Abstract
Butenolides are a family of lactones containing a double bond and have been frequently found in the extracts of Streptomyces bacterial species with various pharmacological activities. In this study, seven butenolides (1–7) were discovered and isolated from the culture broth of a marine-derived Streptomyces sp. 13G036 based on a molecular networking analysis. Among the seven isolates, compound 7 was first isolated as a natural product in this study. The structures of compounds 1–7 were determined by combined analysis of 1D/2D Nuclear Magnetic Resonance (NMR) spectra, Mass Spectrometry (MS) spectra and electronic circular dichroism (ECD) data. Compounds 1–6 showed potential anti-inflammatory activities by inhibiting the production of nitric oxide (NO), tumor necrosis factor-α (TNF-α) and interleukine-6 (IL-6) in lipopolysaccharide-stimulated macrophages.
1. Introduction
Acute and chronic inflammation is a complex biological response of the body to various external stimuli including pathogens, damaged cells and toxins, resulting in various diseases, such as allergies, inflammatory bowel diseases, sepsis, cancer and Alzheimer’s disease [1,2,3]. Many studies have reported several signaling pathways that are associated with the initiation and progression of inflammation in canonical/non-canonical fashions and that are essential factors for successful therapy of inflammation-related diseases [4].
Even though many attempts have been made to discover potent anti-inflammatory agents in small molecule drugs or natural compounds, the therapeutic outcomes are still not satisfactory owing to limitations and potential risks. Thus, the development of new anti-inflammatory agents without adverse effects is urgently required for clinical application.
Microbial natural products are regarded as one of the major sources of bioactive compounds and have been used as potential drug leads. Marine-derived actinobacteria is the most prolific group of microorganisms that provide bioactive natural products [5,6,7]. Particularly, cyclomarins and salinamides have been reported as marine actinobacteria-derived anti-inflammatory natural products [8,9].
The increasing rediscovery rate of natural products and a time-consuming dereplication process have become bottlenecks that retard the progress of natural product-derived drug discovery. With the advent of new techniques, there are commonly used natural product databases, such as “MarineLit”, “Antibase” and the “Dictionary of Natural Products”, which relay the MS information of samples to dereplicate known natural products [10,11,12]. These databases, however, cannot enable automatic spectral mining of large experiments with up to thousands of samples within few hours [13].
The tandem mass spectrometry (MS/MS)-based dereplication platform, called ‘Global Natural Products Social Molecular Networking (GNPS)’, is widely used today to shorten the dereplication time and investigate the molecular groups in the mixture samples, such as crude extracts and fractions [13,14,15,16,17]. GNPS provides the molecular annotation results if the observed MS/MS data of the chemical components in samples are found in the GNPS-database. Although the acquired MS/MS data in this study were not found from the GNPS-database, the GNPS platform provided the molecular networking map showing the molecular families with their associated compound nudes.
As the part of our goal to discover anti-inflammatory natural products, Streptomyces sp. 13G036 was cultured in SYP broth medium. Molecular networking analysis on the crude extract of Streptomyces sp. 13G036 using the GNPS platform showed several molecular clusters without molecular annotation results. Among the molecular clusters, the Molecular Family-1 (MF-1) cluster was found to consist of molecules mostly possessing parent ion mass values (m/z) between 200 and 250 amu. Further LC-ESI-MS analysis on the subfractions of the extract showed that members of the MF-1 cluster were mostly found in the subfractions B–D.
Further LC-ESI-MS-guided isolation led to the purification of compounds 1–7 possessing butanolide structures (Figure 1). The natural occurrence of compound 7 has not been reported previously. Importantly, compounds 1–6 showed potential anti-inflammatory effects by inhibiting the production of NO, TNF-α and IL-6 in LPS-stimulated macrophages via down-regulation of NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) activity.
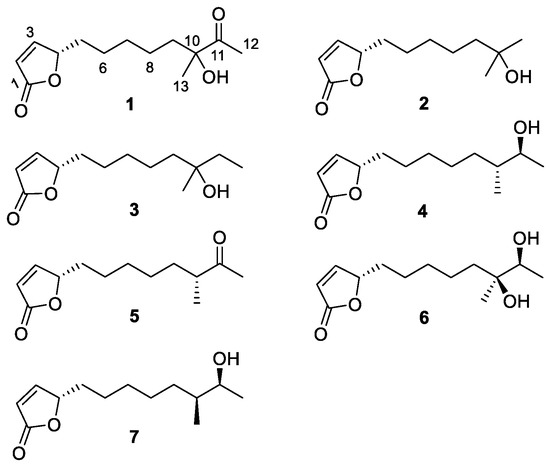
Figure 1.
The structures of compounds 1–7.
