Comparative Analysis of Single Bands, Vegetation Indices, and Their Combination in Predicting Grass Species Nitrogen in a Protected Mountainous Area
Abstract
:1. Introduction
2. Study Area
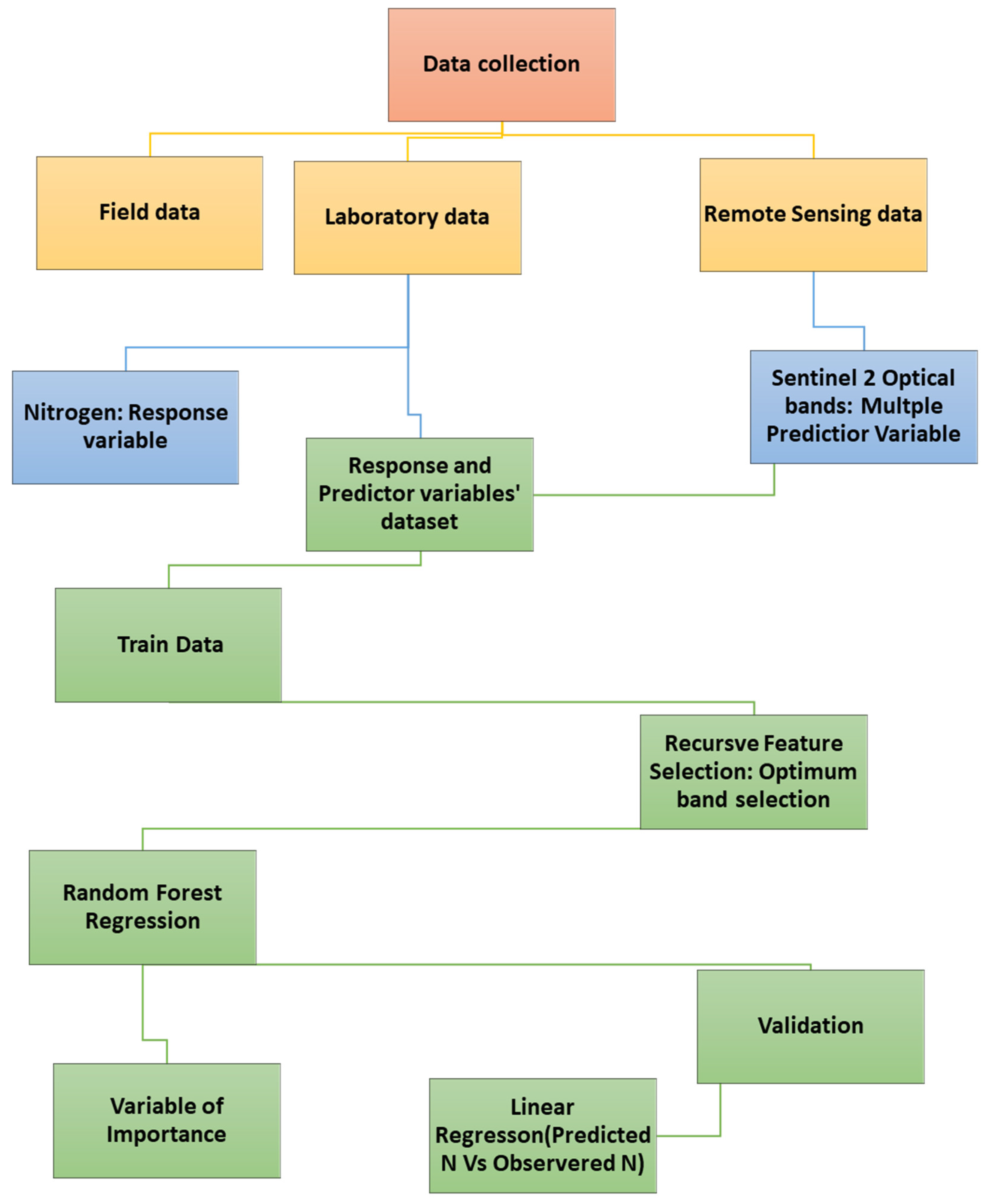
3. Data Collection and Sampling
4. Data Analysis
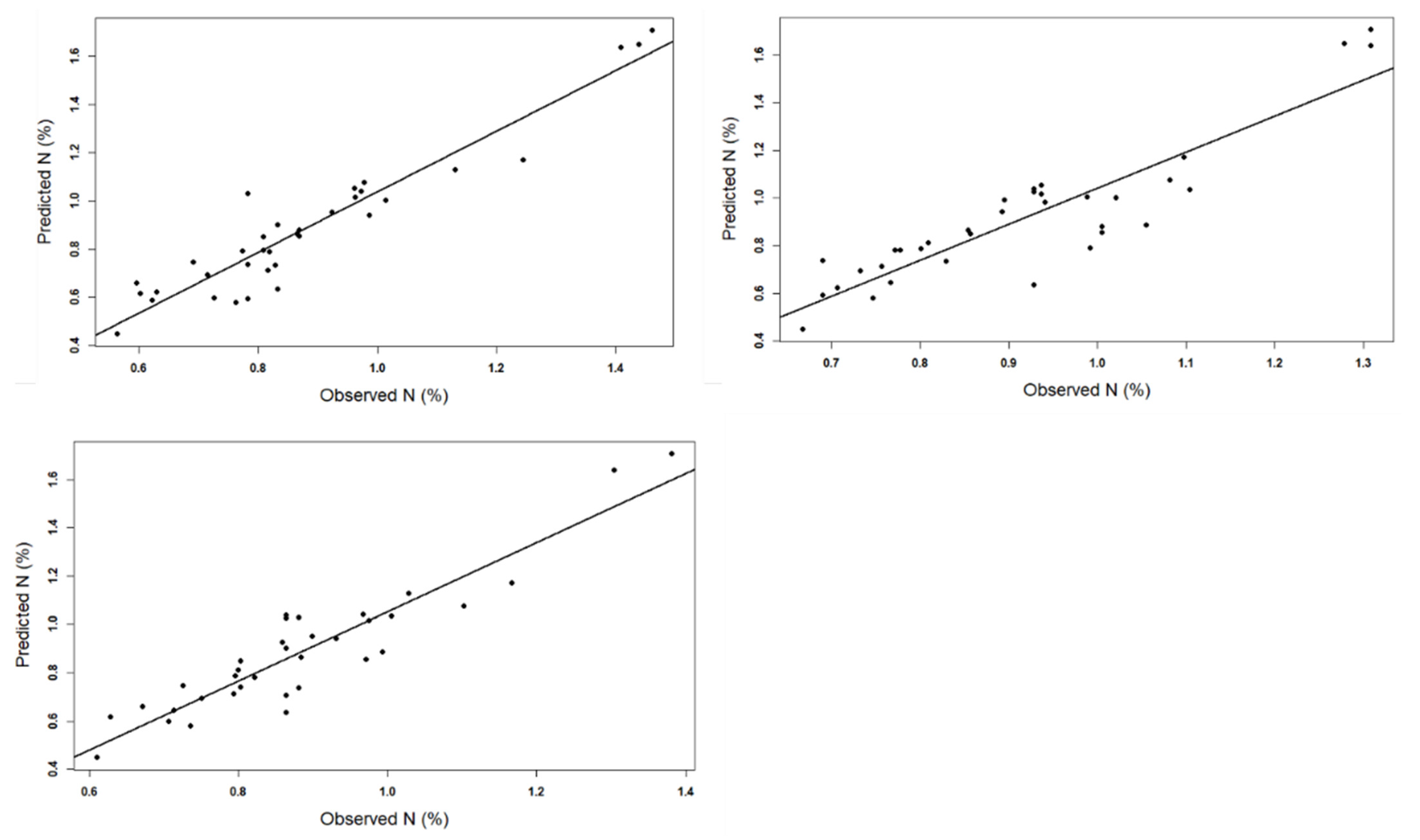
Model Performance (Validation)
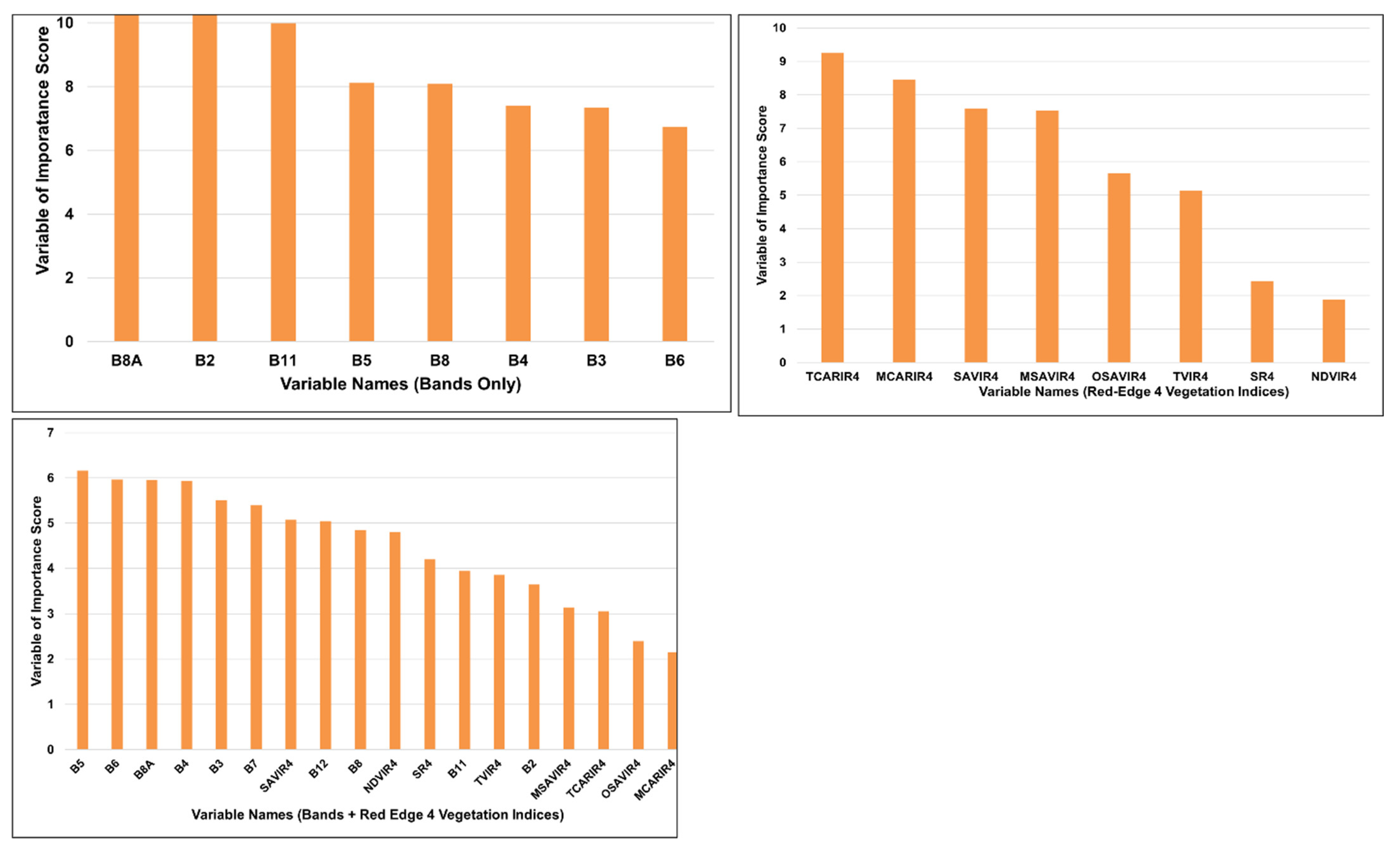
5. Results
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cardinale, B.J.; Duffy, J.E.; Gonzalez, A.; Hooper, D.U.; Perrings, C.; Venail, P.; Narwani, A.; Mace, G.M.; Tilman, D.; Wardle, D.A.; et al. Biodiversity loss and its impact on humanity. Nature 2012, 486, 59–67. [Google Scholar] [CrossRef] [Green Version]
- Hooper, D.U.; Adair, E.C.; Cardinale, B.J.; Byrnes, J.E.K.; Hungate, B.A.; Matulich, K.L.; Gonzalez, A.; Duffy, J.E.; Gamfeldt, L.; O’Connor, M.I. A global synthesis reveals biodiversity loss as a major driver of ecosystem change. Nature 2012, 486, 105–108. [Google Scholar] [CrossRef]
- Tilman, D.; Wedin, D.; Knops, J. Productivity and sustainability in grassland ecosystems. Nature 1996, 379, 718–720. [Google Scholar] [CrossRef]
- Tilman, D.; Reich, P.B.; Isbell, F. Biodiversity impacts ecosystem productivity as much as resources, disturbance, or herbivory. Proc. Natl. Acad. Sci. USA 2012, 109, 10394–10397. [Google Scholar] [CrossRef] [PubMed]
- Mashiane, K.; Ramoelo, A.; Adelabu, S. Diversifying modeling techniques to disentangle the complex patterns of species richness and diversity in the protected Afromontane grasslands. Biodivers. Conserv. 2023, 32, 1423–1436. [Google Scholar] [CrossRef]
- van Oijen, M.; Bellocchi, G.; Höglind, M. Effects of Climate Change on Grassland Biodiversity and Productivity: The Need for a Diversity of Models. Agronomy 2018, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- McNaughton, S.J. Mineral nutrition and spatial concentrations of African ungulates. Nature 1988, 334, 343–345. [Google Scholar] [CrossRef]
