Teeth Segmentation in Panoramic Dental X-ray Using Mask Regional Convolutional Neural Network
Abstract
Featured Application
Abstract
1. Introduction
2. Materials and Methods
2.1. Data
2.2. Data Annotation
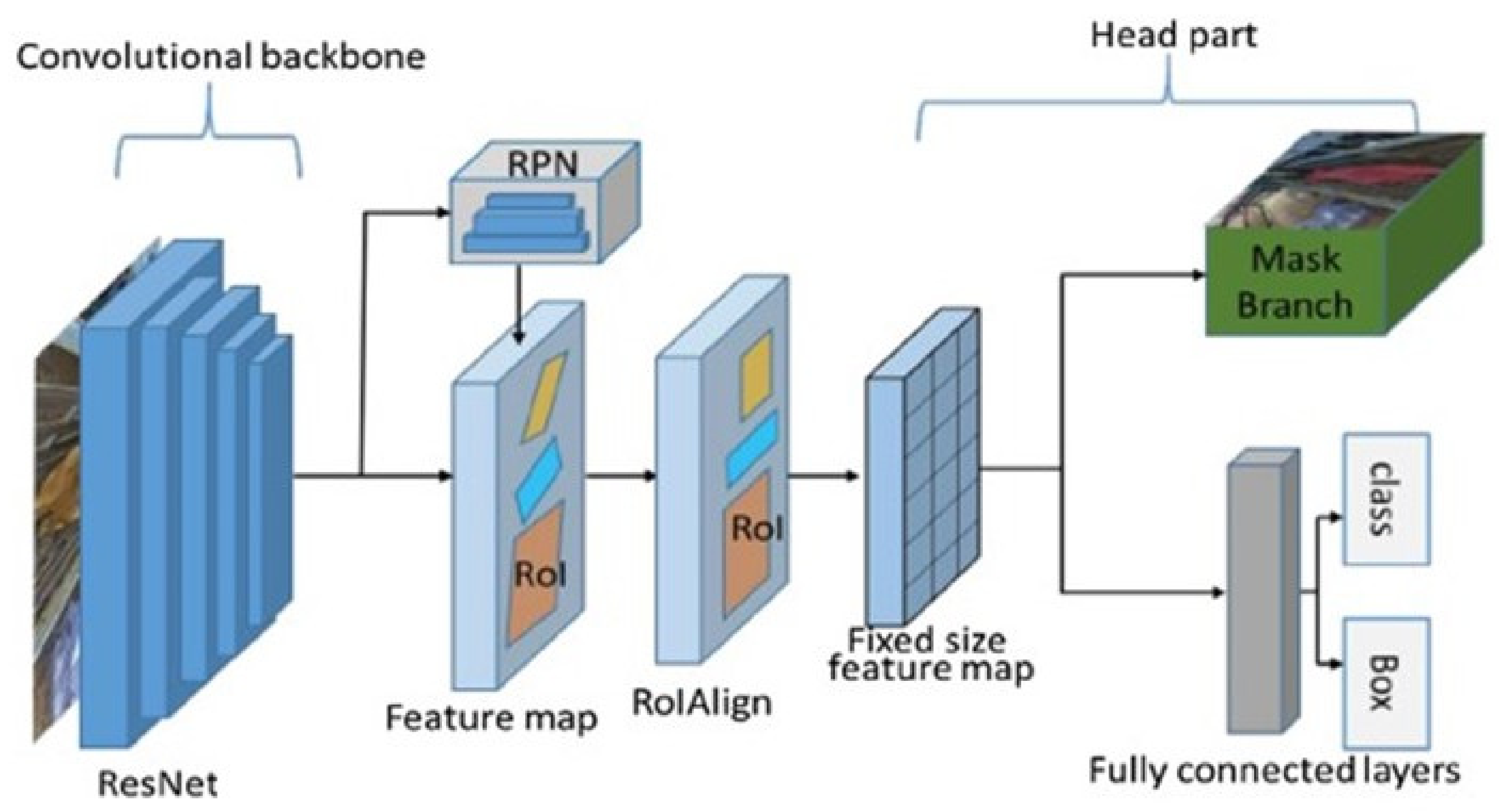
2.3. The Model
2.4. Training
2.5. Validation and Testing
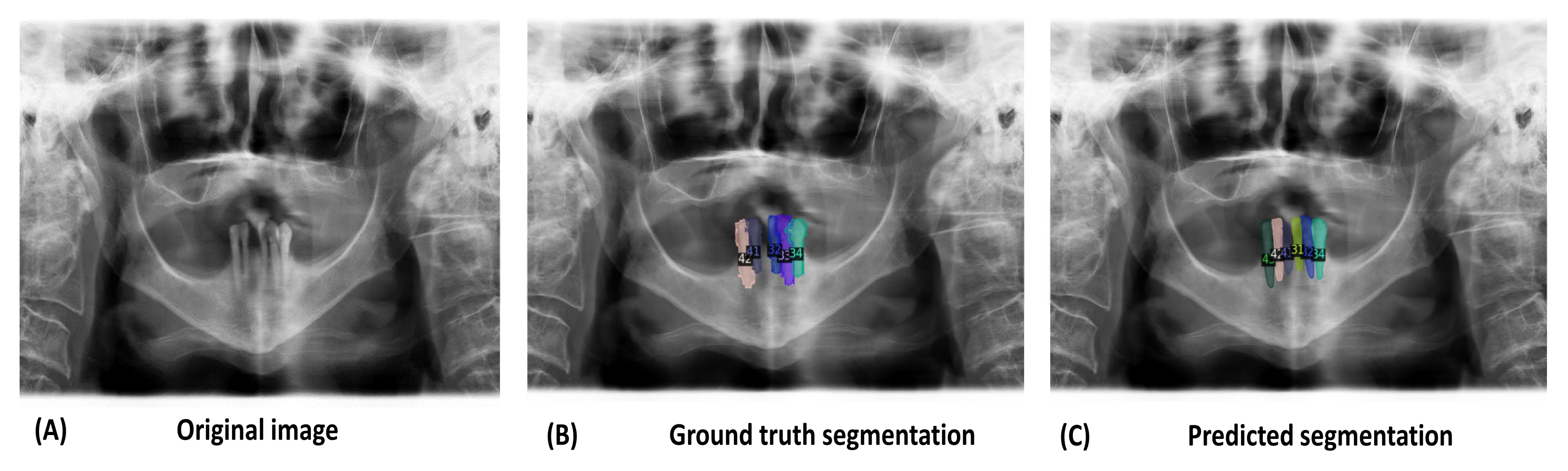
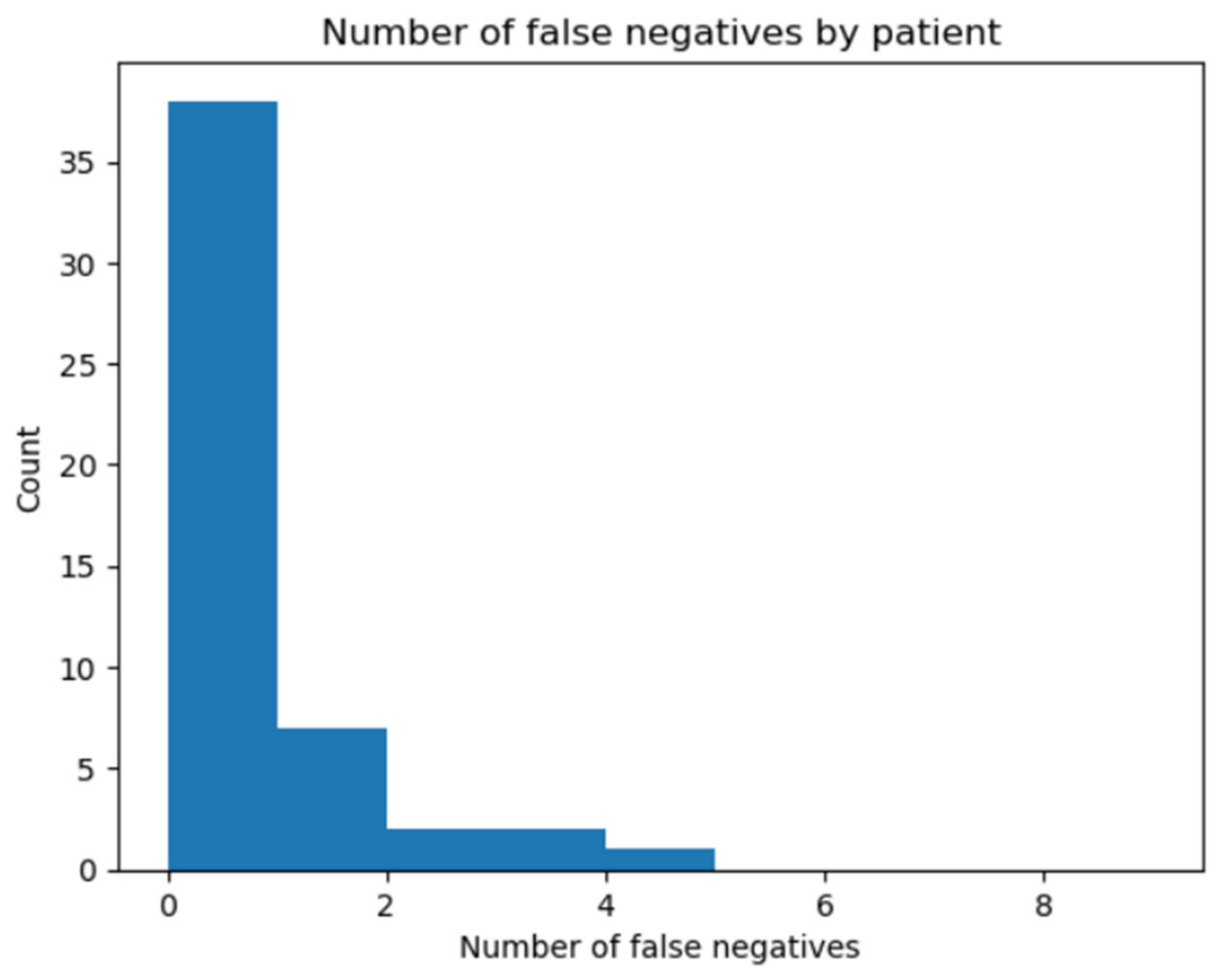
3. Results
4. Discussion
5. Limitations and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamano, H.; Sasahara, H.; Kitahara, K.; Kubota, S.; Kawada, M.; Takashima, T. The Orthopantomography-Its Basic Images. J. Nihon Univ. Sch. Dent. 1973, 15, 44–51. [Google Scholar] [CrossRef]
- Karatas, O.H.; Toy, E. Three-Dimensional Imaging Techniques: A Literature Review. Eur. J. Dent. 2014, 8, 132–140. [Google Scholar] [CrossRef]
- Dammann, F.; Bootz, F.; Cohnen, M.; Haßfeld, S.; Tatagiba, M.; Kösling, S. Diagnostic Imaging Modalities in Head and Neck Disease. Dtsch. Arztebl. Int. 2014, 111, 417–423. [Google Scholar] [CrossRef]
- Caloro, E.; Cè, M.; Gibelli, D.; Palamenghi, A.; Martinenghi, C.; Oliva, G.; Cellina, M. Artificial Intelligence (AI)-Based Systems for Automatic Skeletal Maturity Assessment through Bone and Teeth Analysis: A Revolution in the Radiological Workflow? Appl. Sci. 2023, 13, 3860. [Google Scholar] [CrossRef]
- Malik, S.; Pillai, J.; Malik, U. Forensic Genetics: Scope and Application from Forensic Odontology Perspective. J. Oral Maxillofac. Pathol. 2022, 26, 558. [Google Scholar] [CrossRef] [PubMed]
