Predicting and Evaluating Decoring Behavior of Inorganically Bound Sand Cores, Using XGBoost and Artificial Neural Networks
Abstract
:1. Introduction
1.1. Motivation and Context
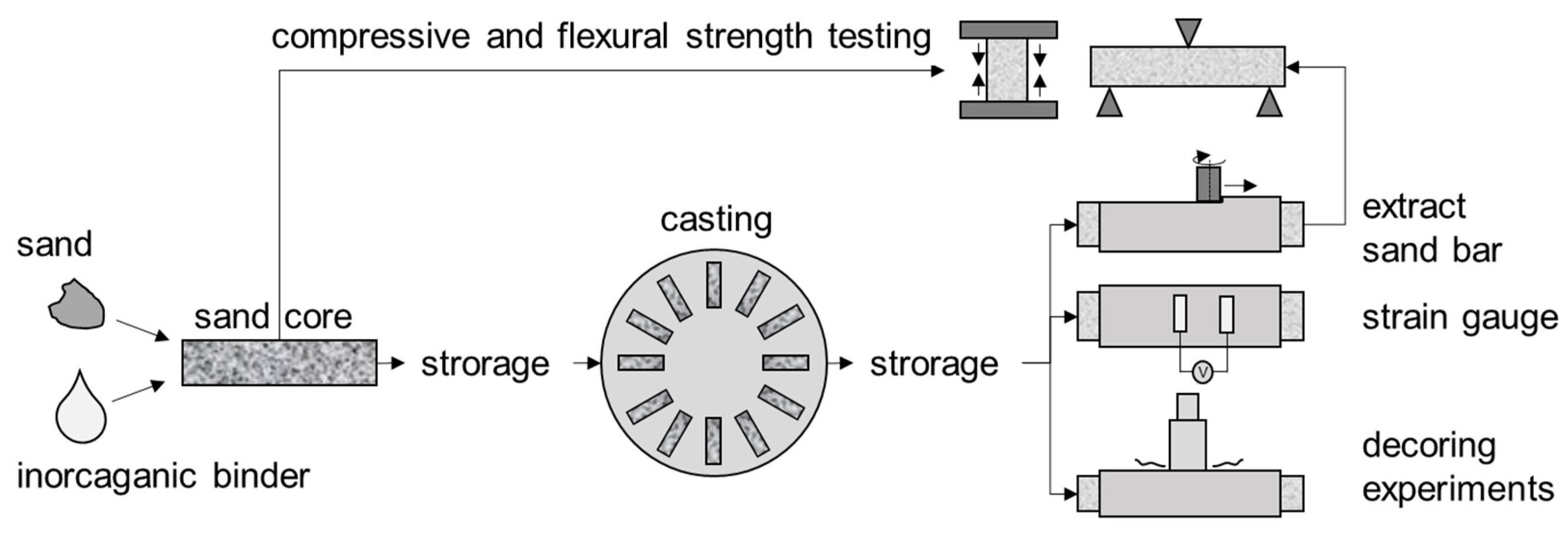
1.2. Decoring of Inorganically Bound Sandcores
- The assumption of a pressure-dependent material model to describe sand-binder systems is valid. It can be seen as essential for an accurate characterization of the mechanical behavior of decoring.
- The decoring behavior of inorganically bound sand-binder systems is significantly influenced by the molding material and the binder system used. The molding material dominates the influence on the decoring behavior.
- Larger binder quantities can lead to higher residual strengths and correspondingly poorer decoring behavior for the same molding material.
- Assuming constant decoring energy, the angle phi and the cohesion of the remaining residual strength after casting are the two main influencing variables for the decoring behavior.
1.3. Machine Learning Models
1.4. Aim of This Work
- Is it possible to train robust machine learning models to predict the decoring behavior of sand-binder systems using the given data?
- Which features are important for the decoring behavior according to the machine learning models, and are those the same as described in the previous work?
- Is it possible to extract further, possibly new insights from the data using machine learning models?
- Categorical features such as the name of the experimental series contain information beyond the collected data, for example, a different room temperature level during the decoring process or slight but systematical differences in the clamping of the specimen. Including these features in the model training will improve the model quality but reduce the interpretability due to the aggregation of information into a single variable.