- McNaughton, S.J.; Banyikwa, F.F.; McNaughton, M.M. Promotion of the Cycling of Diet-Enhancing Nutrients by African Grazers. Science 1997, 278, 1798–1800. [Google Scholar] [CrossRef]
- Ramoelo, A.; Cho, M.A.; Mathieu, R.; Madonsela, S.; van de Kerchove, R.; Kaszta, Z.; Wolff, E. Monitoring grass nutrients and biomass as indicators of rangeland quality and quantity using random forest modelling and WorldView-2 data. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 43–54. [Google Scholar] [CrossRef]
- Adjorlolo, C.; Botha, J.O. Integration of remote sensing and conventional models for modeling grazing/browsing capacity in southern African savannas. J. Appl. Remote Sens. 2015, 9, 096041. [Google Scholar] [CrossRef]
- Mcnaughton, S.J. Ecology of a Grazing Ecosystem: The Serengeti. Ecol. Monogr. 1985, 55, 259–294. [Google Scholar] [CrossRef]
- Stock, W.D.; Bond, W.J.; van de Vijver, C.A.D.M. Herbivore and nutrient control of lawn and bunch grass distributions in a southern African savanna. Plant Ecol. 2010, 206, 15–27. [Google Scholar] [CrossRef]
- McNaughton, S.J. Mineral nutrition and seasonal movements of African migratory ungulates. Nature 1990, 345, 613–615. [Google Scholar] [CrossRef]
- Ungar, E.D. Perspectives on the concept of rangeland carrying capacity, and their exploration by means of Noy-Meir’s two-function model. Agric. Syst. 2019, 173, 403–413. [Google Scholar] [CrossRef]
- McCord, S.E.; Buenemann, M.; Karl, J.W.; Browning, D.M.; Hadley, B.C. Integrating Remotely Sensed Imagery and Existing Multiscale Field Data to Derive Rangeland Indicators: Application of Bayesian Additive Regression Trees. Rangel. Ecol. Manag. 2017, 70, 644–655. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Red edge shift and biochemical content in grass canopies. ISPRS J. Photogramm. Remote Sens. 2007, 62, 34–42. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Ramoelo, A.; Skidmore, A.K.; Cho, M.A.; Schlerf, M.; Mathieu, R.; Heitkönig, I.M.A. Regional estimation of savanna grass nitrogen using the red-edge band of the spaceborne rapideye sensor. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 151–162. [Google Scholar] [CrossRef]
- Chabalala, Y.; Adam, E.; Oumar, Z.; Ramoelo, A. Exploiting the capabilities of Sentinel-2 and RapidEye for predicting grass nitrogen across different grass communities in a protected area. Appl. Geomat. 2020, 12, 379–395. [Google Scholar] [CrossRef]
- Xulu, S.; Mbatha, N.; Peerbhay, K. Burned area mapping over the southern cape forestry region, South Africa using sentinel data within gee cloud platform. ISPRS Int. J. Geo-Inf. 2021, 10, 511. [Google Scholar] [CrossRef]
- Semela, M.; Ramoelo, A.; Adelabu, S. Testing and Comparing the Applicability of Sentinel-2 and Landsat 8 Reflectance Data in Estimating Mountainous Herbaceous Biomass before and after Fire Using Random Forest Modelling. In Proceedings of the IGARSS 2020-2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 4493–4496. [Google Scholar] [CrossRef]
- Zhang, X.; Liang, T.; Gao, J.; Zhang, D.; Liu, J.; Feng, Q.; Wu, C.; Wang, Z. Mapping the forage nitrogen, phosphorus, and potassium contents of alpine grasslands by integrating Sentinel-2 and Tiangong-2 data. Plant Methods 2023, 19, 48. [Google Scholar] [CrossRef]
- Adagbasa, E.G.; Adelabu, S.A.; Okello, T.W. Application of deep learning with stratified K-fold for vegetation species discrimation in a protected mountainous region using Sentinel-2 image. Geocarto Int. 2022, 37, 142–162. [Google Scholar] [CrossRef]
- SANParks Golden Gate Highlands National Park Park Management Plan; SANParks: Pretoria, South Africa, 2020.