- Jader, G.; Fontineli, J.; Ruiz, M.; Abdalla, K.; Pithon, M.; Oliveira, L. Deep Instance Segmentation of Teeth in Panoramic X-ray Images. In Proceedings of the 2018 31st SIBGRAPI Conference on Graphics, Patterns and Images (SIBGRAPI), Paraná, Brazil, 29 October–1 November 2018; pp. 400–407. [Google Scholar]
- Ariji, Y.; Yanashita, Y.; Kutsuna, S.; Muramatsu, C.; Fukuda, M.; Kise, Y.; Nozawa, M.; Kuwada, C.; Fujita, H.; Katsumata, A.; et al. Automatic Detection and Classification of Radiolucent Lesions in the Mandible on Panoramic Radiographs Using a Deep Learning Object Detection Technique. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 128, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Han, S.-S.; Kim, Y.H.; Lee, C.; Kim, I. Application of a Fully Deep Convolutional Neural Network to the Automation of Tooth Segmentation on Panoramic Radiographs. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 129, 635–642. [Google Scholar] [CrossRef] [PubMed]
- Schwendicke, F.; Elhennawy, K.; Paris, S.; Friebertshäuser, P.; Krois, J. Deep Learning for Caries Lesion Detection in Near-Infrared Light Transillumination Images: A Pilot Study. J. Dent. 2020, 92, 103260. [Google Scholar] [CrossRef]
- Litjens, G.; Kooi, T.; Bejnordi, B.E.; Setio, A.A.A.; Ciompi, F.; Ghafoorian, M.; van der Laak, J.A.W.M.; van Ginneken, B.; Sánchez, C.I. A Survey on Deep Learning in Medical Image Analysis. Med. Image Anal. 2017, 42, 60–88. [Google Scholar] [CrossRef] [PubMed]
- Irmici, G.; Cè, M.; Caloro, E.; Khenkina, N.; Della Pepa, G.; Ascenti, V.; Martinenghi, C.; Papa, S.; Oliva, G.; Cellina, M. Chest X-ray in Emergency Radiology: What Artificial Intelligence Applications Are Available? Diagnostics 2023, 13, 216. [Google Scholar] [CrossRef]