2. Materials and Methods
2.1. General Experimental Procedures
Optical rotations were measured by using the DIP 1000 Digital Polarimeter (JASCO Corporation, Tokyo, Japan). 1D and 2D NMR data were obtained by using the Bruker AVANCE 250 spectrometer (Bruker BioSpin Corporation, Billerica, MA, USA) with TMS as the internal standard. LC-LR-ESI-MS was conducted using the Agilent 6120 quadrupole MSD system consisting of a 1260 Infinity pump, a 1260 Infinity autosampler, a 1260 Infinity DAD detector. LC-ESI-MS/MS data for molecular networking analysis were acquired on an Agilent 6530 q-TOF MS and Kinetex C18 column (150 × 2.1 mm, 1.7 µm) with a flow rate of 0.7 mL/min with mobile phase as A (0.1% formic acid in deionized water) and B (0.1% formic acid in acetonitrile) and with solvent system 0 to 3 min (B 5%), 3 to 13 min (B 100%), 13 to 18 min (B 100%), 18 to 19 min (5%) and 19 to 23 min (B 5%).
MS/MS settings were as follows: isolation width (4 m/z), gas temp (300 °C), drying gas (11 L/min), nebulizer (35 psi), MS-TOF fragmentor (100 V), skimmer (65 V), OCT1RFV (750 V), source collision energy (0 V), capillary voltage (3000 V), capillary voltage (0.037 µA), chamber (10.98 µA), data dependent MS/MS acquisition (three most intense peaks selection), MS/MS collision E (35), profile type detection. HR-FAB-MS spectra of compounds were measured on a JEOL JMS-600W (Tokyo, Japan) mass spectrometer. Preparative HPLC was achieved by using the GILSON LC system equipped with a 321 pump and a UV/Vis 155 detector with a Phenomenex Luna (Phenomenex, Torrance, CA, USA) C18 column (250 × 21 mm, 10 μm).
Analytical HPLC was performed on a Waters system (Waters 1525 Pump and Waters 996 PDA; Waters, Milford, MA, USA) with a Phenomenex Luna C18(2) column (250 × 4.6 mm, 5 μm). The ECD experiments were conducted using the J810 Circular Dichroism (CD) Spectropolarimeter (JASCO Corporation, Tokyo, Japan) at the Core Research Support Center for Natural Products and Medical Materials (CRCNM). The vacuum liquid chromatography (VLC) column (80 mm diameter with 50 mm height) was equipped with a silica gel (0.040–0.063 mm, 230–400 mesh) and was used for fractionation of crude extract. Levels of NO, TNF-α and IL-6 were measured with ELISA kits using a microplate reader (BioTek, Santa Clara, CA, USA), separately.
2.2. Materials and Reagents
The HPLC grade solvents (MeOH and acetonitrile) were purchased from Fisher Scientific (Hampton, NH, USA). The analytical solvents (ethyl acetate and methylene chloride) were purchased from Duksan Pure Chemicals Co., Ltd. (Ansan-si, Korea). Silica gel 60 (0.040–0.063 mm, 230–400 mesh) used for VLC was purchased from Merck & Co., Inc. (Kenilworth, NJ, USA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Corning (Manassas, VA, USA). NO, TNF-α and IL-6 ELISA kits were purchased from R & D Systems (Minneapolis, MN, USA), while d-luciferin was provided by PerkinElmer (Waltham, MA, USA).
2.3. Isolation and Identification of Streptomyces sp. 13G036
The marine sediment, collected from the coastal waters of Jeju Island in 2013, was transferred to the lab and dried at room temperature for two days and then heated at 55 °C for 5 min. The sediment sample was stamped on a solid SYP SW medium plate (soluble starch, 10 g/L; yeast extract, 4 g/L; peptone, 2 g/L; agar, 16 g/L; sterilized and filtered sea water, 1 L) for one month, and newly observed colonies were picked up and sub-cultured on new SYP SW plates. 16S rRNA sequencing was performed, and 13G036 was identified as Streptomyces champavatii strain NRRL B-5682 with 99.80% similarity.
2.4. Cultivation of Streptomyces sp. 13G036
The bacterial strain Streptomyces sp. 13G036 was seed cultured in 10 mL SYP SW liquid medium (soluble starch, 10 g/L; yeast extract, 4 g/L; peptone, 2 g/L; sterilized and filtered sea water, 1 L) for three days at 25 °C in a shaking incubator (150 rpm). The seed cultures were transferred to 2.5 L culture flasks containing 1 L SYP SW broth medium (total 37 L) at 25 °C for seven days in shaking incubators (150 rpm) to obtain the secondary metabolites.
2.5. Extraction and Fractionation
After seven days of cultivation, the culture broths were extracted twice with the same volume of ethyl acetate (EtOAc) (37 L × 2). The EtOAc layers were combined and dried under reduced pressure to give 4.2 g of crude extract. The crude extract was subjected to NP VLC using the step gradient method (MC:MeOH ratios = 100:0, 100:1, 50:1, 20:1, 10:1 and 1:1; MeOH:H2O ratios = 100:0, 30:1 and 7:3; 700 mL each). Each eluate was evaporated and nine fractions were prepared (fraction A: 298.3 mg; fraction B: 101.7 mg; fraction C: 280.7 mg; fraction D: 208.8 mg; fraction E: 385.3 mg; fraction F: 1317.6 mg; fraction G: 406.7 mg; fraction H: 140.0 mg; and fraction I: 732.8 mg).
