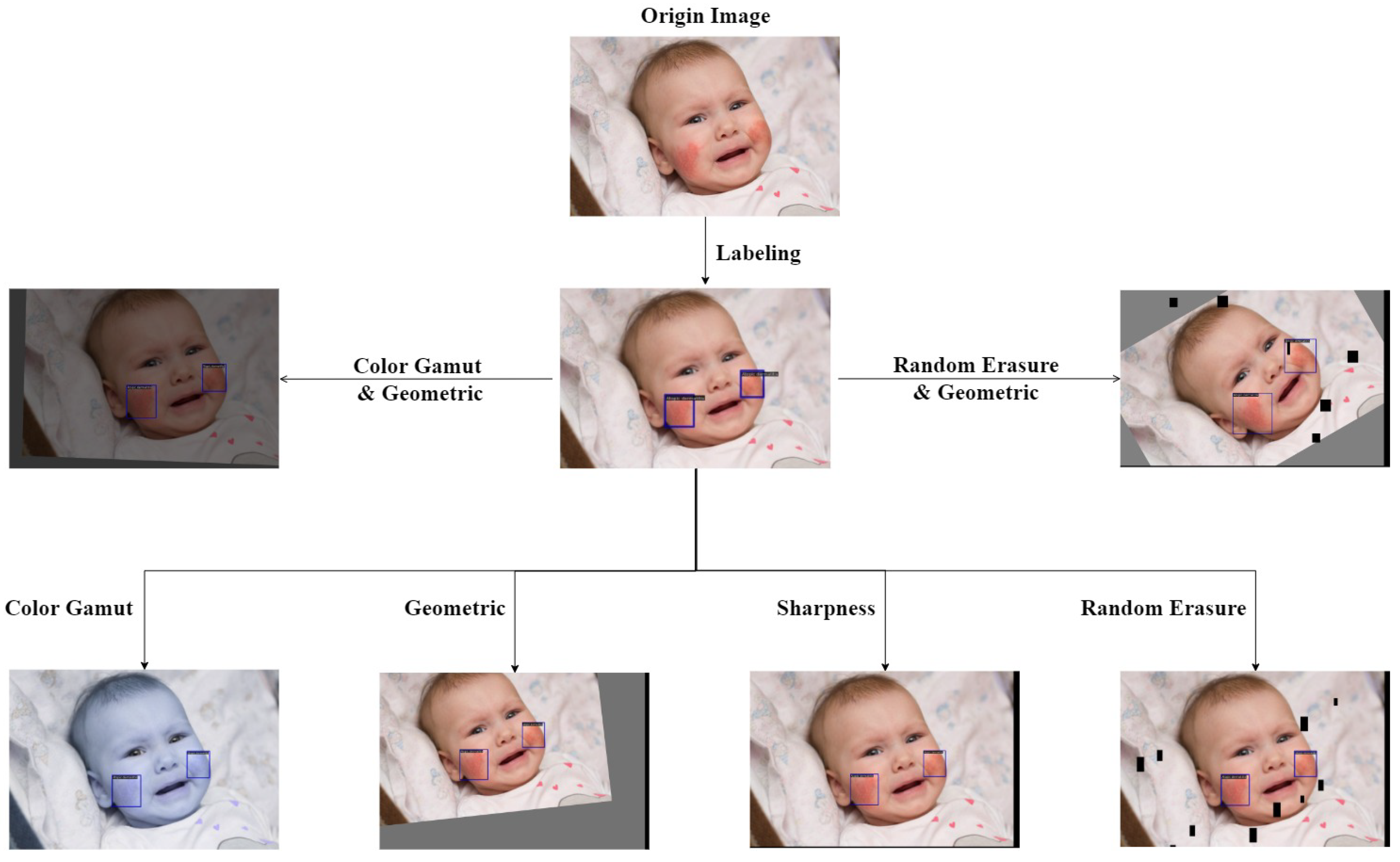
5.4.1. Random Online Data Augmentation
In this experiment, we use ResNet-50, ConvNeXt-T, Swin-T, Res2Net-101, ResNeXt-101, and PVTv2-B2, as the feature extraction networks, based on the Faster R-CNN algorithm. In training on the CPD-10 dataset, we observe varying degrees of overfitting and inadequate generalization ability. This is primarily due to the tiny data size. In this paper, we employ the Random Online Data Augmentation preprocessing method.