- The acceleration data contain important information for predicting the decoring behavior. Including it in the model training will improve the model quality.
- Different processing methods of the acceleration data are differently suited for machine learning models.
- Including all features in the model training will reduce the model quality due to a high data complexity without more information than in a reduced dataset. Reducing the data complexity in multiple steps using a feature selection method will improve the model quality for each dataset. At a given threshold, the model quality will drop significantly and abruptly. All features included at that threshold can be considered significantly important for the decoring behavior.
1.5. Approach and Big Picture
2. Materials and Methods
2.1. Data Origin
2.2. Used Model Quality Criteria and Their Interpretation
2.3. Model Training Sequence
3. Results
3.1. Generated Experimental Database and Time-Series Processing
3.2. Data Composition of the Twelve Datasets
3.3. Basic Model Scores for Unfiltered and Filtered Datasets
3.4. Feature Importance Values
3.5. Model Scores for Varying ANN Complexity
3.6. Comparing ANN and XGB over Varying Filter Fractions
4. Discussion
4.1. Global Model Results
4.2. Comparison to Decoring Theory
4.3. Discussion of the Model Training and Model Behavior
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Used Programming Libraries
| System Feature Name | Unit | Composition Feature | Extended Feature | Abbreviated Name |
|---|---|---|---|---|
| binder name | - | X | binder_name_he | |
| sand name | - | X | sand_name_he | |
| binder content | %wt | X | binder_wt | |
| sand-binder system name | - | X | system_name_he | |
| sieving percentage | % | sieving_pct | ||
| mean grain diameter | µm | X | mean_grain_d_microm | |
| flexural strength according to the manufacturer | MPa | flex_manuf | ||
| flexural strength at 25 °C | MPa | flex_raw | ||
| compressive strength at 25 °C | MPa | comp_raw | ||
| phi at 25 °C | ° | phi_raw_grad | ||
| cohesion at 25 °C | - | cohesion_raw | ||
| flexural strength at 400 °C | MPa | X | flex_400_grC | |
| compressive strength at 400 °C | MPa | X | comp_400_grC | |
| phi at 400 °C | ° | X | phi_400_grC_grad | |
| cohesion at 400 °C | - | X | cohesion_400_grC | |
| flexural strength after casting | MPa | X | flex_casted | |
| compressive strength after casting | MPa | X | comp_casted | |
| phi after casting | ° | X | phi_casted_grad | |
| cohesion after casting | - | X | cohesion_casted | |
| tension in the middle, perpendicular | MPa | X | tension_pp_middle | |
| tension in the middle, lengthwise | MPa | X | tension_lw_middle | |
| tension near inflow, perpendicular | MPa | X | tension_pp_inflow | |
| relative flexural strength after casting, according to the manufacturer | % | X | flex_residual_pct | |
| flexural strength after casting, according to the manufacturer | MPa | X | flex_residual | |
