Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond
Abstract
:Featured Application
Abstract
1. Introduction
2. Materials and Methods
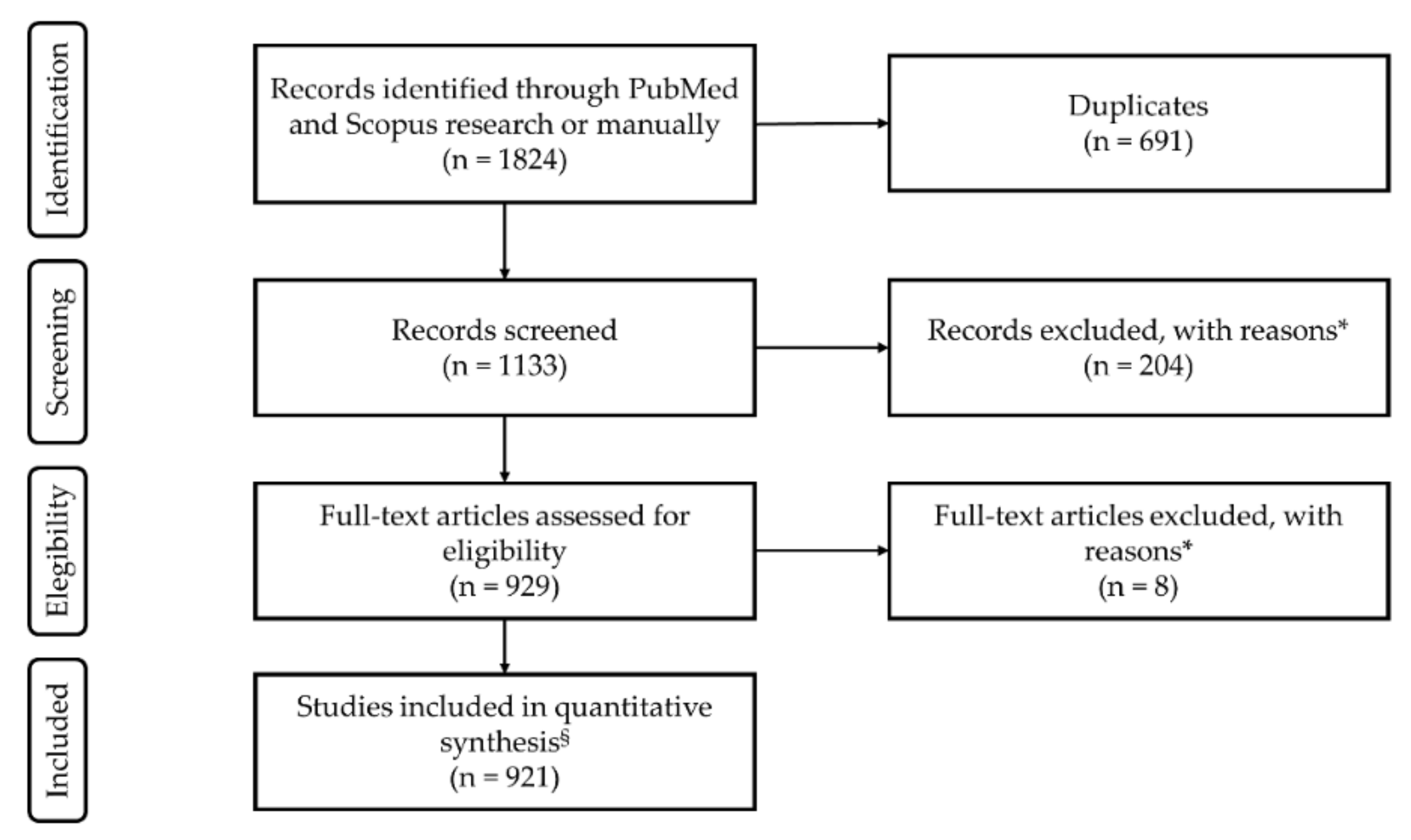
2.1. Literature Search Strategy
2.2. Study Selection
3. Results and Discussion
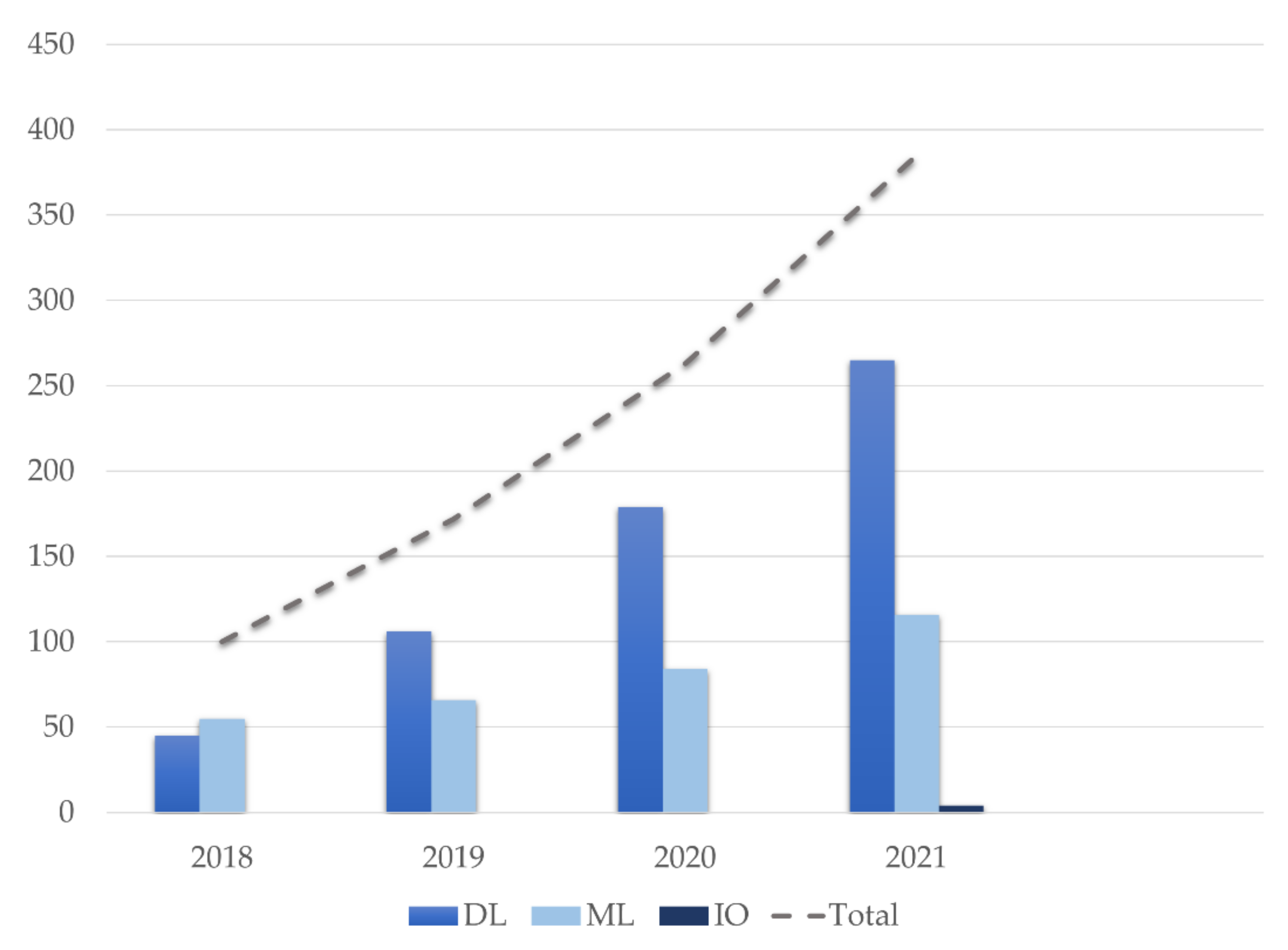
3.1. Search Inclusion Criteria and Study Description
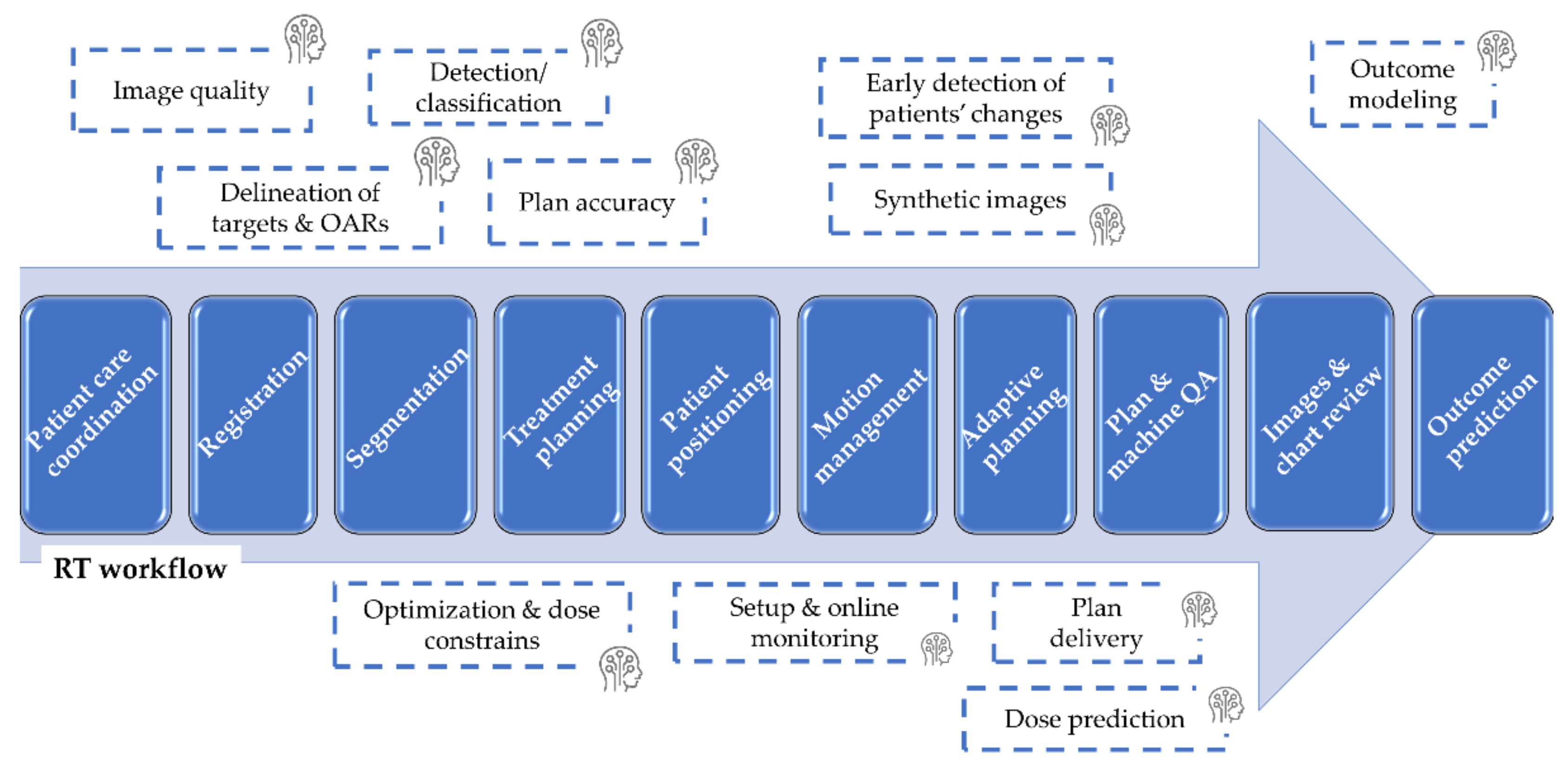
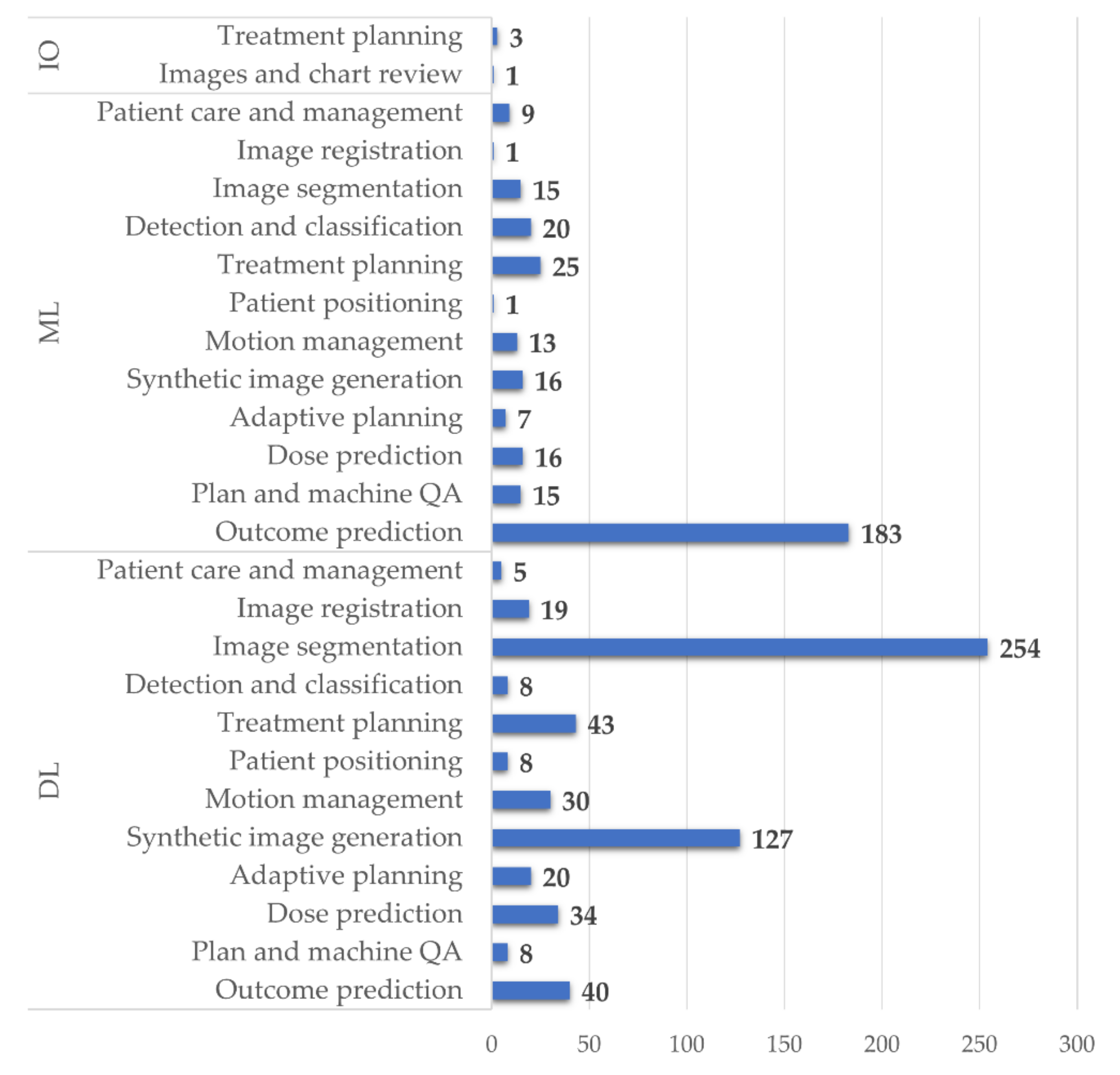
3.2. Reported Application of AI to RT
3.2.1. Patient Care Coordination and Optimization
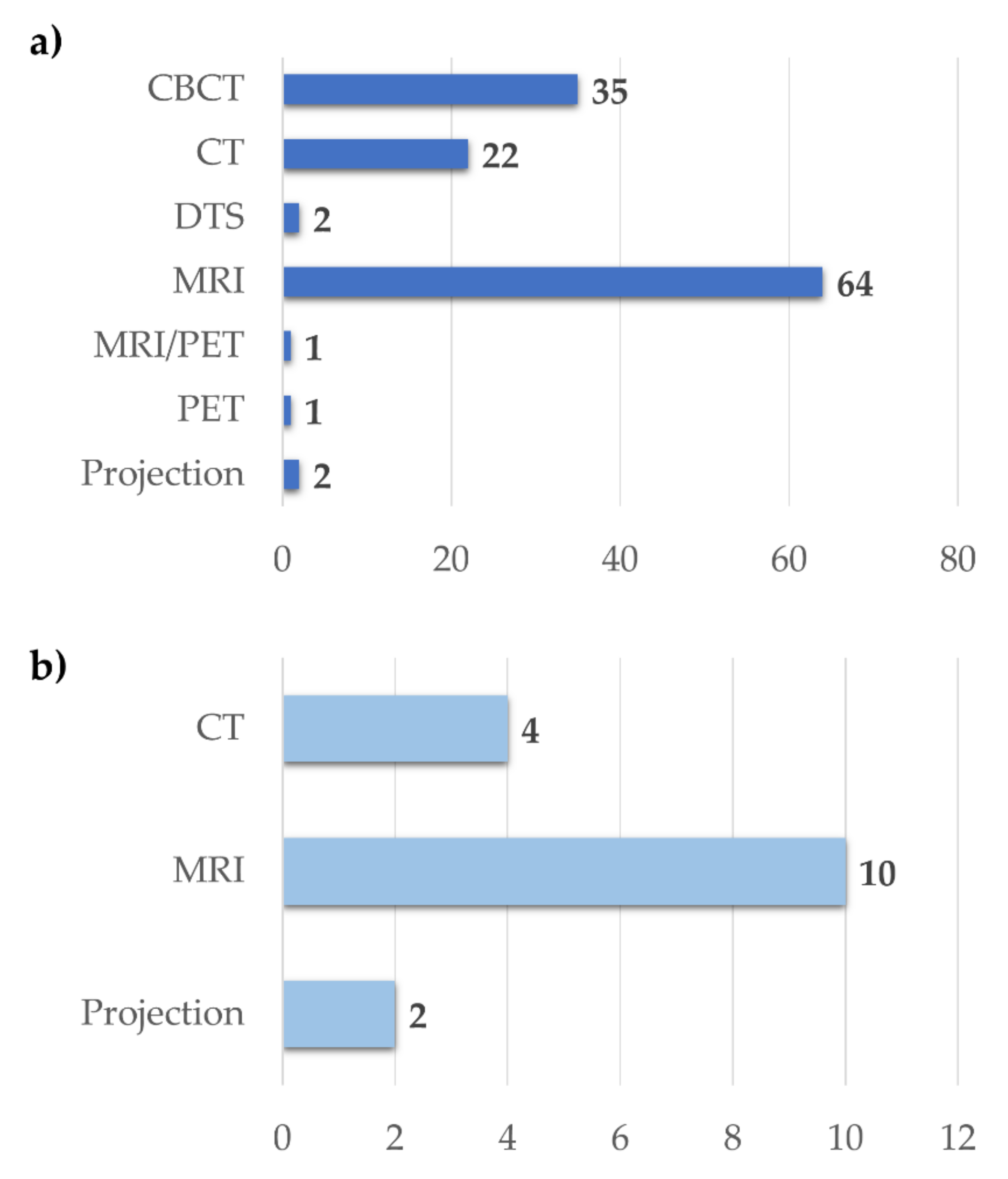
3.2.2. Image Registration
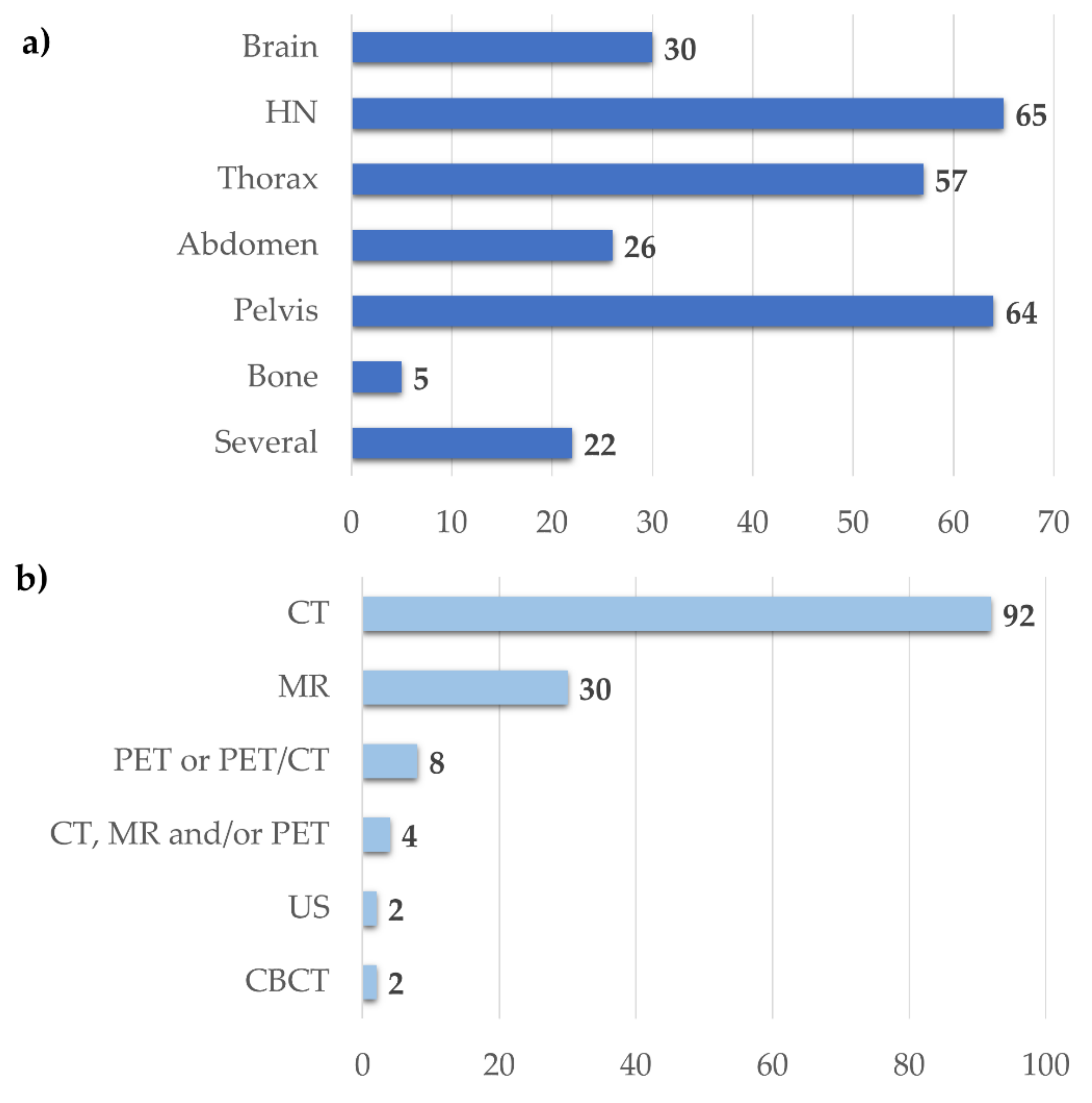
3.2.3. Image Segmentation
3.2.4. Synthetic Image Generation
3.2.5. Treatment Planning
3.2.6. Patient Positioning and Monitoring and Adaptive Planning
3.2.7. Planning Quality Assurance, Commissioning, and Machine Performance Checks
3.2.8. Outcome Prediction
3.3. Skill and Concern
3.4. Ethics
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hussein, M.; Heijmen, B.J.M.; Verellen, D.; Nisbet, A. Automation in intensity modulated radiotherapy treatment planning—A review of recent innovations. Br. J. Radiol. 2018, 91, 20180270. [Google Scholar] [CrossRef]
- Barragán-Montero, A.; Javaid, U.; Valdés, G.; Nguyen, D.; Desbordes, P.; Macq, B.; Willems, S.; Vandewinckele, L.; Holmström, M.; Löfman, F.; et al. Artificial intelligence and machine learning for medical imaging: A technology review. Phys. Med. 2021, 83, 242–256. [Google Scholar] [CrossRef]
- Manco, L.; Maffei, N.; Strolin, S.; Vichi, S.; Bottazzi, L.; Strigari, L. Basic of machine learning and deep learning in imaging for medical physicists. Phys. Med. 2021, 83, 194–205. [Google Scholar] [CrossRef]