Table 6 displays a comparison of model mAP values, with RDA denoting the Random Online Data Augmentation method.
It is apparent that, in the classification and identification of natural images of common pediatric dermatoses, PVTv2-B2 has a superior feature extraction ability for disease representation in comparison to the convolutional networks ResNet-50, Res2Net-101, ResNeXt-101, and ConvNeXt-T, which possess local feature extraction ability, and the Swin-T network, which disregards the local feature continuity of the image. This is primarily due to the fact that the PVTv2-B2 backbone network is not only capable of extracting global information, but also contains more local continuity of the image, allowing it to effectively extract the overall characteristics of the disease representation. No matter which backbone network is used, however, there is overfitting in training due to the small scope of the dataset and the data’s homogeneity.
To address this issue, we employ the combined Random Online Augmentation method, which can improve the model’s mAP by more than
, with the AP of small, medium, and large objects all being improved to varying degrees. This is primarily because stochastic data augmentation can increase the diversity of training data, thereby alleviating the overfitting problem during training, making the model more robust and generalizable, and decreasing the leakage detection rate. However, the randomness of the data augmentation strategy causes instability in training. As shown in
Table 6, although the overall performance of the model was improved after using the Random Online Data Augmentation method, the accuracy for small objects decreased instead with the ResNet-50, Swin-T, and Res2Net-101 networks, which may be because some small lesion representations are impacted by the stochastic augmentation strategy, e.g., chickenpox discrimination may be weakened under increasing illumination. To prove it, using ResNet-50 as the backbone network, we conducted repeated experiments with the Random Online Data Augmentation method, with RDA denoting the Random Online Data Augmentation method, the results of which are shown in
Table 7.
It can be seen from
Table 7 that, while the overall performance of the model was improved in repeated experiments, small target detection accuracy enhancement was not stable, which is inextricably related to the randomness augmentation strategy, as well as the sensitivity of the network on small objects. In addition, although random online data enhancement improves the detection accuracy, the model’s precision is relatively low. This is due, in part, to the low resolution and subpar quality of the majority of the original images, which makes training difficult.
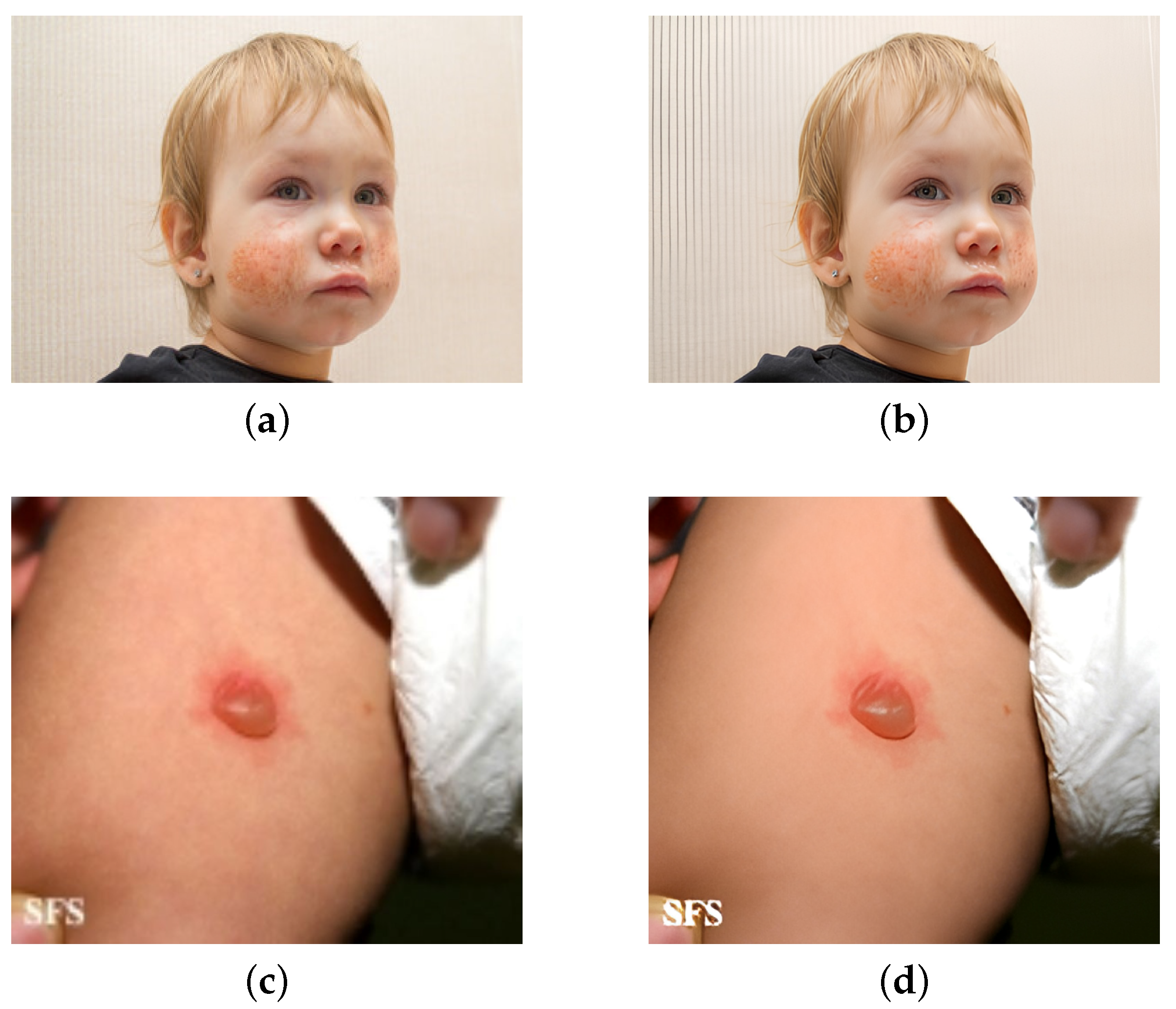
5.4.2. Selective Super-Resolution Reconstruction