- Kay, C.; Bredenkamp, G.J.; Theron, G.K. The plant communities of the Golden Gate Highlands National Park in the north-eastern Orange Free State. S. Afr. J. Bot. 1993, 59, 442–449. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, R.A.; Rexroad, P.R. Comparison of LECO FP-228 “nitrogen determinator” with AOAC copper catalyst Kjeldahl method for crude protein. J. Assoc. Off. Anal. Chem. 1987, 70, 1028–1030. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Hyperspectral band depth analysis for a better estimation of grass biomass (Cenchrus ciliaris) measured under controlled laboratory conditions. Int. J. Appl. Earth Obs. Geoinf. 2004, 5, 87–96. [Google Scholar] [CrossRef]
- Daughtry, C. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Rouse, J.W.; Hass, R.H.; Schell, J.A.; Deering, D.W.; Harlan, J.C. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; NASA Technical Reports Server: Cleveland, OH, USA, 1974. [Google Scholar]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Huete, A. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Jordan, C.F. Derivation of Leaf-Area Index from Quality of Light on the Forest Floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Haboudane, D. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Broge, N.; Leblanc, E. Comparing prediction power and stability of broadband and hyperspectral vegetation indices for estimation of green leaf area index and canopy chlorophyll density. Remote Sens. Environ. 2001, 76, 156–172. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.; Corsi, F.; van Wieren, S.E.; Sobhan, I. Estimation of green grass/herb biomass from airborne hyperspectral imagery using spectral indices and partial least squares regression. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 414–424. [Google Scholar] [CrossRef]