- Price, W.N., II. Regulating Black-Box Medicine. Mich. Law Rev. 2017, 116, 421–474. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Lewis, P.J.; Court, L.E.; Lievens, Y.; Aggarwal, A. Structure and Processes of Existing Practice in Radiotherapy Peer Review: A Systematic Review of the Literature. Clin. Oncol. 2021, 33, 248–260. [Google Scholar] [CrossRef]
- Francolini, G.; Desideri, I.; Stocchi, G.; Salvestrini, V.; Ciccone, L.P.; Garlatti, P.; Loi, M.; Livi, L. Artificial Intelligence in radiotherapy: State of the art and future directions. Med. Oncol. 2020, 37, 50. [Google Scholar] [CrossRef]
- Kusters, J.; Bzdusek, K.; Kumar, P.; van Kollenburg, P.G.M.; Kunze-Busch, M.C.; Wendling, M.; Dijkema, T.; Kaanders, J. Automated IMRT planning in Pinnacle: A study in head-and-neck cancer. Strahlenther. Onkol. 2017, 193, 1031–1038. [Google Scholar] [CrossRef]
- Marazzi, F.; Tagliaferri, L.; Masiello, V.; Moschella, F.; Colloca, G.F.; Corvari, B.; Sanchez, A.M.; Capocchiano, N.D.; Pastorino, R.; Iacomini, C.; et al. GENERATOR Breast DataMart-The Novel Breast Cancer Data Discovery System for Research and Monitoring: Preliminary Results and Future Perspectives. J. Pers. Med. 2021, 11, 65. [Google Scholar] [CrossRef]
- Field, M.; Vinod, S.; Aherne, N.; Carolan, M.; Dekker, A.; Delaney, G.; Greenham, S.; Hau, E.; Lehmann, J.; Ludbrook, J.; et al. Implementation of the Australian Computer-Assisted Theragnostics (AusCAT) network for radiation oncology data extraction, reporting and distributed learning. J. Med. Imaging Radiat. Oncol. 2021, 65, 627–636. [Google Scholar] [CrossRef]