2.6. Molecular Networking Analysis
The extract and subfractions A–I were analyzed by using the LC-ESI-MS/MS spectrometer. The acquired MS/MS data of each fraction was analyzed by molecular networking using the GNPS platform (https://gnps.ucsd.edu, accessed on 18 January 2018) to identify the molecular families (MFs) with similar MS/MS features, using the following parameters: Min Pairs Cos of 0.6, Min Matched Peaks of 4 and Minimum Cluster Size of 2.
2.7. Extraction and Isolation
The subfractions B and C were subjected to a GILSON RP HPLC equipped with a Luna preparative column (250 × 21.2 mm, 10 μm) with gradient conditions (30% B to 48% B for 45 min and to 100% B after 10 min and then washed with 100% B for 20 min at a flow rate of 6 mL/min; A: deionized water, B: acetonitrile) to obtain compounds 1 (Rt 27.5 min, 3.2 mg), 2 (Rt 32.1 min, 8.3 mg), 3 (Rt 43.2 min, 13.6 mg) and 5 (Rt 52.8 min, 93.0 mg). The subfraction D was also subjected to the same RP HPLC using the method (isocratic 24% B for 40 min and to 100% B after 2 min and then washed with 100% B for 20 min at a flow rate of 6 mL/min; A deionized water, B: acetonitrile) to obtain compounds 6 (Rt 30.1 min, 3.3 mg).
Compounds 4 and 7 were obtained in the form of a mixture, which was confirmed by the appearance of two adjacent peaks in a ratio of 2:1 in the 13C NMR spectrum. Thus, compounds 4 and 7 were further purified by using the Waters RP HPLC equipped with a analytical Luna column (250 × 4.6 mm, 5 μm) with 40% B in A at the flow rate of 1 mL/min to obtain compounds 4 (Rt 25.1 min, 1.5 mg) and 7 (Rt 25.6 min, 0.7 mg) (A: deionized water, B: acetonitrile).
2.8. Cell Culture
Raw264.7 cells were cultured in DMEM medium supplemented with 10% heat-inactivated fetal bovine serum, 1% penicillin/streptomycin (100 mg/mL) at 37 °C under a humidified 5% CO2 atmosphere.
2.9. Cell Proliferation Assay
A cell proliferation assay was performed using a Cell Counting Kit (CCK-8, Dojindo Laboratories, Tokyo, Japan). Raw264.7 cells were seeded at 1 × 104 cells per well in 96-well plates, followed by incubation of various concentration of respective compounds for 24 h. Ten microliters of CCK-8 solution were added to each well at 24 h after compound treatment, and then the plate was incubated at 37 °C for 1 h. The absorbance at 450 nm was measured using a microplate reader (BioTek).
2.10. NO and Cytokine Production
Raw264.7 cell were treated with compounds 1–6 (10 μM) for 2 h, followed by 1 μg/mL LPS treatment for 24 h. The supernatant was collected and used to measure the level of TNF-α, IL-6 and NO. TNF-α and IL-6 levels in the supernatant were determined using the Quantikine ELISA mouse TNF-α kit (Cat No. MTA00B, R&D Systems, Minneapolis, MN, USA) and Quantikine ELISA mouse IL-6 kit (Cat No. M6000B, R&D Systems, Minneapolis, MN, USA) according to the manufacturer’s instructions.
The NO concentration was determined by using the Griess assay kit (Promega, Madison, WI, USA). Briefly, the supernatant was reacted with Griess reagent (sulfanilamide solution and N-1-naphthylethylenediamide solution) at a ratio of 1:1, followed by 10 min incubation in the dark. The absorbance of the final product was then measured at 540 nm using a microplate reader (BioTek).
2.11. NF-κB Reporter Assay
A firefly luciferase gene driven by four copies of the NF-κB response element located upstream of the minimal TATA promoter was introduced into Raw264.7 via lentiviral infection. Luciferase expressing cells were selected with 2 ug/mL puromycin for 2 weeks. NF-κB-reporter expressing Raw264.7 cells were pretreated with compounds 1–6 (10 μM) for 2 h and then treated with 1 μg/mL LPS for 10 h. D-Luciferin, dissolved in PBS (pH 7.5), was added to respective cells, and bioluminescent signals were determined using the IVIS imaging system (PerkinElmer).
Grayscale photographic images and bioluminescent color images were superimposed using the LIVINGIMAGE (version 2.12, Waltham, MA, USA, PerkinElmer) and IGOR Image Analysis FX softwares (WaveMetrics, Lake Oswego, OR, USA). The BLI signals were expressed in units of photons per cm2 per second per steradian (p/cm2/s/sr). After imaging acquisition, the Cell Counting Kit (CCK)-8 (Dojindo Laboratories, Tokyo, Japan) was used to assess cell viability. We added 10 μL of the cell count solution to each well, and the plates were incubated at 37 °C for 3 h. The absorbance was measured at 450 nm using a microplate reader (BioTek). NF-κB luciferase activity was normalized to the absorbance of the CCK8 assay of each sample.
2.12. Statistical Analysis
Data analysis was conducted using GraphPad Prism 9 (San Diego, CA, USA). All data are presented as the means ± standard deviations from at least three representative experiments. Statistical significance was determined using the unpaired Student’s t-test. p-Values < 0.05 were considered statistically significant.