In order to increase the Precision of disease symptom recognition, it is important to address the hardware equipment limitations and other issues that result in fuzziness, poor quality, and insignificant interest regions. SwinIR was used to conduct super-resolution reconstruction on some low-quality original images that had been filtered-out based on the findings of Random Online Data Augmentation. The experimental results are shown in
Table 8.
The experimental results demonstrate that using Super Resolution Reconstruction not only improves image quality, but also increases the accuracy of model detection and the mAP. The PVTv2-B2 feature extraction network, which achieves the greatest detection effect at present, was chosen to compare the precision of detecting 10 types of diseases.
Figure 9 depicts the estimation of the effect of Selective Super Resolution Reconstruction.
It is evident that, after selective picture super-resolution reconstruction, the model’s disease detection accuracy increased. This is primarily due to the fact that, following image Super-Resolution Reconstruction, can more effectively enhance the clarity of the lesion area, sharpen the edge features, and reduce the impact of image noise, improving the accuracy of model detection.
5.4.3. DK_Loss
There are 10 diseases involved in the CPD-10 dataset, and it has similarities and variations within each class that affect how challenging it is to diagnose different diseases. We developed the DK_Loss loss function to address this issue and enable the two-stage object recognition algorithm to concentrate more on the difficult-to-classify samples. We compare DK_Loss with the frequently used cross-entropy loss function in the two-stage object identification algorithm. The evaluation is conducted over the CPD-10 original dataset and the CPD-10 augmented dataset, which was created through online data augmentation and Selective Super-Resolution Reconstruction, respectively. When
, only the impact of the coefficients
is verified. Various values for
are established and compared, and the optimal parameter values for
are then determined. In
Table 9, the testing findings are displayed.
Combining the experimental results of DK Loss on the original CPD-10 dataset and the augmented dataset in
Table 9, we can observe that when
, i.e., only increasing the coefficient
, the mAP of the model can be improved, which has a positive influence on training. This is because the corresponding score
of a sample’s true category can, to some extent, reflect the classification difficulty of a sample. The closer
is to 1, the more easily the sample can be considered a simple sample, and the closer the coefficient
is to 0, the easier the sample is. In contrast, the coefficient
is closer to 1 than to 0. By adding coefficients
, the loss contribution of the samples that are simple to classify can be reduced, allowing the model to focus on the learning of samples that are difficult to classify.