- Galofaro, E.; Malizia, C.; Ammendolia, I.; Galuppi, A.; Guido, A.; Ntreta, M.; Siepe, G.; Tolento, G.; Veraldi, A.; Scirocco, E.; et al. COVID-19 Pandemic-Adapted Radiotherapy Guidelines: Are They Really Followed? Curr. Oncol. 2021, 28, 288. [Google Scholar] [CrossRef]
- Haskins, G.; Kruecker, J.; Kruger, U.; Xu, S.; Pinto, P.A.; Wood, B.J.; Yan, P. Learning deep similarity metric for 3D MR-TRUS image registration. Int. J. Comput. Assist. Radiol. Surg. 2019, 14, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Cao, X.; Yang, J.; Wang, L.; Xue, Z.; Wang, Q.; Shen, D. Deep Learning based Inter-Modality Image Registration Supervised by Intra-Modality Similarity. Mach. Learn. Med. Imaging 2018, 11046, 55–63. [Google Scholar] [CrossRef]
- Oh, S.; Kim, S. Deformable image registration in radiation therapy. Radiat. Oncol. J. 2017, 35, 101–111. [Google Scholar] [CrossRef]
- Brock, K.K.; Mutic, S.; McNutt, T.R.; Li, H.; Kessler, M.L. Use of image registration and fusion algorithms and techniques in radiotherapy: Report of the AAPM Radiation Therapy Committee Task Group No. 132. Med. Phys. 2017, 44, e43–e76. [Google Scholar] [CrossRef] [Green Version]
- Weppler, S.; Schinkel, C.; Kirkby, C.; Smith, W. Lasso logistic regression to derive workflow-specific algorithm performance requirements as demonstrated for head and neck cancer deformable image registration in adaptive radiation therapy. Phys. Med. Biol. 2020, 65, 195013. [Google Scholar] [CrossRef]
- Kumarasiri, A.; Siddiqui, F.; Liu, C.; Yechieli, R.; Shah, M.; Pradhan, D.; Zhong, H.; Chetty, I.J.; Kim, J. Deformable image registration based automatic CT-to-CT contour propagation for head and neck adaptive radiotherapy in the routine clinical setting. Med. Phys. 2014, 41, 121712. [Google Scholar] [CrossRef]