- Litjens, G.; Ciompi, F.; Wolterink, J.M.; de Vos, B.D.; Leiner, T.; Teuwen, J.; Išgum, I. State-of-the-Art Deep Learning in Cardiovascular Image Analysis. JACC Cardiovasc. Imaging 2019, 12, 1549–1565. [Google Scholar] [CrossRef]
- Schwendicke, F.; Golla, T.; Dreher, M.; Krois, J. Convolutional Neural Networks for Dental Image Diagnostics: A Scoping Review. J. Dent. 2019, 91, 103226. [Google Scholar] [CrossRef] [PubMed]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Khenkina, N.; Toto-Brocchi, M.; Martinenghi, C.; Papa, S.; Carrafiello, G. Artificial Intelligence in Lung Cancer Imaging: Unfolding the Future. Diagnostics 2022, 12, 2644. [Google Scholar] [CrossRef]
- Cellina, M.; Cè, M.; Irmici, G.; Ascenti, V.; Caloro, E.; Bianchi, L.; Pellegrino, G.; D’Amico, N.; Papa, S.; Carrafiello, G. Artificial Intelligence in Emergency Radiology: Where Are We Going? Diagnostics 2022, 12, 3223. [Google Scholar] [CrossRef] [PubMed]
- Bilgir, E.; Bayrakdar, İ.Ş.; Çelik, Ö.; Orhan, K.; Akkoca, F.; Sağlam, H.; Odabaş, A.; Aslan, A.F.; Ozcetin, C.; Kıllı, M.; et al. An Artifıcial Intelligence Approach to Automatic Tooth Detection and Numbering in Panoramic Radiographs. BMC Med. Imaging 2021, 21, 124. [Google Scholar] [CrossRef]
- Vinayahalingam, S.; Xi, T.; Bergé, S.; Maal, T.; de Jong, G. Automated Detection of Third Molars and Mandibular Nerve by Deep Learning. Sci. Rep. 2019, 9, 9007. [Google Scholar] [CrossRef] [PubMed]
- Jader, G.; Oliveira, L.; Pithon, M. Automatic Segmenting Teeth in X-Ray Images: Trends, a Novel Data Set, Benchmarking and Future Perspectives. Expert Syst. Appl. 2018, 107, 15–31. [Google Scholar]