Meanwhile, by varying , we can also determine, for multiple detection boxes that can be identified as detecting the same lesion region, the predicted number of distinct categories , which play a complementary role to the coefficient . On the original CPD-10 dataset, the model mAP can be enhanced by more than 2%. When or , i.e., , it not only reduces the influence of anomalous samples within a certain range, but also improves the model’s mAP. By increasing the contribution of loss to hard samples, the model focuses on learning the majority of normal hard samples. In contrast, model training is degraded when , i.e., , is influenced by the anomalous samples.
In addition, there is a significant disparity between the effect of model enhancement on the original CPD-10 dataset and the augmented dataset. When training on the CPD-10 original dataset, the PVTv2-B2 backbone network has the highest degree of overfitting and the most apparent improvement. This is primarily due to overfitting of varying degrees. It can be seen that the DK_Loss loss function not only addresses the problem of unbalanced hard and easy samples from the dataset, but also reduces overfitting during training, which is more pertinent to object detection on small datasets, for which data collection is more challenging.
Random Online Data Augmentation has a destabilizing effect on the experimental outcomes of the CPD-10 augmented dataset. To more accurately determine the value of , we conduct experiments on the PASCAL VOC dataset, utilizing the VOC2007 training set, and evaluate the results on the VOC2007 test set. Additionally, we demonstrate the efficacy of the DK_Loss loss function.
As shown in
Table 10, when the parameter
is set in the DK_Loss loss function, the mAP improves by
for the Faster R-CNN,
for the Cascade R-CNN, and
for the Dynamic R-CNN, which is the current optimal, but taken as 4, the performance decreases instead, primarily because the DK_Loss loss function makes the model focus more on learning difficult samples, by increasing the loss contribution of difficult samples. If the value of
is not restricted with
, the focus of the model will always be on the abnormal hard-to-classify samples, which may not be correctly identified for other reasons, and mislead the subsequent optimization direction of the model. Therefore,
is not as large as possible, but depends on the number of misclassification categories of most normal samples in the dataset, making the model focus on the global situation of the distribution of normal difficult samples rather than extreme data. Additionally, the
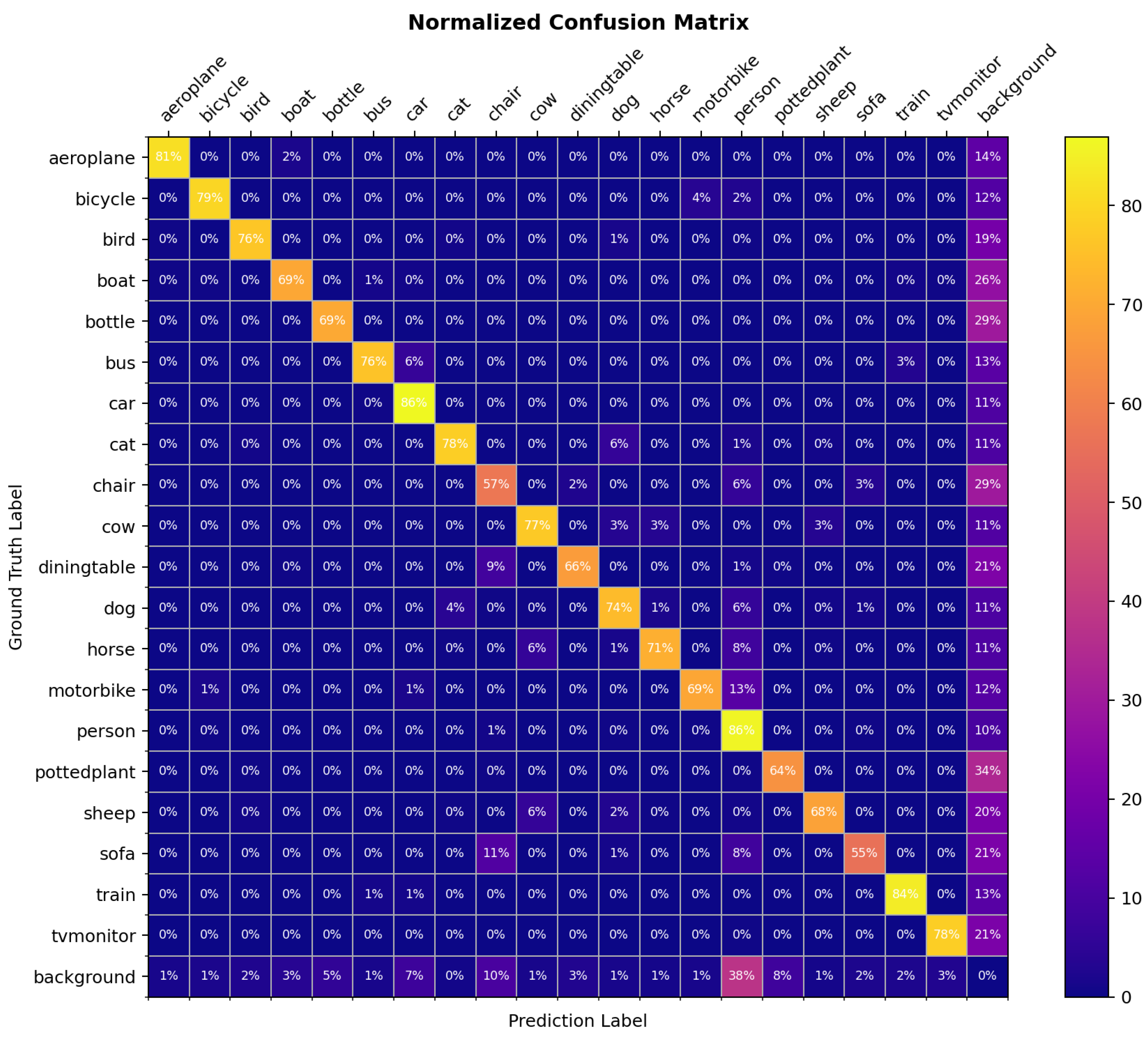
’s value mainly depends on the likelihood of the misclassification category number for most samples. For example, for the PASCAL VOC2007 dataset, the confusion matrix of the model trained using the Cascade R-CNN is shown in