- Liang, X.; Bibault, J.E.; Leroy, T.; Escande, A.; Zhao, W.; Chen, Y.; Buyyounouski, M.K.; Hancock, S.L.; Bagshaw, H.; Xing, L. Automated contour propagation of the prostate from pCT to CBCT images via deep unsupervised learning. Med. Phys. 2021, 48, 1764–1770. [Google Scholar] [CrossRef]
- Hoffmann, C.; Krause, S.; Stoiber, E.M.; Mohr, A.; Rieken, S.; Schramm, O.; Debus, J.; Sterzing, F.; Bendl, R.; Giske, K. Accuracy quantification of a deformable image registration tool applied in a clinical setting. J. Appl. Clin. Med. Phys. 2014, 15, 4564. [Google Scholar] [CrossRef]
- Ramadaan, I.S.; Peick, K.; Hamilton, D.A.; Evans, J.; Iupati, D.; Nicholson, A.; Greig, L.; Louwe, R.J. Validation of Varian’s SmartAdapt® deformable image registration algorithm for clinical application. Radiat. Oncol. 2015, 10, 73. [Google Scholar] [CrossRef] [Green Version]
- Pukala, J.; Johnson, P.B.; Shah, A.P.; Langen, K.M.; Bova, F.J.; Staton, R.J.; Mañon, R.R.; Kelly, P.; Meeks, S.L. Benchmarking of five commercial deformable image registration algorithms for head and neck patients. J. Appl Clin. Med. Phys. 2016, 17, 25–40. [Google Scholar] [CrossRef]
- Loi, G.; Fusella, M.; Lanzi, E.; Cagni, E.; Garibaldi, C.; Iacoviello, G.; Lucio, F.; Menghi, E.; Miceli, R.; Orlandini, L.C.; et al. Performance of commercially available deformable image registration platforms for contour propagation using patient-based computational phantoms: A multi-institutional study. Med. Phys. 2018, 45, 748–757. [Google Scholar] [CrossRef]