- Wirtz, A.; Mirashi, S.G.; Wesarg, S. Automatic Teeth Segmentation in Panoramic X-Ray Images Using a Coupled Shape Model in Combination with a Neural Network. In Medical Image Computing and Computer Assisted Intervention, Proceedings of the MICCAI 2018: 21st International Conference, Granada, Spain, 16–20 September 2018; Springer International Publishing: Cham, Switzerland, 2018; pp. 712–719. [Google Scholar]
- Koch, T.L.; Perslev, M.; Igel, C.; Brandt, S.S. Accurate Segmentation of Dental Panoramic Radiographs with U-NETS. In Proceedings of the 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019), Venice, Italy, 8–11 April 2019; pp. 15–19. [Google Scholar]
- Kim, C.; Kim, D.; Jeong, H.; Yoon, S.-J.; Youm, S. Automatic Tooth Detection and Numbering Using a Combination of a CNN and Heuristic Algorithm. Appl. Sci. 2020, 10, 5624. [Google Scholar] [CrossRef]
- Tuzoff, D.V.; Tuzova, L.N.; Bornstein, M.M.; Krasnov, A.S.; Kharchenko, M.A.; Nikolenko, S.I.; Sveshnikov, M.M.; Bednenko, G.B. Tooth Detection and Numbering in Panoramic Radiographs Using Convolutional Neural Networks. Dentomaxillofacial Radiol. 2019, 48, 20180051. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, C.; Morishita, T.; Takahashi, R.; Hayashi, T.; Nishiyama, W.; Ariji, Y.; Zhou, X.; Hara, T.; Katsumata, A.; Ariji, E.; et al. Tooth Detection and Classification on Panoramic Radiographs for Automatic Dental Chart Filing: Improved Classification by Multi-Sized Input Data. Oral Radiol. 2021, 37, 13–19. [Google Scholar] [CrossRef]
- Leite, A.F.; Van Gerven, A.; Willems, H.; Beznik, T.; Lahoud, P.; Gaêta-Araujo, H.; Vranckx, M.; Jacobs, R. Artificial Intelligence-Driven Novel Tool for Tooth Detection and Segmentation on Panoramic Radiographs. Clin. Oral Investig. 2021, 25, 2257–2267. [Google Scholar] [CrossRef]
