564. [Google Scholar] [CrossRef]
- Hubbard, G.P.; Atwal, K.; Graham, L.; Narayanan, S.; Cooke, L.; Casewell, C.; Denton, S.A.; Gavin, J.; Browne, R.M.; Kinnear, F.J.; et al. Synbiotic containing extensively hydrolyzed formula improves gastrointestinal and atopic symptom severity, growth, caregiver quality of life, and hospital-related healthcare use in infants with cow’s milk allergy. Immun. Inflamm. Dis. 2022, 10, e636. [Google Scholar] [CrossRef]
- Effendi, R.M.R.A.; Anshory, M.; Kalim, H.; Dwiyana, R.F.; Suwarsa, O.; Pardo, L.M.; Nijsten, T.E.C.; Thio, H.B. Akkermansia muciniphila and Faecalibacterium prausnitzii in Immune-Related Diseases. Microorganisms 2022, 10, 2382. [Google Scholar] [CrossRef]
- Koh, L.F.; Ong, R.Y.; Common, J.E. Skin microbiome of atopic dermatitis. Allergol. Int. 2022, 71, 31–39. [Google Scholar] [CrossRef]
- Wan, P.; Chen, J. A Calm, Dispassionate Look at Skin Microbiota in Atopic Dermatitis: An Integrative Literature Review. Dermatol. Ther. 2020, 10, 53–61. [Google Scholar] [CrossRef]
- Panthee, B.; Gyawali, S.; Panthee, P.; Techato, K. Environmental and Human Microbiome for Health. Life 2022, 12, 456. [Google Scholar] [CrossRef]
| Region | Luminal Concentration | Main Composition |
|---|---|---|
| Stomach | 0–102 | Lactobacillus |
| Candida | ||
| Streptococcus | ||
| Helicobacter pylori | ||
| Peptostreptococcus | ||
| Duodenum | 102 | Lactobacillus |
| Streptococcus | ||
| Jejuno | 102 | Lactobacillus |
| Streptococcus | ||
| Proximal ileum | 102 | Lactobacillus |
| Streptococcus | ||
| Distal ileum | 107–108 | Clostridium |
| Steptococcus | ||
| Bacteroides | ||
| Actinomycinae | ||
| Corynebacteria | ||
| Colon | 1011–1012 | Bacteroides |
| Clostridium | ||
| Bifidobacterium | ||
| Enterobacteriaceae |
| Therapy | Definition | Examples | Reference |
|---|---|---|---|
| Probiotics | Beneficial living microorganisms capable of establishing themselves in the human gut to foster or reinstate a well-balanced gut microbiota composition | Lactobaccillus strains L. reuteri (microencapsulated in yogurt) L. plantarum (capsules) | [136,137,138] |
| Prebiotics | Nutritional elements that nourish and encourage the development of a thriving gut microbiota composition | Plant polyphenols Fruits and vegetables (e.g., apples) Dietary fructans Foods high in inulin and/or oligofructose | [138,139,140,141,142,143,144] |
| Diet intervention | Ingestion of dietary components serving as sources of energy and nutrients for microbial growth, shaping the microbial community composition | Diets rich in fiber and vegetables | [145,146,147] |
| Fecal microbiota transplantation | Transference of fecal matter, containing a mixture of beneficial microorganisms, from a healthy donor to the gastrointestinal tract of a recipient | Restore the microbial balance in the intestine | [148,149,150] |
| Therapy | Examples | References |
|---|---|---|
| Probiotics | Lactobacillus, Bifidobaterium | [152,158,159] |
| Prebiotics | Inulin, resistant starch, polydextrose, pectic oligosaccharides derived from pectin (fructooligosaccharides (FOS) and galactooligosaccharides (GOS)) | [153,164,165,166] |
| Synbiotics | Bifidobacterium and galactooligosaccharide | [167,168,169,170] |
| Improving nutritional status | Fibers, fruit, and Mediterranean diet consumption | [152] |
| Short-chain fatty acids (SCFAs) | Acetate (C2), propionate (C3), and butyrate (C4) | [160] |
| Phage therapy | Staphylococcal phage, SaGU1; phage endolysin, Staphefekt SA.100 | [161,162,163,171] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferraz, M.P. An Overview of the Relevance of Human Gut and Skin Microbiome in Disease: The Influence on Atopic Dermatitis. Appl. Sci. 2023, 13, 10540. https://doi.org/10.3390/app131810540
Ferraz MP. An Overview of the Relevance of Human Gut and Skin Microbiome in Disease: The Influence on Atopic Dermatitis. Applied Sciences. 2023; 13(18):10540. https://doi.org/10.3390/app131810540
Chicago/Turabian StyleFerraz, Maria Pia. 2023. "An Overview of the Relevance of Human Gut and Skin Microbiome in Disease: The Influence on Atopic Dermatitis" Applied Sciences 13, no. 18: 10540. https://doi.org/10.3390/app131810540
APA StyleFerraz, M. P. (2023). An Overview of the Relevance of Human Gut and Skin Microbiome in Disease: The Influence on Atopic Dermatitis. Applied Sciences, 13(18), 10540. https://doi.org/10.3390/app131810540