- Rosen, B.S.; Hawkins, P.G.; Polan, D.F.; Balter, J.M.; Brock, K.K.; Kamp, J.D.; Lockhart, C.M.; Eisbruch, A.; Mierzwa, M.L.; Ten Haken, R.K.; et al. Early Changes in Serial CBCT-Measured Parotid Gland Biomarkers Predict Chronic Xerostomia After Head and Neck Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2018, 102, 1319–1329. [Google Scholar] [CrossRef]
- Li, X.; Chen, H.; Qi, X.; Dou, Q.; Fu, C.W.; Heng, P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation From CT Volumes. IEEE Trans. Med. Imaging 2018, 37, 2663–2674. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Bentley, P.; Mori, K.; Misawa, K.; Fujiwara, M.; Rueckert, D. DRINet for Medical Image Segmentation. IEEE Trans. Med. Imaging 2018, 37, 2453–2462. [Google Scholar] [CrossRef]
- Tong, N.; Gou, S.; Yang, S.; Ruan, D.; Sheng, K. Fully automatic multi-organ segmentation for head and neck cancer radiotherapy using shape representation model constrained fully convolutional neural networks. Med. Phys. 2018, 45, 4558–4567. [Google Scholar] [CrossRef] [Green Version]
- Tong, N.; Gou, S.; Yang, S.; Cao, M.; Sheng, K. Shape constrained fully convolutional DenseNet with adversarial training for multiorgan segmentation on head and neck CT and low-field MR images. Med. Phys. 2019, 46, 2669–2682. [Google Scholar] [CrossRef]
- Nelms, B.E.; Tomé, W.A.; Robinson, G.; Wheeler, J. Variations in the contouring of organs at risk: Test case from a patient with oropharyngeal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 368–378. [Google Scholar] [CrossRef]
- Sibolt, P.; Andersson, L.M.; Calmels, L.; Sjostrom, D.; Bjelkengren, U.; Geertsen, P.; Behrens, C.F. Clinical implementation of artificial intelligence-driven cone-beam computed tomography-guided online adaptive radiotherapy in the pelvic region. Phys. Imaging Radiat. Oncol. 2021, 17, 1–7. [Google Scholar] [CrossRef]