- Panetta, K.; Rajendran, R.; Ramesh, A.; Rao, S.; Agaian, S. Tufts Dental Database: A Multimodal Panoramic X-Ray Dataset for Benchmarking Diagnostic Systems. IEEE J. Biomed. Health Inform. 2022, 26, 1650–1659. [Google Scholar] [CrossRef]
- Kaggle. Available online: https://www.kaggle.com/datasets/deepologylab/tufts-dental-database (accessed on 20 May 2023).
- ISO 3950:2016; Dentistry—Designation System for Teeth and Areas of the Oral Cavity. ISO: Geneva, Switzerland, 2016. Available online: https://www.iso.org/standard/68292.html (accessed on 20 May 2023).
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017. [Google Scholar]
- Github/Detectron2. Available online: https://github.com/facebookresearch/detectron2 (accessed on 20 May 2023).
- De Angelis, D.; Gibelli, D.; Merelli, V.; Botto, M.; Ventura, F.; Cattaneo, C. Application of Age Estimation Methods Based on Teeth Eruption: How Easy Is Olze Method to Use? Int. J. Leg. Med. 2014, 128, 841–844. [Google Scholar] [CrossRef]
- Gibelli, D.; De Angelis, D.; Riboli, F.; Dolci, C.; Cattaneo, C.; Sforza, C. Quantification of Odontological Differences of the Upper First and Second Molar by 3D-3D Superimposition: A Novel Method to Assess Anatomical Matches. Forensic Sci. Med. Pathol. 2019, 15, 570–573. [Google Scholar] [CrossRef]
- El Bsat, A.R.; Shammas, E.; Asmar, D.; Sakr, G.E.; Zeno, K.G.; Macari, A.T.; Ghafari, J.G. Semantic Segmentation of Maxillary Teeth and Palatal Rugae in Two-Dimensional Images. Diagnostics 2022, 12, 2176. [Google Scholar] [CrossRef] [PubMed]