| relative drop of flexural strength after casting, according to the manufacturer | % | X | flex_residual_drop_pct |
| Interval Feature Name | Interval Phase | Unit | Abbreviated Name |
|---|---|---|---|
| decored mass target feature “DM” | end | g | 06_i_m_progress |
| mass of specimen | start | g | 00_i_s_m_total |
| already decored distance | start | cm | 01_i_s_d_progress |
| sand mass of specimen | start | g | 02_i_s_m_sand |
| already decored mass | start | g | 03_i_s_m_progress |
| ratio of sand mass to metal mass | start | - | 04_i_s_m_sand_r_metall |
| ratio of sand mass to total mass | start | - | 05_i_s_m_sand_r_total |
| number of impacts during the interval | 20_i_n_hammerblows |
| Preprocessing | No. of Features | Names |
|---|---|---|
| Fast Fourier Transformation 4800 bins of 4 Hz each | 4800 | fft_00004_Hz to fft_19200_hz |
| Mel Frequency Cepstral Coefficient Analysis mean, median, stdev, max, and min for five coefficients | 25 | mean_mfcc01…05 median_mfcc01…05 stdev_mfcc01…05 max_mfcc01…05 min_mfcc01…05 |
| Time-Based Analysis a: absolute acceleration sum: sum over interval (i) or bar (b), max: maximum; pos: positive; neg: negative Example: 21_i_a_sum_max_posneg means “feature 21, absolute sum over all positive and negative maxima over all hammer blows of the examined interval” | 7 | 21_i_a_sum_max_posneg 22_i_a_sum_max_pos 23_i_a_sum_max_neg 24_i_a_max_pos 25_i_a_max_pos 26_i_i_a_mean_max_pos 27_i_a_mean_max_neg |
| Filter Fractions in Percent: | 100, 90, 80, 70, 60, 50, 40, 30, 20, 10, 7, 4, 1 |
| XGB-parameter | value(s) |
| n_estimators | 2000 |
| early_stopping_rounds | 50 |
| eta | 0.1 |
| parallel tree | 1, 3, 5, 7 |
| max depth | 2, 4, 6 |
| min child weight | 1 |
| subsample | 0.6 |
| colsample_bytree | 0.7 |
| colsample_bylevel | 0.7 |
| colsample_bynode | 0.7 |
| alpha | 0.0001 |
| lambda | 1 |
| gamma | 0 |
| random seed | 3 |
| ANN-parameter | value(s) |
| max iter | 2000 |
| n iter no change | 50 |
| layers | (10, 10), (20, 20), (30, 30), (40, 40), (50, 50), (100, 100), (200, 200), (400, 400), (800, 800), (10, 5), (20, 10), (30, 15), (40, 20), (50, 25), (100, 50), (200, 100), (400, 200), (800, 400), (10, 10, 5), (20, 20, 10), (30, 30, 15), (40, 40, 20), (50, 50, 25), (100, 100, 50), (200, 200, 100), (400, 400, 200), (800, 800, 400) |
| alpha | 0.0001 |
| batch size | 30 |
| learning_rate_init | 0.0001 |
| tol | 1 × 10−6 |
| validation fraction | 0.1 |
| activation | relu |
| beta 1 | 0.9 |
| beta 2 | 0.999 |
| epsilon | 1 × 10−8 |
| max fun | 15,000 |
| learning rate | constant |
| shuffle | true |
| random state | 3 |
| Dataset | DS 9 | DS 1 | DS 11 | DS 3 | DS 7 | DS 5 | DS 12 | DS 10 | DS 4 | DS 2 | DS 8 | DS 6 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global Rank | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 |
| mean RMSE (-) | 0.093 | 0.098 | 0.098 | 0.099 | 0.114 | 0.117 | 0.127 | 0.133 | 0.138 | 0.142 | 0.147 | 0.153 |
| best RMSE (-) | 0.069 | 0.070 | 0.070 | 0.073 | 0.081 | 0.090 | 0.080 | 0.077 | 0.079 | 0.080 | 0.122 | 0.126 |