Figure 10. It can be seen that most of the category confusion category numbers are less than or equal to 3, Consistent with the experimental results, so the proposed value of
is set to 3.
The similarity between the conclusion and the CPD-10 dataset results validates the validity and generalizability of the DK_Loss loss function. Noting that the specific value of may vary across datasets, when is not applicable, you can observe the rough interval of value through the confusion matrix, based on the likelihood of the misclassification category number for most samples, and then experimentally set the optimal value.
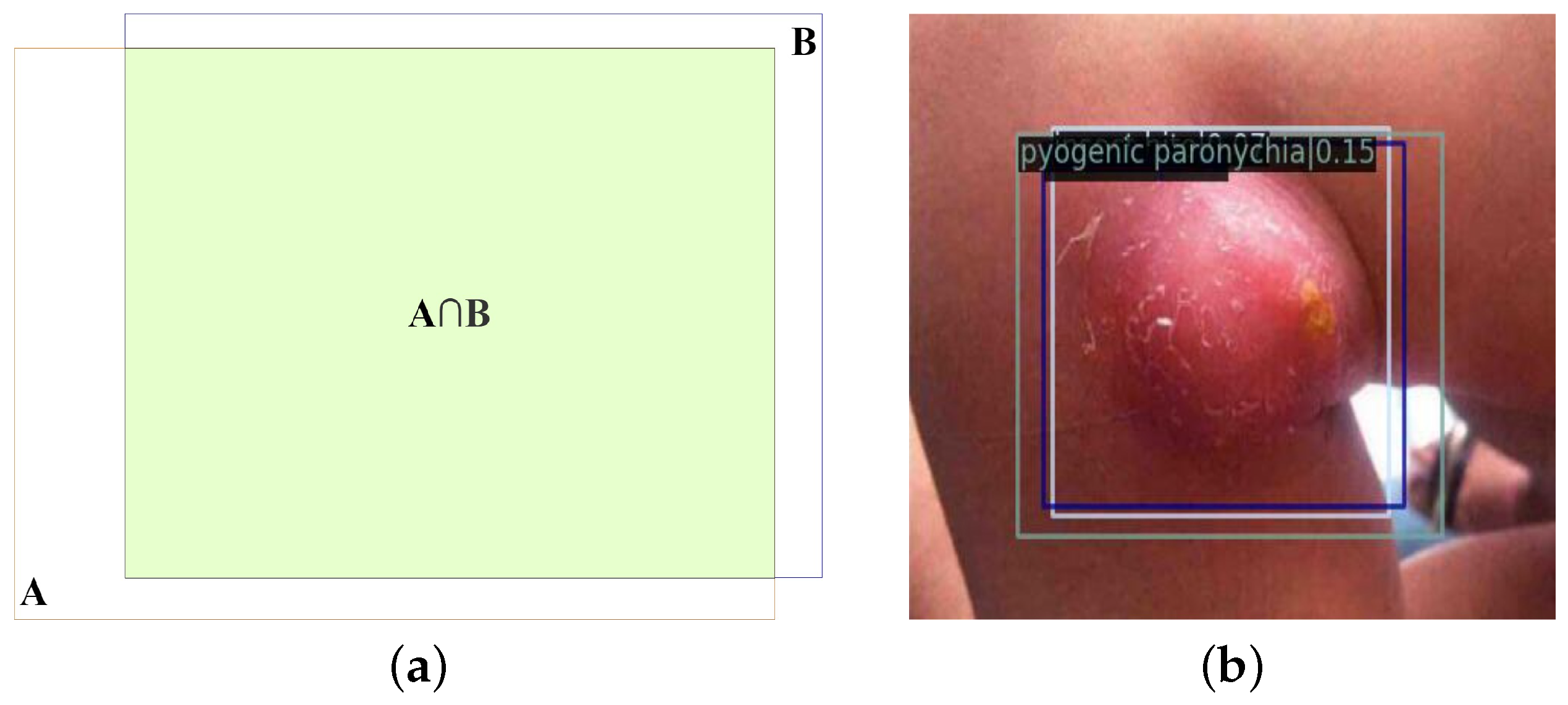
5.4.4. Fliter_nms
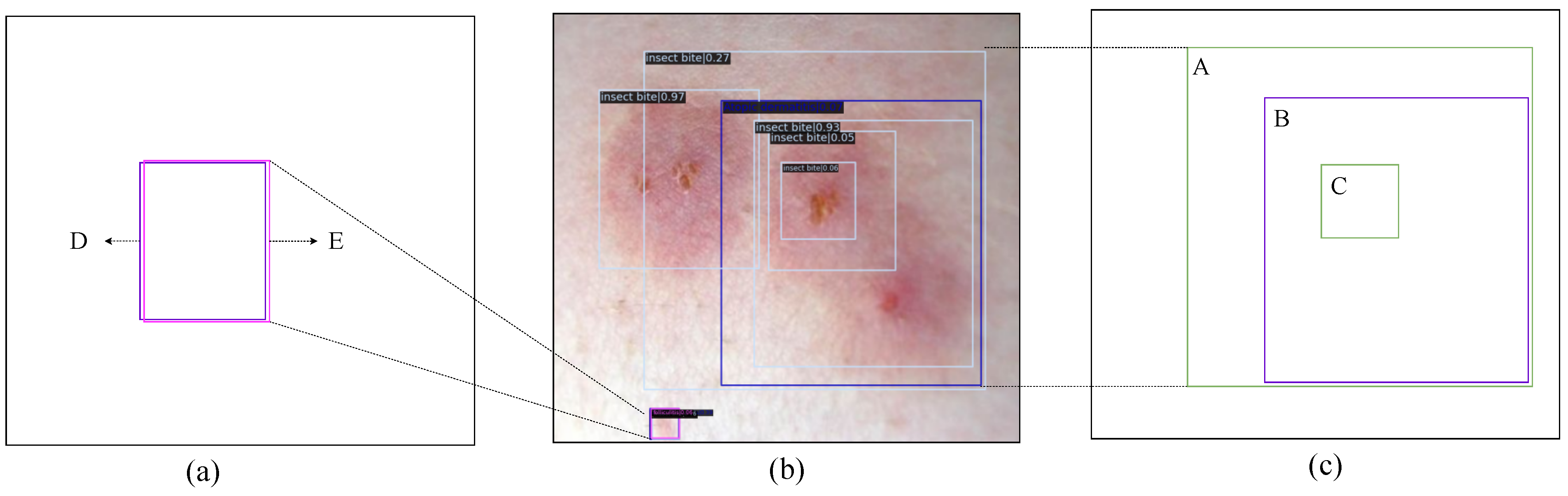
The experiments in this section are based on the outcomes of the DK_Loss experiments conducted in the previous section. The evaluation employs the PVTv2-B2 backbone network with different con_thr and cro_thr threshold parameters. First, eliminate the detection boxes for inclusion and contained relationships, and have distinct prediction categories and greater than con_thr confidence score differences. Then, the detection boxes that can be identified as detecting the same lesion region, have distinct prediction categories, and where the difference in confidence scores is greater than the set value cro_thr are deleted. The results of the experiments are presented in
Table 11.