- Feng, C.H.; Cornell, M.; Moore, K.L.; Karunamuni, R.; Seibert, T.M. Automated contouring and planning pipeline for hippocampal-avoidant whole-brain radiotherapy. Radiat. Oncol. 2020, 15, 251. [Google Scholar] [CrossRef]
- Pan, K.; Zhao, L.; Gu, S.; Tang, Y.; Wang, J.; Yu, W.; Zhu, L.; Feng, Q.; Su, R.; Xu, Z.; et al. Deep learning-based automatic delineation of the hippocampus by MRI: Geometric and dosimetric evaluation. Radiat. Oncol. 2021, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lei, Y.; Fu, Y.; Wang, T.; Zhou, J.; Jiang, X.; McDonald, M.; Beitler, J.J.; Curran, W.J.; Liu, T.; et al. Head and neck multi-organ auto-segmentation on CT images aided by synthetic MRI. Med. Phys. 2020, 47, 4294–4302. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, Y.; Fang, Y.; Wang, J.; Hu, W. A Preliminary Experience of Implementing Deep-Learning Based Auto-Segmentation in Head and Neck Cancer: A Study on Real-World Clinical Cases. Front. Oncol. 2021, 11, 638197. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Huang, Y.; Zeng, L.; Chen, X.; Liu, Y.; Qian, Z.; Du, N.; Fan, W.; Xie, X. AnatomyNet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Med. Phys. 2018, 46, 576–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Li, Y.; Dyer, B.A.; Feng, X.; Rao, S.; Benedict, S.H.; Chen, Q.; Rong, Y.A.-O. Deep learning vs. atlas-based models for fast auto-segmentation of the masticatory muscles on head and neck CT images. Radiat. Oncol. 2020, 15, 176. [Google Scholar] [CrossRef]