| mean MAE (g) | 5.78 | 5.83 | 5.83 | 5.85 | 7.26 | 7.44 | 8.41 | 9.07 | 9.23 | 9.59 | 9.25 | 10.1 |
| best MAE (g) | 4.90 | 4.39 | 4.39 | 4.84 | 5.35 | 5.74 | 5.66 | 6.06 | 5.77 | 5.75 | 8.12 | 6.73 |
| mean DM (g) (of 10% test data) | 24.7 | 24.7 | 24.7 | 24.8 | 24.8 | 24.8 | 28.8 | 28.1 | 29.0 | 29.0 | 29.8 | 30.2 |
| mean MAE, relative (%) | 23 | 24 | 24 | 30 | 30 | 29 | 32 | 32 | 32 | 33 | 31 | 34 |
| best MAE, relative (%) | 19 | 18 | 18 | 20 | 22 | 23 | 21 | 22 | 21 | 21 | 26 | 25 |
| filter fraction (%) | 40 | 90 | 50 | 100 | 7 | 4 | 30 | 30 | 30 | 30 | 4 | 1 |
| extended features | x | x | x | x | x | x | ||||||
| composition features | x | x | ||||||||||
| signal processing | mfcc | none | time- based | none | FFT | all | time- based | mfcc | none | none | FFT | all |
| Target Feature: Decored Mass in Interval (DM) | ||||||||
| Test Dataset | Best XGBoost | Best ANN | ||||||
| R2 (-) | RMSE (-) | MAE (g) | Rank | R2 (-) | RMSE (-) | MAE (g) | Rank | |
| DS 1 | 0.75 | 0.099 | 5.79 | 1/2 | 0.73 | 0.105 | 6.30 | 1 |
| DS 2 | 0.56 | 0.145 | 10.2 | 8 | 0.57 | 0.148 | 9.98 | 6 |
| DS 3 | 0.75 | 0.099 | 5.85 | 1/2 | 0.71 | 0.108 | 6.38 | 2 |
| DS 4 | 0.53 | 0.149 | 10.4 | 9 | 0.55 | 0.146 | 9.60 | 5 |
| DS 5 | 0.61 | 0.127 | 7.85 | 5/6 | 0.27 | 0.172 | 10.9 | 9 |
| DS 6 | 0.32 | 0.179 | 11.3 | 12 | −0.25 | 0.248 | 16.1 | 12 |
| DS 7 | 0.61 | 0.127 | 7.91 | 5/6 | 0.27 | 0.173 | 10.9 | 10 |
| DS 8 | 0.35 | 0.174 | 11.3 | 11 | −0.12 | 0.229 | 14.6 | 11 |
| DS 9 | 0.73 | 0.105 | 6.74 | 3 | 0.66 | 0.118 | 7.51 | 4 |
| DS 10 | 0.52 | 0.151 | 10.0 | 10 | 0.48 | 0.160 | 10.6 | 8 |
| DS 11 | 0.70 | 0.110 | 6.60 | 4 | 0.68 | 0.113 | 6.85 | 3 |
| DS 12 | 0.62 | 0.138 | 9.29 | 7 | 0.54 | 0.149 | 9.81 | 7 |
| Val Dataset | Same XGB Model as Above | Same ANN Model as Above | ||||||
| R2 (-) | RMSE (-) | MAE (g) | Rank | R2 (-) | RMSE (-) | MAE (g) | Rank | |
| DS 1 | −1.47 | 0.261 | 25.0 | 4 | −2.47 | 0.31 | 28.8 | 5 |
| DS 2 | −2.54 | 0.330 | 33.4 | 7 | −1380 | 6.12 | 100,409 | 12 |
| DS 3 | −1.54 | 0.266 | 25.4 | 5 | −2.83 | 0.33 | 30.4 | 6 |
| DS 4 | −3.50 | 0.371 | 36.8 | 9 | −894 | 4.16 | 100,239 | 11 |
| DS 5 | −0.90 | 0.230 | 22.3 | 1/2 | −1.52 | 0.26 | 25.1 | 3 |
| DS 6 | −4.54 | 0.411 | 40.0 | 12 | −7.54 | 0.47 | 44.5 | 7 |
| DS 7 | −0.90 | 0.230 | 22.3 | 1/2 | −1.19 | 0.25 | 23.7 | 2 |
| DS 8 | −4.17 | 0.397 | 38.8 | 10 | −29.7 | 0.82 | 71.2 | 8 |
| DS 9 | −1.21 | 0.247 | 23.8 | 3 | −0.64 | 0.21 | 20.9 | 1 |
| DS 10 | −4.34 | 0.403 | 39.4 | 11 | −1188 | 5.33 | 427 | 9 |
| DS 11 | −1.59 | 0.269 | 25.6 | 6 | −1.80 | 0.28 | 26.2 | 4 |
| DS 12 | −2.66 | 0.335 | 33.8 | 8 | −2803 | 7.51 | 400,179 | 10 |
| Target Feature: Decored Mass | ||||||||||
| Test Dataset | Best XGBoost | Best ANN | ||||||||
| R2 (-) | RMSE (-) | MAE (g) | Filter Fraction (%) | Rank | R2 (-) | RMSE (-) | MAE (g) | Filter Fraction (%) | Rank | |
| DS 1 | 0.76 | 0.098 | 5.83 | 90 | 2/3 | 0.73 | 0.105 | 6.30 | 100 | 3/4 |
| DS 2 | 0.56 | 0.145 | 10.2 | 100 | 9 | 0.59 | 0.142 | 9.59 | 30 | 9 |