3. Results
3.1. Molecular Networking Analysis
The LC-ESI-MS/MS data of crude extracts of Streptomyces sp. 13G036 and the fractions A–I were analyzed using the GNPS platform (http://gnps.ucsd.edu, accessed on 29 January 2018), obtaining several molecular clusters (Figure 2). The MF-1 cluster consisted of the molecules with relatively low m/z values below 300. The compounds in MF-1 were not annotated in GNPS results. Most of the compounds in the MF-1 cluster were found in the subfractions B–D. Further LC-LR-ESI-MS-guided isolation was performed to obtain pure compounds from the MF-1 cluster.
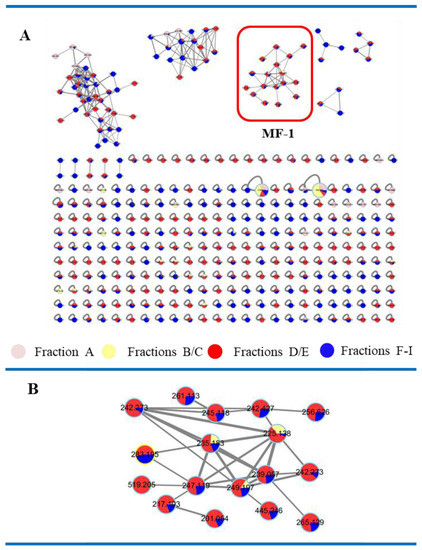
Figure 2.
The molecular networking map of the fractions A–I. (A) A molecular networking map. (B) Expansion of the MF-1 cluster.
3.2. LC-ESI-MS Guided Isolation
Combined analysis of molecular networking data and LC-ESI-MS data of the fractions A–I indicated that the compounds found in the MF-1 cluster were also observed in the fractions B–D. Thus, these three fractions were further isolated by HPLC and seven butenolides (compounds 1–7) (1, 3.2 mg; 2, 8.3 mg; 3, 13.6 mg; 4, 1.5 mg; 5, 93.0 mg; 6, 3.3 mg; and 7, 0.7 mg) were obtained. Compounds 7 and 8 are natural products isolated in this study for the first time, although they were previously synthesized for absolute configuration determination of the butenolides [18,19,20,21].
3.3. Structure Elucidation of the Isolated Compounds
Compound 1 was isolated as yellowish colored oil and its molecular formula was deduced as C13H20O4 on the basis of HR-FAB-MS [M + H]+ 241.1440 (Figure S8 in the Supplementary Materials), indicating four unsaturation numbers. Analysis of 1H NMR spectrum showed two olefinic signals at δH 7.42 (CH-3) and δH 6.10 (CH-2), one heteroatomic bond CH at δH 5.02 and two singlet methyls at δH 2.21 and δH 1.36.
Interpretation of the 13C NMR spectrum of compound 1 showed one ketone (C-11, δC 212.21), one ester carbonyl (C-1, δC 173.05), one olefinic (C-2, δC 121.85; C-3, δC 156.12), two oxygen bonded carbons (C-4, δC 83.35; C-10, δC 78.89) and seven alkyl groups (C-5, δC 33.21; C-6, δC 25.59; C-7, δC 29.57; C-8, δC 23.72; C-9, δC 39.39; C-12, δC 23.29; C-13, δC 24.97). Combined analysis of 1H and 13C NMR data suggested that compound 1 could be a butenolide, which is a family of α, β-unsaturated lactones [16,17].
The following COSY correlations were observed: δH 6.10 (C-2, CH)/δH 7.42 (C-3, CH), δH 7.42 (C-3, CH)/δH 5.02 (C-4, CH) and δH 5.02 (C-4, CH)/δH 1.68 (C-5, CH2). HMBC correlations (H-13/C-9, C-10, C-11; H-12/C-11; C-10-OH/C-9, C-11, C-13) were also observed as shown in Figure 3. The planar structure of compound 1 was confirmed as (4S)-4,10-dihydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide (mycenolide A) by comparing the NMR data with published data from the literature [22].
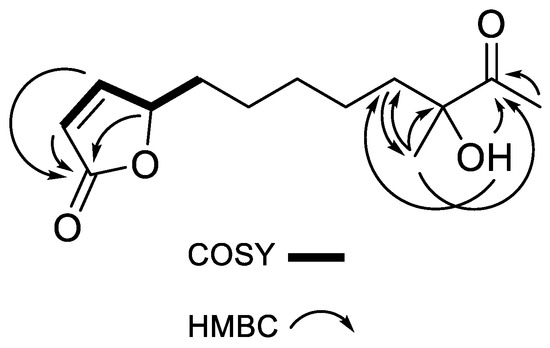
Figure 3.
Key COSY and HMBC correlations of compound 1.
Likewise, the planar structures of compounds 2–7 were deduced by comparison of the NMR spectroscopic data with the published data from the literature. The absolute configuration of the C-4 in compounds 1–7 was assigned by ECD absorption around 210 nm corresponding to positive π-π * Cotton effect (Figure 4) [18].