- Arora, S.; Tripathy, S.K.; Gupta, R.; Srivastava, R. Exploiting Multimodal CNN Architecture for Automated Teeth Segmentation on Dental Panoramic X-Ray Images. Proc. Inst. Mech. Eng. Part H J. Eng. Med. 2023, 237, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Adnan, N.; Khalid, W.B.; Umer, F. An Artificial Intelligence Model for Teeth Segmentation and Numbering on Orthopantomograms. Int. J. Comput. Dent. 2023. [Google Scholar] [CrossRef]
- Hou, S.; Zhou, T.; Liu, Y.; Dang, P.; Lu, H.; Shi, H. Teeth U-Net: A Segmentation Model of Dental Panoramic X-Ray Images for Context Semantics and Contrast Enhancement. Comput. Biol. Med. 2023, 152, 106296. [Google Scholar] [CrossRef]
- Gardiyanoğlu, E.; Ünsal, G.; Akkaya, N.; Aksoy, S.; Orhan, K. Automatic Segmentation of Teeth, Crown–Bridge Restorations, Dental Implants, Restorative Fillings, Dental Caries, Residual Roots, and Root Canal Fillings on Orthopantomographs: Convenience and Pitfalls. Diagnostics 2023, 13, 1487. [Google Scholar] [CrossRef] [PubMed]
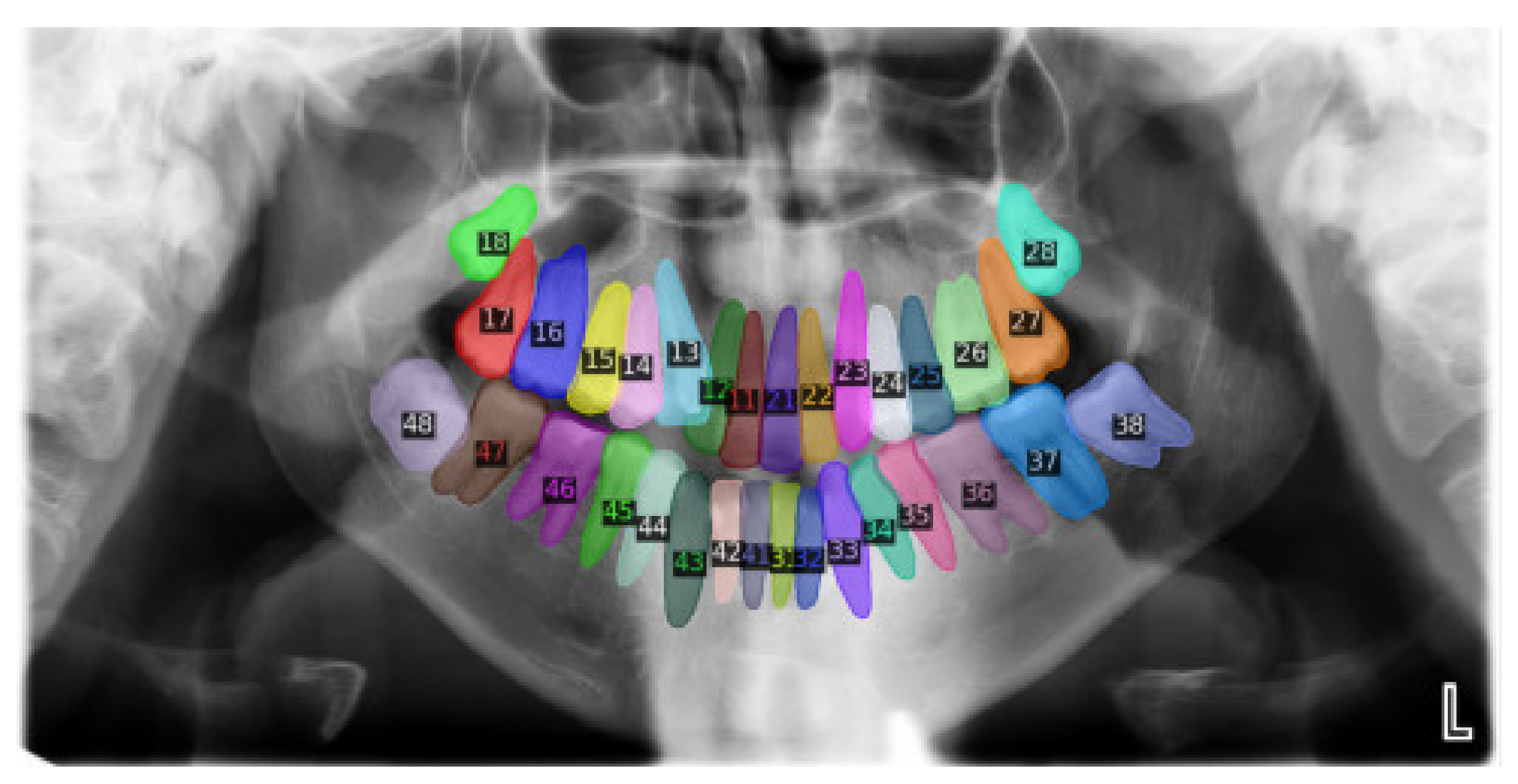

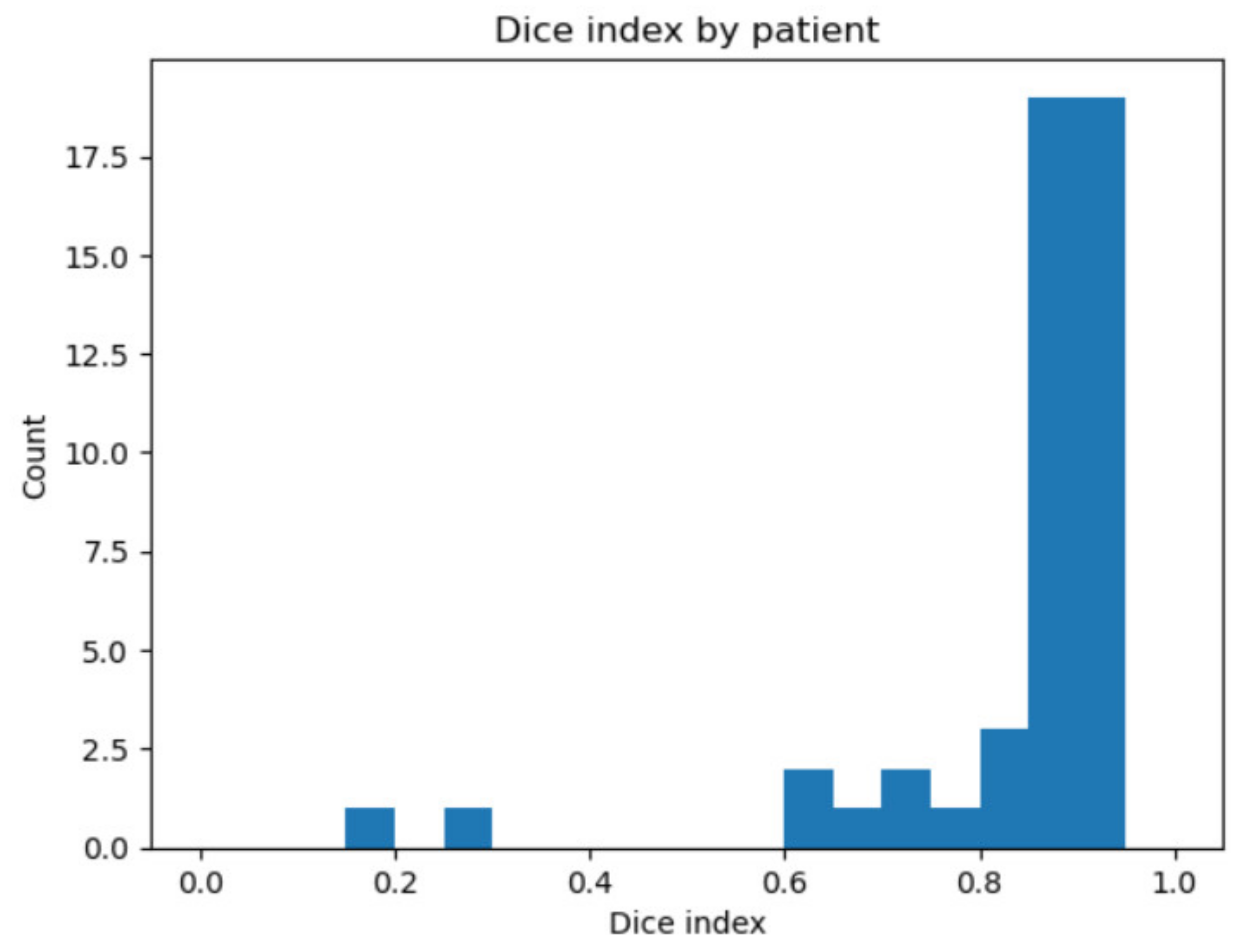
| ISO 3950 Code | Name | Dice Index | Accuracy (%) |
|---|---|---|---|
| 11 | Maxillary central incisor (R) | 0.78 | 86.67 |
| 12 | Maxillary lateral incisor (R) | 0.89 | 100 |
| 13 | Maxillary canine (R) | 0.88 | 100 |
| 14 | Maxillary first premolar (R) | 0.85 | 100 |
| 15 | Maxillary second premolar (R) | 0.85 | 97.61 |
| 16 | Maxillary first molar (R) | 0.88 | 100 |
| 17 | Maxillary second molar (R) | 0.88 | 100 |
| 18 | Maxillary third molar (R) | 0.90 | 100 |
| 21 | Maxillary central incisor (L) | 0.90 | 100 |
| 22 | Maxillary lateral incisor (L) | 0.89 | 100 |
| 23 | Maxillary canine (L) | 0.90 | 100 |
| 24 | Maxillary first premolar (L) | 0.88 | 100 |