| DS 3 | 0.75 | 0.099 | 5.85 | 100 | 4 | 0.73 | 0.104 | 6.05 | 60 | 2 |
| DS 4 | 0.55 | 0.146 | 10.2 | 90 | 10 | 0.61 | 0.138 | 9.23 | 30 | 5 |
| DS 5 | 0.67 | 0.117 | 7.44 | 4 | 6 | 0.54 | 0.137 | 8.78 | 4 | 6 |
| DS 6 | 0.47 | 0.153 | 10.1 | 1 | 12 | 0.42 | 0.157 | 10.4 | 4 | 11 |
| DS 7 | 0.68 | 0.114 | 7.26 | 7 | 5 | 0.51 | 0.141 | 9.03 | 4 | 8 |
| DS 8 | 0.50 | 0.147 | 9.25 | 4 | 11 | 0.43 | 0.161 | 10.7 | 4 | 12 |
| DS 9 | 0.78 | 0.093 | 5.78 | 40 | 1 | 0.75 | 0.100 | 6.19 | 20 | 1 |
| DS 10 | 0.64 | 0.133 | 9.07 | 30 | 8 | 0.56 | 0.143 | 9.47 | 30 | 10 |
| DS 11 | 0.76 | 0.098 | 5.83 | 50 | 2/3 | 0.73 | 0.105 | 6.37 | 50 | 3/4 |
| DS 12 | 0.67 | 0.127 | 8.41 | 30 | 7 | 0.62 | 0.139 | 9.19 | 70 | 7 |
| Val. Dataset | Same XGB Model as Above | Same ANN Model as Above | ||||||||
| R2 (-) | RMSE (-) | MAE (g) | Filter Fraction (%) | Rank | R2 (-) | RMSE (-) | MAE (g) | Filter Fraction (%) | Rank | |
| DS 1 | −1.51 | 0.264 | 25.2 | 90 | 4/5 | −2.5 | 0.308 | 28.8 | 100 | 6 |
| DS 2 | −2.54 | 0.330 | 33.4 | 100 | 8 | −3.8 | 0.382 | 37.5 | 30 | 7 |
| DS 3 | −1.54 | 0.266 | 25.4 | 100 | 6 | −2.0 | 0.288 | 27.2 | 60 | 5 |
| DS 4 | −3.33 | 0.364 | 36.2 | 90 | 10 | −54 | 0.932 | 79.1 | 30 | 10 |
| DS 5 | −1.00 | 0.236 | 22.9 | 4 | 1 | −0.6 | 0.213 | 20.9 | 4 | 3 |
| DS 6 | −5.01 | 0.428 | 41.3 | 1 | 12 | −5.1 | 0.429 | 41.4 | 4 | 9 |
| DS 7 | −1.12 | 0.243 | 23.4 | 7 | 2 | −1.0 | 0.236 | 22.9 | 4 | 4 |
| DS 8 | −3.94 | 0.388 | 38.1 | 4 | 11 | −4.4 | 0.404 | 39.4 | 4 | 8 |
| DS 9 | −1.15 | 0.245 | 23.6 | 40 | 3 | −0.3 | 0.190 | 19.0 | 20 | 1 |
| DS 10 | −2.66 | 0.335 | 33.8 | 30 | 9 | −44 | 1.002 | 87.0 | 30 | 11 |
| DS 11 | −1.51 | 0.264 | 25.2 | 50 | 4/5 | −0.5 | 0.205 | 20.3 | 50 | 2 |
| DS 12 | −2.40 | 0.322 | 32.8 | 30 | 7 | −1260 | 5309 | 2.00 × 105 | 70 | 12 |
References
- Holtzer, M. Mold and Core Sands in Metalcasting. Sustainable Development; Springer International Publishing AG: Cham, Switzerland, 2020; pp. 219–221. [Google Scholar] [CrossRef]
- Ettemeyer, F.; Schweinefuß, M.; Lechner, P.; Stahl, J.; Greß, T.; Kaindl, J.; Durach, L.; Volk, W.; Günther, D. Characterisation of the decoring behaviour of inorganically bound cast-in sand cores for light metal casting. J. Mater. Process. Technol. 2021, 296, 117201. [Google Scholar] [CrossRef]
- Herfurth, K.; Scharf, S. Casting. In Springer Handbook of Mechanical Engineering; Karl-Heinrich, G., Hamid, H., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 98, pp. 325–356. [Google Scholar] [CrossRef]
- Xin, F.H.; Liu, W.H.; Song, L.; Li, Y.M. Modification of inorganic binder used for sand core-making in foundry practice. China Foundry 2020, 17, 341–346. [Google Scholar] [CrossRef]
- Stauder, B.J.; Berbic, M.; Schumacher, P. Mohr-Coulomb failure criterion from unidirectional mechanical testing of sand cores after thermal exposure. J. Mater. Process. Technol. 2019, 274, 116274. [Google Scholar] [CrossRef]