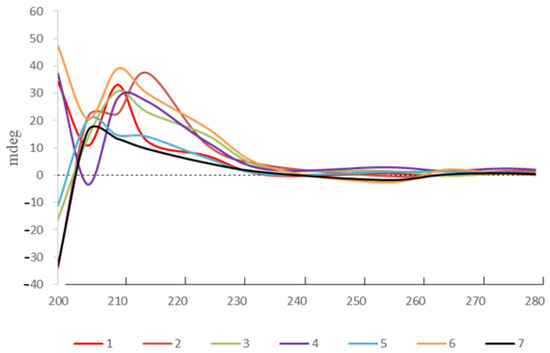
Figure 4.
ECD spectra of compounds 1–7.
The chiral centers at C-10/C-11 in compounds 4 and 7 were confirmed as 10R/11S and 10S/11S by the comparison of the NMR data and specific rotations with the previously reported data of synthetic butenolides [18,20]. The relative configuration at C-10/C-11 in compound 6 was elucidated as a threo (10S *, 11S *) by comparison with the partial structure according to the reported data; however, the absolute configuration has not yet been determined [23,24].
Thus, the structures of compounds 1–7 were confirmed as follows: 1, (4S)-4,10-dihydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide; 2, (4S)-4,10-dihydroxy-10 -methyl-undec-2-en-1,4-olide [25]; 3, (4S)-4,10-dihydroxy-10-methyl-dodec-2-en-1, 4-olide [25]; 4, (4S,10R,11S)-4,11-dihydroxy-10-methyl-dodec-2-en-1,4-olide; 5, (4S)-4-hydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide [20]; 6, (4S,10S,11S)-4,10,11-trihydroxy-10-methyl-dodec-2-en-1,4-olide [25]; 7, (4S,10S,11S)-4,11-dihydroxy-10-methyl-dodec-2-en-1,4-olide. Due to the limitation of compound supply or inactiveness of the stereogenic carbinols of compounds 1, 3 and 6, the absolute configuration of stereogenic centers, except for C-4, could not be determined.
3.4. Evaluation of Anti-Inflammatory Effects of Compounds 1–6
For successful applications in clinical situation, novel therapeutic agents should not induce toxicity in vitro and in vivo. Therefore, the cytotoxicity of compounds 1–6 at various concentrations was inspected in Raw264.7 macrophages. As shown in Figure 5, no differences were seen in the cell viability across vehicle-treated cells and compounds 1–6-treated cells.
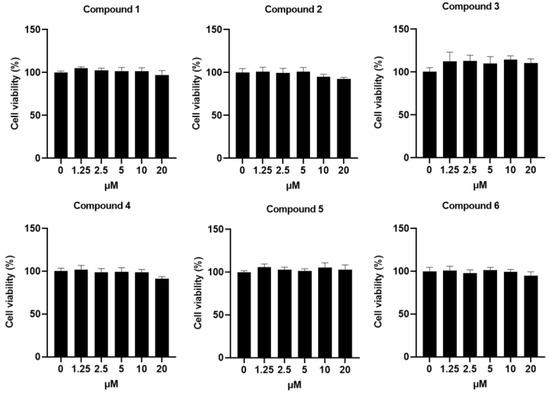
Figure 5.
Effects of the compounds 1–6 on cell viability of Raw264.7 cells. Cell proliferation in Raw264.7, CHO and L929 cells treated with or without the addition of compounds 1–6.
External stimuli, such as LPS, lead to phosphorylation of IkB and subsequently translocation of NF-κB to the nucleus, thereby resulting in the transcription upregulation of inflammatory genes and finally drastic production of the proinflammatory cytokine and other inflammatory mediators [26]. Therefore, we investigated the effects of compounds 1–6 on NF-κB signaling using the NF-κB reporter Raw264.7 cells that contain the firefly luciferase gene under the NF-κB response element and allow for monitoring of the NF-κB transduction pathway by measuring the bioluminescent activity.
Dexamethasone (DEX) was used in this study as a positive anti-inflammatory agent. As expected, LPS induced drastic increase of bioluminescent signals indicating NF-κB activation in macrophages, which was attenuated by treatment with DEX (Figure 6A,B). Similar to DEX, compounds 1–6 significantly decreased the bioluminescent signals from LPS-stimulated macrophages, revealing the potential inhibitory effects of compounds 1–6 on the activated NF-κB signaling.
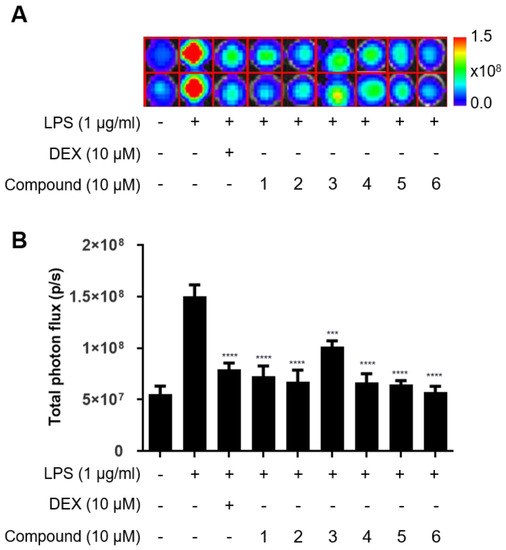
Figure 6.