According to the experiment results, the mAP of the model with various con_thr and cro_thr threshold settings is relatively smooth. In addition, when
and
are set, the best filtering effect is achieved for the misclassified boxes. Setting
and
and utilizing various post-processing algorithms,
Figure 11 compares the detection precision of 10 disease representations with
and
.
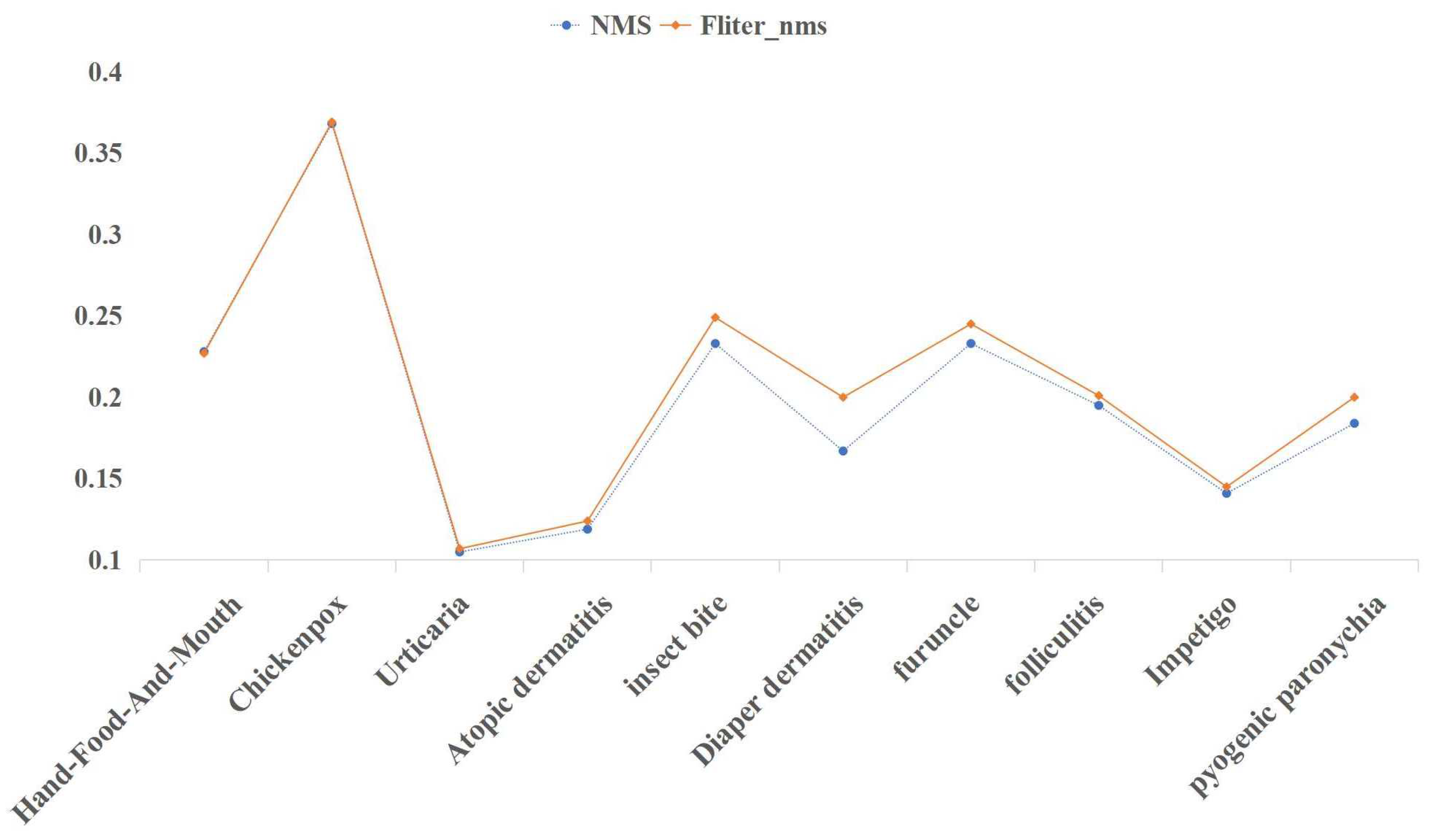
In
Figure 11, it is clear that the Fliter_nms post-processing algorithm can increase accuracy while largely maintaining mAP. The Fliter_nms algorithm can reduce the interference caused by the messy detection boxes and satisfy the requirement of natural image object detection of common pediatric dermatoses, which takes precision as the first criterion, by filtering parts of the detection boxes that are obviously incorrect in prediction categories.