- Lechner, P.; Stahl, J.; Hartmann, C.; Ettemeyer, F.; Volk, W. Mohr–Coulomb characterisation of inorganically-bound core materials. J. Mater. Process. Technol. 2021, 296, 117214. [Google Scholar] [CrossRef]
- Lee, K.; Ayyasamy, M.V.; Ji, Y.; Balachandran, P.V. A comparison of explainable artificial intelligence methods in the phase classification of multi-principal element alloys. Sci. Rep. 2022, 12, 11591. [Google Scholar] [CrossRef]
- Stadter, C.; Kick, M.K.; Schmoeller, M.; Zaeh, M.F. Correlation analysis between the beam propagation and the vapor capillary geometry by machine learning. Procedia CIRP 2020, 94, 742–747. [Google Scholar] [CrossRef]
- Wang, P.; Fan, Z.; Kazmer, D.O.; Gao, R.X. Orthogonal Analysis of Multisensor Data Fusion for Improved Quality Control. J. Manuf. Sci. Eng. 2017, 139, 5. [Google Scholar] [CrossRef]
- Meng, Y.; Yang, N.; Qian, Z.; Zhang, G. What Makes an Online Review More Helpful: An Interpretation Framework Using XGBoost and SHAP Values. J. Theor. Appl. Electron. Commer. Res. 2021, 16, 466–490. [Google Scholar] [CrossRef]
- Philine, K.; Johannes, G.; Dierk, H. Analyse von Gießereidaten mit Methoden des Maschinellen Lernens—Teil 2. Giess.-Prax. 2018, 69, 9–15. Available online: https://www.giesserei-praxis.de/news-artikel/artikel/analyse-von-giessereidaten-mit-methoden-des-maschinellen-lernens-teil-2 (accessed on 16 June 2023).
- Nasiri, H.; Kheyroddin, G.; Dorrigiv, M.; Esmaeili, M.; Nafchi, A.R.; Ghorbani, M.H.; Zarkesh-Ha, P. Classification of COVID-19 in Chest X-ray Images Using Fusion of Deep Features and LightGBM. In Proceedings of the 2022 IEEE World AI IoT Congress (AIIoT), Seattle, WA, USA, 6–9 June 2022; IEEE: New York, NY, USA, 2022; pp. 201–206. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery (KDD ’16): New York, NY, USA, 2016; pp. 785–794. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Algahtani, M.; Kaewunruen, S. Energy Forecasting in a Public Building: A Benchmarking Analysis on Long Short-Term Memory (LSTM), Support Vector Regression (SVR), and Extreme Gradient Boosting (XGBoost) Networks. Appl. Sci. 2022, 12, 9788. [Google Scholar] [CrossRef]
- Chelgani, S.C.; Nasiri, H.; Tohry, A. Modeling of particle sizes for industrial HPGR products by a unique explainable AI tool-A “Conscious Lab” development. Adv. Powder Technol. 2021, 32, 4141–4148. [Google Scholar] [CrossRef]
- Fatahi, R.; Nasiri, H.; Homafar, A.; Khosravi, R.; Siavoshi, H.; Chehreh Chelgani, S. Modeling operational cement rotary kiln variables with explainable artificial intelligence methods—A “conscious lab” development. Part. Sci. Technol. 2023, 41, 715–724. [Google Scholar] [CrossRef]
- Lundberg, S.M.; Lee, S.-I. A Unified Approach to Interpreting Model Predictions. In Advances in Neural Information Processing Systems 30; Guyon, I., Luxburg, U.V., Bengio, S., Wallach, H., Fergus, R., Vishwanathan, S., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2017; pp. 4765–4774. Available online: http://papers.nips.cc/paper/7062-a-unified-approach-to-interpreting-model-predictions.pdf (accessed on 16 June 2023).