| 25 | Maxillary second premolar (L) | 0.85 | 95 |
| 26 | Maxillary first molar (L) | 0.91 | 100 |
| 27 | Maxillary second molar (L) | 0.85 | 97.61 |
| 28 | Maxillary third molar (L) | 0.77 | 86.67 |
| 31 | Mandibular central incisor (L) | 0.91 | 100 |
| 32 | Mandibular lateral incisor (L) | 0.91 | 100 |
| 33 | Mandibular canine (L) | 0.88 | 94.59 |
| 34 | Mandibular first premolar (L) | 0.88 | 97.5 |
| 35 | Mandibular second premolar (L) | 0.86 | 95.45 |
| 36 | Mandibular first molar (L) | 0.88 | 97.83 |
| 37 | Mandibular second molar (L) | 0.85 | 100 |
| 38 | Mandibular third molar (L) | 0.86 | 100 |
| 41 | Mandibular central incisor (R) | 0.81 | 97.87 |
| 42 | Mandibular lateral incisor (R) | 0.82 | 97.83 |
| 43 | Mandibular canine (R) | 0.88 | 97.78 |
| 44 | Mandibular first premolar (R) | 0.86 | 97.67 |
| 45 | Mandibular second premolar (R) | 0.862 | 95.45 |
| 46 | Mandibular first molar (R) | 0.92 | 100 |
| 47 | Mandibular second molar (R) | 0.93 | 100 |
| 48 | Mandibular third molar (R) | 0.78 | 100 |
| ISO 3950 Code | Name | Dice Index | Accuracy |
|---|---|---|---|
| 51 | Maxillary central incisor (R) | 0.90 | 100 |
| 52 | Maxillary lateral incisor (R) | 0.87 | 100 |
| 53 | Maxillary canine (R) | 0.72 | 100 |
| 54 | Maxillary first molar (R) | n/a | n/a |
| 55 | Maxillary second molar (R) | n/a | n/a |
| 61 | Maxillary central incisor (L) | n/a | n/a |
| 62 | Maxillary lateral incisor (L) | n/a | n/a |
| 63 | Maxillary canine (L) | 0.86 | 100 |
| 64 | Maxillary first molar (L) | 0.87 | 100 |
| 65 | Maxillary second molar (L) | 0.92 | 100 |
| 71 | Mandibular central incisor (L) | 0.87 | 100 |
| 72 | Mandibular lateral incisor (L) | 0.84 | 100 |