Simultaneously, we can observe an improvement in the precision of disease detection; however, compared to four diseases, namely insect bite, diaper dermatitis, furuncle, and pyogenic paronychia, there is no significant improvement. This is because of the fact that, for the screened detection boxes with evident prediction errors, the disease features in the prediction region cannot share a high degree of similarity with other disease features. Otherwise, it is highly probable that multiple prediction categories and their corresponding probabilities do not differ significantly, thereby failing to meet the threshold requirement. There are similarities between hand-foot-and-mouth disease, varicella, and folliculitis, as well as urticaria, atopic dermatitis, and impetigo, among the 10 diseases in the CPD-10 dataset. Consequently, the detection precision of these six diseases has not substantially improved. Real scenarios of natural images of common pediatric dermatoses after different post-processing methods are evaluated in
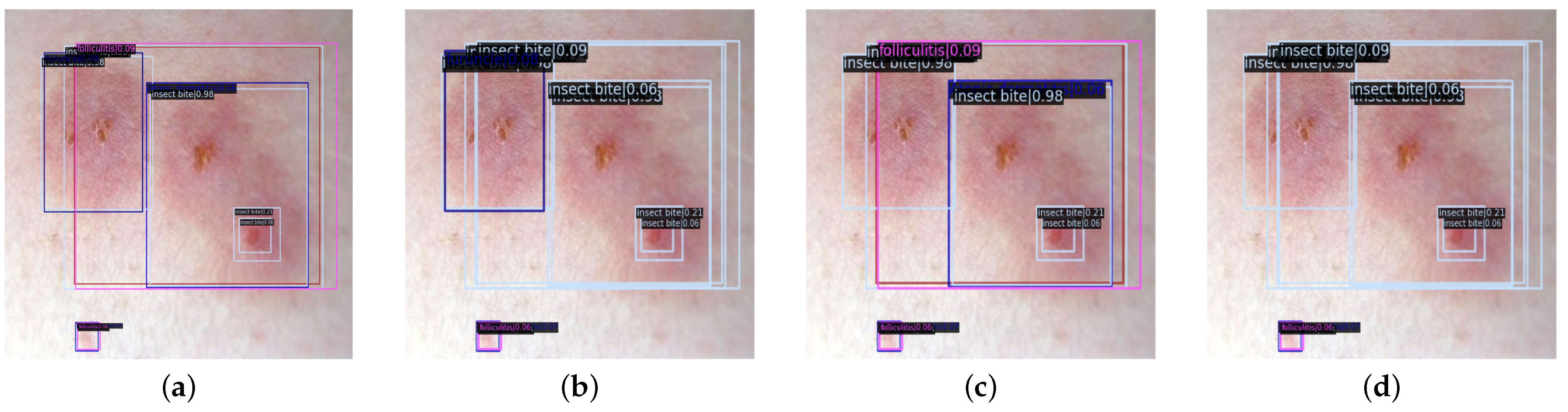
Figure 12.