- Volk, W.; Groche, P.; Brosius, A.; Ghiotti, A.; Kinsey, B.L.; Liewald, M.; Junying, M.; Jun, Y. Models and modelling for process limits in metal forming. CIRP Annals 2019, 68, 775–798. [Google Scholar] [CrossRef]
- Chehreh Chelgani, S.; Nasiri, H.; Tohry, A.; Heidari, H.R. Modeling industrial hydrocyclone operational variables by SHAP-CatBoost—A “conscious lab” approach. Powder Technol. 2023, 420, 118416. [Google Scholar] [CrossRef]
- Nasiri, H.; Ebadzadeh, M.M. MFRFNN: Multi-Functional Recurrent Fuzzy Neural Network for Chaotic Time Series Prediction. Neurocomputing 2022, 507, 292–310. [Google Scholar] [CrossRef]
- Chang, A.M.; Freeze, J.G.; Batista, V.S. Hammett neural networks: Prediction of frontier orbital energies of tungsten-benzylidyne photoredox complexes. Chem. Sci. 2019, 10, 6844–6854. [Google Scholar] [CrossRef] [Green Version]
- Lechner, P.; Heinle, P.; Hartmann, C.; Bauer, C.; Kirchebner, B.; Dobmeier, F.; Volk, W. Feasibility of Acoustic Print Head Monitoring for Binder Jetting Processes with Artificial Neural Networks. Appl. Sci. 2021, 11, 10672. [Google Scholar] [CrossRef]
- Rossum, G.; Drake, F.L., Jr. Python reference manual: Centrum voor Wiskunde en Informatica Amsterdam. 1995. Available online: https://docs.python.org/3/reference/index.html (accessed on 16 June 2023).
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. Available online: https://www.jmlr.org/papers/v12/pedregosa11a.html (accessed on 16 June 2023).
- Wes, M. Data Structures for Statistical Computing in Python. In Proceedings of the 9th Python in Science Conference, Austin, TX, USA, 28 June–3 July 2010; pp. 56–61. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, S.M.; Erion, G.; Chen, H.; DeGrave, A.; Prutkin, J.M.; Nair, B.; Katz, R.; Himmelfarb, J.; Bansal, N.; Lee, S.-I. From local explanations to global understanding with explainable AI for trees. Nat. Mach. Intell. 2020, 2, 2522–5839. [Google Scholar] [CrossRef] [PubMed]
- Hunter, J.D. Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 2007, 9, 90–95. [Google Scholar] [CrossRef]
- McFee, B.; Raffel, C.; Liang, D.; Ellis, D.P.; McVicar, M.; Battenberg, E.; Nieto, O. Librosa: Audio and music signal analysis in python. In Proceedings of the 14th Python in Science Conference, Austin, TX, USA, 6–12 July 2015; Volume 8. [Google Scholar] [CrossRef] [Green Version]
- Harris, C.R.; Millman, K.J.; Van Der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef] [PubMed]





















| Sand-Binder Systems | ||||||||
|---|---|---|---|---|---|---|---|---|
| Feature | A | B | D | L | M | N | P | Q |
| binder name | R | R_low | R | R | R_low | R_low | R_low | R |
| binder content | 1.9 | 1.5 | 2.25 | 1.9 | 1.9 | 1.9 | 1.9 | 2.25 |
| sand name | H32 | H32 | F34 | F34 | H32 | W65 | F34 | H32 |
| number of specimens tested | 17 | 12 | 2 | 16 | 12 | 6 | 5 | 5 |
| total number of intervals conducted | 64 | 17 | 23 | 48 | 54 | 8 | 36 | 53 |
| extended features | yes | yes | yes | no | no | no | no | no |