- Cho, M.A.; Skidmore, A.K. A new technique for extracting the red edge position from hyperspectral data: The linear extrapolation method. Remote Sens. Environ. 2006, 101, 181–193. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; De Jong, S.M.; Epema, G.F.; Van Der Meer, F.D.; Bakker, W.H.; Skidmore, A.K.; Scholte, K.H. Derivation of the red edge index using the MERIS standard band setting. Int. J. Remote Sens. 2002, 23, 3169–3184. [Google Scholar] [CrossRef] [Green Version]

| Band Number | Band Name | Wavelength | Description |
|---|---|---|---|
| Band 1 | B1 | 443.9 nm/442.3 nm | Aerosols |
| Band 2 | B2 | 496.6 nm/492.1 nm | Blue |
| Band 3 | B3 | 560 nm/559 nm | Green |
| Band 4 | B4 | 664.5 nm/665 nm | Red |
| Band 5 | B5 | 703.9 nm/703.8 nm | Red edge 1 |
| Band 6 | B6 | 740.2 nm/739.1 nm | Red edge 2 |
| Band 7 | B7 | 782.5 nm/779.7 nm | Red edge 3 |
| Band 8 | B8 | 835.1 nm/833 nm | Near infrared |
| Band 9 | B8A | 864.8 nm/864 nm | Red edge 4 |
| Band 10 | B9 | 945 nm/943.2 nm | Water vapor |
| Band 11 | B11 | 1613.7 nm/1610.4 nm | Shortwave infrared 1 |
| Band 12 | B12 | 2202.4 nm/2185 nm | Shortwave infrared 2 |
| Index | Used Formulae | Reference |
|---|---|---|
| MCARIR4 | ((NIR − Red edge 4) − 0.2*(NIR-Red edge 4))*(NIR/Red edge 4) | [28] |
| MSAVIR4 | 0.5*(2*NIR + 1 − SQRT((2*NIR + 1) − 8(NIR − Red edge 4))) | [29] |
| NDVIR4 | (NIR − Red edge 4)/(NIR + Red edge 4) | [30] |
| OSAVIR4 | (1 + 0.6)*(NIR − Red edge 4)/(NIR − Red edge 4 + 0.16) | [31] |
| RDVIR4 | (NIE − Red edge 4)/SQRT(NIR + Red edge 4) | [28] |
| SAVIR4 | (2.5*NIR − Red edge 4)/((NIR + Red edge 4) + 2) | [32] |
| SR4 | NIR/Red edge 4 | [33] |
| TCARIR4 | 3*(NIR − Red edge 4) − 0.2*(NIR − Red edge 4)*(NIR/Red edge 4) | [34] |
| TVIR4 | 0.5*(120*(NIR − Red edge 4) − 200*(NIR − Red edge 4)) | [35] |
| Datasets | N | Min | Max | Mean | SD | CV (%) |