- Kim, N.A.-O.; Chun, J.; Chang, J.A.-O.; Lee, C.G.; Keum, K.C.; Kim, J.S. Feasibility of Continual Deep Learning-Based Segmentation for Personalized Adaptive Radiation Therapy in Head and Neck Area. Cancers 2021, 13, 702. [Google Scholar] [CrossRef]
- Van Rooij, W.; Dahele, M.; Nijhuis, H.; Slotman, B.J.; Verbakel, W.F. Strategies to improve deep learning-based salivary gland segmentation. Radiat. Oncol. 2020, 15, 272. [Google Scholar] [CrossRef]
- Nikolov, S.; Blackwell, S.; Zverovitch, A.; Mendes, R.; Livne, M.; De Fauw, J.; Patel, Y.; Meyer, C.; Askham, H.; Romera-Paredes, B.; et al. Clinically Applicable Segmentation of Head and Neck Anatomy for Radiotherapy: Deep Learning Algorithm Development and Validation Study. J. Med. Internet Res. 2021, 23, e26151. [Google Scholar] [CrossRef]
- Dai, X.; Lei, Y.; Wang, T.; Zhou, J.; Roper, J.; McDonald, M.; Beitler, J.J.; Curran, W.J.; Liu, T.; Yang, X. Automated delineation of head and neck organs at risk using synthetic MRI-aided mask scoring regional convolutional neural network. Med. Phys. 2021, 48, 5862–5873. [Google Scholar] [CrossRef]
- Nemoto, T.; Futakami, N.; Yagi, M.; Kumabe, A.; Takeda, A.; Kunieda, E.; Shigematsu, N. Efficacy evaluation of 2D, 3D U-Net semantic segmentation and atlas-based segmentation of normal lungs excluding the trachea and main bronchi. J. Radiat. Res. 2020, 61, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Wong, J.; Huang, V.; Giambattista, J.A.; Teke, T.; Kolbeck, C.; Giambattista, J.; Atrchian, S. Training and Validation of Deep Learning-Based Auto-Segmentation Models for Lung Stereotactic Ablative Radiotherapy Using Retrospective Radiotherapy Planning Contours. Front. Oncol. 2021, 11, 626499. [Google Scholar] [CrossRef] [PubMed]