| Datasets | Base Features | Extended Features | Aggregated Composition Features | Signal Features | No. of Features | Number of Datapoints |
|---|---|---|---|---|---|---|
| DS 1 | X | none | 10 | 280 | ||
| DS 2 | X | X | none | 24 | 81 | |
| DS 3 | X | X | none | 24 | 280 | |
| DS 4 | X | X | X | none | 32 | 81 |
| DS 5 | X | all | 4842 | 280 | ||
| DS 6 | X | X | all | 4856 | 81 | |
| DS 7 | X | FFT | 4810 | 280 | ||
| DS 8 | X | X | FFT | 4824 | 81 | |
| DS 9 | X | mfcc | 35 | 280 | ||
| DS 10 | X | X | mfcc | 49 | 81 | |
| DS 11 | X | time-based | 17 | 280 | ||
| DS 12 | X | X | time-based | 31 | 81 |
| All 507 Parameter Sets | 20 Best Parameter Sets of Each Dataset (According to Mean RMSE) | |||||
|---|---|---|---|---|---|---|
| Datasets | Average | Standard Deviation | p-Value | Average | Standard Deviation | p-Value |
| DS 9 | 0.128 | 6.07 × 10−4 | - | 0.096 | 1.24 × 10−6 | - |
| DS 1 | 0.138 | 6.12 × 10−4 | 3.08 × 10−10 | 0.100 | 2.76 × 10−7 | 1.97 × 10−14 |
| DS 11 | 0.138 | 9.57 × 10−4 | 1.82 × 10−8 | 0.101 | 4.98 × 10−7 | 4.31 × 10−18 |
| DS 3 | 0.134 | 8.85 × 10−4 | 6.63 × 10−4 | 0.100 | 1.49 × 10−7 | 2.15 × 10−14 |
| DS 7 | 0.161 | 5.84 × 10−4 | 2.43 × 10−86 | 0.117 | 1.68 × 10−6 | 1.43 × 10−37 |
| DS 5 | 0.161 | 5.50 × 10−4 | 1.67 × 10−84 | 0.119 | 1.23 × 10−6 | 7.83 × 10−41 |
| DS 12 | 0.174 | 8.28 × 10−4 | 9.63 × 10−123 | 0.132 | 8.68 × 10−6 | 3.11 × 10−26 |
| DS 10 | 0.179 | 6.70 × 10−4 | 2.97 × 10−157 | 0.137 | 5.05 × 10−6 | 8.84 × 10−34 |
| DS 4 | 0.176 | 6.40 × 10−4 | 4.81 × 10−147 | 0.147 | 6.46 × 10−6 | 5.84 × 10−33 |
| DS 2 | 0.177 | 5.92 × 10−4 | 3.43 × 10−152 | 0.148 | 3.77 × 10−6 | 2.65 × 10−40 |
| DS 8 | 0.218 | 1.73 × 10−3 | 2.57 × 10−206 | 0.152 | 1.20 × 10−5 | 3.85 × 10−28 |
| DS 6 | 0.220 | 1.56 × 10−3 | 1.61 × 10−222 | 0.156 | 3.29 × 10−6 | 6.30 × 10−45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dobmeier, F.; Li, R.; Ettemeyer, F.; Mariadass, M.; Lechner, P.; Volk, W.; Günther, D. Predicting and Evaluating Decoring Behavior of Inorganically Bound Sand Cores, Using XGBoost and Artificial Neural Networks. Appl. Sci. 2023, 13, 7948. https://doi.org/10.3390/app13137948
Dobmeier F, Li R, Ettemeyer F, Mariadass M, Lechner P, Volk W, Günther D. Predicting and Evaluating Decoring Behavior of Inorganically Bound Sand Cores, Using XGBoost and Artificial Neural Networks. Applied Sciences. 2023; 13(13):7948. https://doi.org/10.3390/app13137948
Chicago/Turabian StyleDobmeier, Fabian, Rui Li, Florian Ettemeyer, Melvin Mariadass, Philipp Lechner, Wolfram Volk, and Daniel Günther. 2023. "Predicting and Evaluating Decoring Behavior of Inorganically Bound Sand Cores, Using XGBoost and Artificial Neural Networks" Applied Sciences 13, no. 13: 7948. https://doi.org/10.3390/app13137948
APA StyleDobmeier, F., Li, R., Ettemeyer, F., Mariadass, M., Lechner, P., Volk, W., & Günther, D. (2023). Predicting and Evaluating Decoring Behavior of Inorganically Bound Sand Cores, Using XGBoost and Artificial Neural Networks. Applied Sciences, 13(13), 7948. https://doi.org/10.3390/app13137948