Effects of the compounds 1–6 on LPS-induced NF-κB activity of Raw264.7 cells. (A) In vitro bioluminescent imaging of NF-κB reporter macrophage. (B) Quantification of bioluminescent signals. NF-κB reporter-expressing Raw264.7 cells were preincubated with compounds 1–6 (10 μM) for 2 h and then treated with 1 μg/mL LPS for an additional 10 h. d-Luciferin was added to respective cells and the bioluminescent signal was determined using the IVIS imaging system. *** p < 0.0005 and **** p < 0.0001, compared with the LPS-treated group.
Since effective inhibitory effects of compounds 1–6 on LPS-mediated NF-κB activation were observed, we further examined whether compounds 1–6 decrease the production levels of NO, TNF-α and IL-6 by the LPS-stimulated macrophages. As illustrated in Figure 7A, DEX reduce NO production in LPS-stimulated Raw264.7 cells, while a drastic increase was observed in LPS-stimulated macrophages. Interestingly, significant inhibition of NO production was observed in the macrophages treated with compounds 1–6 but not in the vehicle-treated macrophages. Consistent with the results of the NO assay, the levels of TNF-α and IL-6 were much lower in the macrophages treated with compounds 1–6 compared with the vehicle-treated macrophages, showing the comparable inhibitory effects on cytokine production with DEX and compounds 1–6 (Figure 7B,C).
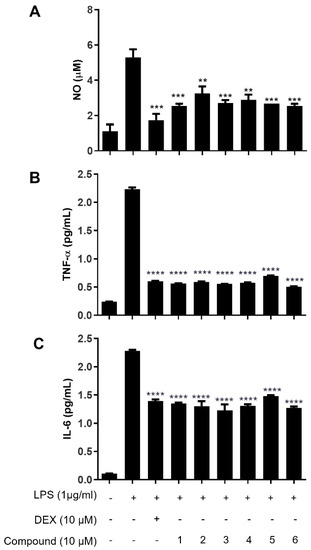
Figure 7.
Effects of compounds 1–6 in LPS-stimulated Raw264.7 cells on the production of (A) NO, (B) TNF-α (a) and (C) IL-6. Cells were pretreated for 2 h with compounds 1–6 (10 μM) and stimulated for 24 h with LPS (1 μg/mL). Effects of compounds 1–6 on NO release and proinflammatory cytokines were determined by Griess reagent assay and TNF-α and IL-6 ELISA kits. Values are expressed as the means ± S.D. of three replicates. ** p < 0.005, *** p < 0.0005 and **** p < 0.0001, compared with the LPS-treated group.
4. Discussion
Culturable marine microorganisms are considered to be the most promising source of novel natural products to overcome the supply problems of marine natural products. Currently, as the rediscovery rate of natural products from terrestrial organisms increases, the discovery process of new drug candidates is delayed even though several new compounds, with various bioactivities, such as antitumor, antibacterial, antifungal and anti-inflammatory activities, have been isolated in recent decades [27,28,29].
In order to speed up the discovery of novel drug candidates, one procedure called ‘dereplication’ is applied in chemical investigations to identify known compounds. The Global Natural Products Social Molecular Networking site (GNPS: http://gnps.ucsd.edu, accessed on 29 January 2018), a platform with open access to MS/MS spectra analysis, provides the ability to analyze a data set and compare it to all publicly available data, thereby, facilitating the discovery rate of novel natural products.
Butenolides, possessing α, β-unsaturated γ-lactone, are commonly discovered from Streptomyces spp. with various biological activities, including antifouling, antifungal, antibacterial, anti-adenoviral, anti-tumor, antibiotic and anti-inflammatory activities [18,20,21,22,30,31].
However, the anti-inflammatory activity of butenolides that possess long chains (over seven carbons) at the γ position of butenolide has not been studied. In this study, Streptomyces sp. 13G036 was selected for the investigation of anti-inflammatory compounds based on the molecular networking analysis of the extract and fractions. LC-ESI-MS guided isolation led to the isolation of seven butenolides (compounds 1–7), including one compound (7) for which its natural occurrence is reported in this study for the first time.