|---|---|---|---|---|---|---|
| Training | 35 | 0.58 | 1.638 | 0.867 | 0.219 | 25 |
| Test | 15 | 0.44 | 1.71 | 0.93 | 0.34 | 36 |
| All combined | 50 | 0.45 | 1.71 | 0.89 | 0.25 | 29 |
| Selected Variables | RMSE | R-Squared | MAE | Selected |
|---|---|---|---|---|
| Bands only | ||||
| Red | 0.2272 | 0.4223 | 0.1727 | |
| Red Edge 4 | 0.2154 | 0.4794 | 0.1672 | * |
| SWIR | 0.2249 | 0.4274 | 0.1721 | |
| Red Edge 4 Indices | ||||
| NDVIR4 | 0.3184 | 0.00025 | 0.2347 | |
| TVIR4 | 0.3156 | 0.0000482 | 0.2284 | * |
| Band + Vegetation Indices | ||||
| Red | 0.2655 | 0.02986 | 0.205 | |
| Red Edge 4 | 0.2473 | 0.10306 | 0.1879 | |
| SR4 | 0.2396 | 0.14878 | 0.1842 | |
| TVIR4 | 0.2387 | 0.15517 | 0.1813 | * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mashiane, K.; Adelabu, S.; Ramoelo, A. Comparative Analysis of Single Bands, Vegetation Indices, and Their Combination in Predicting Grass Species Nitrogen in a Protected Mountainous Area. Appl. Sci. 2023, 13, 7960. https://doi.org/10.3390/app13137960
Mashiane K, Adelabu S, Ramoelo A. Comparative Analysis of Single Bands, Vegetation Indices, and Their Combination in Predicting Grass Species Nitrogen in a Protected Mountainous Area. Applied Sciences. 2023; 13(13):7960. https://doi.org/10.3390/app13137960
Chicago/Turabian StyleMashiane, Katlego, Samuel Adelabu, and Abel Ramoelo. 2023. "Comparative Analysis of Single Bands, Vegetation Indices, and Their Combination in Predicting Grass Species Nitrogen in a Protected Mountainous Area" Applied Sciences 13, no. 13: 7960. https://doi.org/10.3390/app13137960
APA StyleMashiane, K., Adelabu, S., & Ramoelo, A. (2023). Comparative Analysis of Single Bands, Vegetation Indices, and Their Combination in Predicting Grass Species Nitrogen in a Protected Mountainous Area. Applied Sciences, 13(13), 7960. https://doi.org/10.3390/app13137960