| 73 | Mandibular canine (L) | 0.83 | 100 |
| 74 | Mandibular first molar (L) | n/a | n/a |
| 75 | Mandibular second molar (L) | n/a | n/a |
| 81 | Mandibular central incisor (R) | n/a | n/a |
| 82 | Mandibular lateral incisor (R) | 0.89 | 100 |
| 83 | Mandibular canine (R) | 0.91 | 100 |
| 84 | Mandibular first molar (R) | 0.85 | 100 |
| 85 | Mandibular second molar (R) | 0.72 | 83.33 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubiu, G.; Bologna, M.; Cellina, M.; Cè, M.; Sala, D.; Pagani, R.; Mattavelli, E.; Fazzini, D.; Ibba, S.; Papa, S.; et al. Teeth Segmentation in Panoramic Dental X-ray Using Mask Regional Convolutional Neural Network. Appl. Sci. 2023, 13, 7947. https://doi.org/10.3390/app13137947
Rubiu G, Bologna M, Cellina M, Cè M, Sala D, Pagani R, Mattavelli E, Fazzini D, Ibba S, Papa S, et al. Teeth Segmentation in Panoramic Dental X-ray Using Mask Regional Convolutional Neural Network. Applied Sciences. 2023; 13(13):7947. https://doi.org/10.3390/app13137947
Chicago/Turabian StyleRubiu, Giulia, Marco Bologna, Michaela Cellina, Maurizio Cè, Davide Sala, Roberto Pagani, Elisa Mattavelli, Deborah Fazzini, Simona Ibba, Sergio Papa, and et al. 2023. "Teeth Segmentation in Panoramic Dental X-ray Using Mask Regional Convolutional Neural Network" Applied Sciences 13, no. 13: 7947. https://doi.org/10.3390/app13137947
APA StyleRubiu, G., Bologna, M., Cellina, M., Cè, M., Sala, D., Pagani, R., Mattavelli, E., Fazzini, D., Ibba, S., Papa, S., & Alì, M. (2023). Teeth Segmentation in Panoramic Dental X-ray Using Mask Regional Convolutional Neural Network. Applied Sciences, 13(13), 7947. https://doi.org/10.3390/app13137947