Supplementary Materials
The following supporting materials are available online at https://www.mdpi.com/article/10.3390/app12094510/s1, Table S1: Comparison of chemical shifts in 13C NMR data and specific rotation of synthesized butenolides with 4 and 7; Figure S1: The picture of Streptomyces sp 13G036. Figure S2: The assignment of functional group of compound 1. Figure S3: 1H NMR spectrum of compound 1 (CDCl3, 250 MHz); Figure S4: 13C NMR spectrum of compound 1 (CDCl3, 62.5 MHz); Figure S5: COSY spectrum of compound 1 (CDCl3, 250 MHz); Figure S6: HSQC spectrum of compound 1 (CDCl3, 250 MHz); Figure S7: HMBC spectrum of compound 1 (CDCl3, 250 MHz); Figure S8: HR-FAB-MS data of compound 1; Figure S9: LR-ESI-MS data of compound 2; Figure S10: 1H NMR spectrum of compound 2 (CDCl3, 250 MHz); Figure S11: 13C NMR spectrum of compound 2 (CDCl3, 62.5 MHz); Figure S12: LR-ESI-MS data of compound 3; Figure S13: 1H NMR spectrum of compound 3 (CDCl3, 250 MHz); Figure S14: 13C NMR spectrum of compound 3 (CDCl3, 62.5 MHz); Figure S15: LR-ESI-MS data of compound 4; Figure S16: 1H NMR spectrum of compound 4 (CDCl3, 250 MHz); Figure S17: 13C NMR spectrum of compound 4 (CDCl3, 62.5 MHz); Figure S18: LR-ESI-MS data of compound 5; Figure S19: 1H NMR spectrum of compound 5 (CDCl3, 250 MHz); Figure S20: 13C NMR spectrum of compound 5 (CDCl3, 62.5 MHz); Figure S21: LR-ESI-MS data of compound 6; Figure S22: 1H NMR spectrum of compound 6 (CDCl3, 250 MHz); Figure S23: 13C NMR spectrum of compound 6 (CDCl3, 62.5 MHz); Figure S24: LR-ESI-MS data of compound 7. Figure S25: 1H NMR spectrum of compound 7 (CDCl3, 250 MHz); Figure S26: 13C NMR spectrum of compound 7 (CDCl3, 62.5 MHz).
Author Contributions
Conceptualization, J.C., Y.H.J. and H.C.; formal analysis, M.G., S.B.L. and J.-E.L.; investigation, G.J.K., J.M., J.-W.N. and J.-S.B.; writing—original draft preparation, M.G., S.B.L., Y.H.J. and H.C.; writing—review and editing, Y.H.J. and H.C.; visualization, M.G. and S.B.L.; supervision, Y.H.J. and H.C.; project administration, Y.H.J. and H.C.; funding acquisition, H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the 2020 Yeungnam University Research Grants, and the research grants from the Basic Science Research Program through the National Research Foundation of Korea (NRF) (grant number: 2021R1A2C101727, 2020R1A6A1A03044512 and 2022R1A2C1004096).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Medzhitov, R. Inflammation 2010: New adventures of an old flame. Cell 2010, 140, 771–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, S.A. Directing transition from innate to acquired immunity: Defining a role for IL-6. J. Immunol. 2005, 175, 3463–3468. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Haase, I.; Nestle, F.O. Mechanisms regulating skin immunity and inflammation. Nat. Rev. Immunol. 2014, 14, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Yeung, Y.T.; Aziz, F.; Guerrero-Castilla, A.; Arguelles, S. Signaling pathways in inflammation and anti-inflammatory therapies. Curr. Pharm. Des. 2018, 24, 1449–1484. [Google Scholar] [CrossRef]
- Zotchev, S.B. Marine actinomycetes as an emerging resource for the drug development pipelines. J. Biotechnol. 2012, 158, 168–175. [Google Scholar] [CrossRef]
- Prudhomme, J.; McDaniel, E.; Ponts, N.; Bertani, S.; Fenical, W.; Jensen, P.; Le Roch, K. Marine actinomycetes: A new source of compounds against the human malaria parasite. PLoS ONE 2008, 3, 2335–2342. [Google Scholar] [CrossRef]
- Kwon, H.C.; Kauffman, C.A.; Jensen, P.R.; Fenical, W. Marinomycins A–D, antitumor-antibiotics of a new structure class from a marine actinomycete of the recently discovered genus “Marinispora”. J. Am. Chem. Soc. 2006, 128, 1622–1632. [Google Scholar] [CrossRef]
- Trischman, J.A.; Tapiolas, D.M.; Jensen, P.R.; Fenical, W.; McKee, T.C.; Ireland, C.M.; Stout, T.J.; Clardy, J. Salinamides A and B: Anti-inflammatory depsipeptides from a marine Streptomycete. J. Am. Chem. Soc. 1994, 116, 757–758. [Google Scholar] [CrossRef]
- Renner, M.K.; Shen, Y.C.; Cheng, X.C.; Jensen, P.R.; Frankmoelle, W.; Kauffman, C.A.; Fenical, W.; Lobkovsky, E.; Clardy, J. Cyclomarins A–C, new antiinflammatory cyclic peptides produced by a marine bacterium (Streptomyces sp.). J. Am. Chem. Soc. 1999, 121, 11273–11276. [Google Scholar] [CrossRef]
- Munro, M.; Blunt, J.W. MarineLit. Available online: http://pubs.rsc.org/marinlit/ (accessed on 11 March 2015).
- Laatsch, H. AntiBase, a Database for Rapid Dereplication and Structure Determination of Microbial Natural Products; Wiley-VCH: Hoboken, NJ, USA, 2010. [Google Scholar]
- Buckingham, J. Dictionary of Natural Products, Supplement 4; CRC Press: Boca Raton, FL, USA, 1997. [Google Scholar]
- Nothias, L.F.; Nothias-Esposito, M.; Silva, R.; Wang, M.X.; Protsyuk, I.; Zhang, Z.; Sarvepalli, A.; Leyssen, P.; Touboul, D.; Costa, J.; et al. Bioactivity-based molecular networking for the discovery of drug leads in natural product bioassay-guided fractionation. J. Nat. Prod. 2018, 81, 758–767. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.Y.; Sanchez, L.M.; Rath, C.M.; Liu, X.; Boudreau, P.D.; Bruns, N.; Glukhov, E.; Wodtke, A.; Felicio, R.; Fenner, A.; et al. Molecular networking as a dereplication strategy. J. Nat. Prod. 2013, 76, 1686–1699. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef] [Green Version]
- Kurita, K.L.; Glassey, E.; Linington, R.G. Integration of high-content screening and untargeted metabolomics for comprehensive functional annotation of natural product libraries. Proc. Natl. Acad. Sci. USA 2015, 112, 11999–12004. [Google Scholar] [CrossRef] [Green Version]
- Comi, T.J.; Makurath, M.A.; Philip, M.C.; Rubakhin, S.S.; Sweedler, J.V. MALDI MS guided liquid microjunction extraction for capillary electrophoresis–electrospray ionization MS analysis of single pancreatic islet cells. Anal. Chem. 2017, 89, 7765–7772. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, S.; Andersson, F.; Breistein, P.; Hedenström, E. Synthesis of butenolides recently isolated from marine microorganisms. Tetrahedron Lett. 2007, 48, 7878–7881. [Google Scholar] [CrossRef]
- Uchida, M.; Takamatsu, S.; Arima1, S.; Miyamoto, K.T.; Kitani, S.; Nihira, T.; Ikeda, H.; Nagamitsu, T. Total synthesis and absolute configuration of avenolide, extracellular factor in Streptomyces avermitilis. J. Antibiot. 2011, 64, 781–787. [Google Scholar] [CrossRef]
- Mukku, V.J.R.V.; Speitling, M.; Laatsch, H.; Helmke, E. New butenolides from two marine Streptomycetes. J. Nat. Prod. 2000, 63, 1570–1572. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.W.; Lee, H.S.; Rho, J.R.; Kim, T.S.; Mo, S.J.; Shin, J. New lactone-containing metabolites from a marine-derived bacterium of the Genus Streptomyces. J. Nat. Prod. 2001, 64, 664–667. [Google Scholar] [CrossRef]
- de Oliveira, J.A.M.; Williams, D.E.; Andersen, R.J.; Sarragiotto, M.H.; Baldoqui, D.C. Mycenolide A, new butenolide from a marine sediment-derived bacterium Streptomyces sp. 4054. Nat. Prod. Res. 2019, 34, 2986–2989. [Google Scholar] [CrossRef]
- Adam, W.; Nestler, B. Hydroxy-directed regio-diastereoselective ene reaction of singlet oxygen with chiral allylic alcohols. J. Am. Chem. Soc. 1993, 115, 5041–5049. [Google Scholar] [CrossRef]
- Sato, T.; Kaneko, H.; Takahashi, T. Metal-catalyzed organic photoreactions. Titanium (IV) chloride-catalyzed photoreactions of alcohols and formic esters. Chem. Lett. 1981, 10, 1469–1472. [Google Scholar] [CrossRef]
- Zhao, P.J.; Li, G.H.; Shen, Y.M. New chemical constituents from the endophyte Streptomyces Species LR4612 cultivated on Maytenus hookeri. Chem. Biodivers. 2006, 3, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.C. NF-κB signaling in inflammation. Signal Transduct. Target. Ther. 2017, 2, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, C.; Silva, J.; Pinteus, S.; Gaspar, H.; Alpoim, M.C.; Botana, L.M.; Pedrosa, R. From marine origin to therapeutics: The antitumor potential of marine algae-derived compounds. Front. Pharmacol. 2018, 9, 777–801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2019, 36, 122–173. [Google Scholar] [CrossRef] [Green Version]
- Bae, M.H.; Moon, K.H.; Kim, J.H.; Park, H.J.; Lee, S.K.; Shin, J.H.; Oh, D.H. Mohangic acids A–E, p-aminoacetophenonic acids from a marine mudflat derived Streptomyces sp. J. Nat. Prod. 2016, 79, 332–339. [Google Scholar] [CrossRef]
- Strand, M.; Carlsson, M.; Uvell, H.; Islam, K.; Edlund, K.; Cullman, I.; Altermark, B.; Mei, Y.F.; Elofsson, M.; Willassen, N.P.; et al. Isolation and characterization of anti-adenoviral secondary metabolites from marine actinobacteria. Mar. Drugs 2014, 12, 799–821. [Google Scholar] [CrossRef] [Green Version]
- Qi, C.X.; Gao, W.X.; Wang, J.P.; Liu, M.T.; Zhang, J.W.; Chen, C.M.; Hu, Z.X.; Xue, Y.B.; Li, D.Y.; Zhang, Q.; et al. Terrusnolides A-D, new butenolides with anti-inflammatory activities from an endophytic Aspergillus from Tripterygium wilfordii. Fitoterapia 2018, 130, 134–139. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).