- Men, K.; Geng, H.; Biswas, T.; Liao, Z.; Xiao, Y. Automated Quality Assurance of OAR Contouring for Lung Cancer Based on Segmentation With Deep Active Learning. Front. Oncol. 2020, 10, 986. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Gan, W.; Zhang, C.; Feng, A.; Wang, H.; Huang, Y.; Chen, H.; Shao, Y.; Duan, Y.; Xu, Z. A 2D-3D hybrid convolutional neural network for lung lobe auto-segmentation on standard slice thickness computed tomography of patients receiving radiotherapy. Biomed. Eng. Online 2021, 20, 94. [Google Scholar] [CrossRef] [PubMed]
- Lappas, G.; Wolfs, C.J.A.; Staut, N.; Lieuwes, N.G.; Biemans, R.; van Hoof, S.J.; Dubois, L.J.; Verhaegen, F. Automatic contouring of normal tissues with deep learning for preclinical radiation studies. Phys. Med. Biol. 2022, 67, 044001. [Google Scholar] [CrossRef]
- Schreier, J.; Attanasi, F.; Laaksonen, H. Generalization vs. Specificity: In Which Cases Should a Clinic Train its Own Segmentation Models? Front. Oncol. 2020, 10, 675. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, F.; Chen, W.; Liu, X.; Hou, X.; Shen, J.; Guan, H.; Zhen, H.; Wang, S.; Chen, Q.; et al. Automatic Segmentation of Clinical Target Volumes for Post-Modified Radical Mastectomy Radiotherapy Using Convolutional Neural Networks. Front. Oncol. 2020, 10, 581347. [Google Scholar] [CrossRef]
- Choi, M.S.; Choi, B.S.; Chung, S.Y.; Kim, N.; Chun, J.; Kim, Y.B.; Chang, J.S.; Kim, J.S. Clinical evaluation of atlas- and deep learning-based automatic segmentation of multiple organs and clinical target volumes for breast cancer. Radiother. Oncol. 2020, 153, 139–145. [Google Scholar] [CrossRef]
- Liang, F.; Qian, P.; Su, K.H.; Baydoun, A.; Leisser, A.; Van Hedent, S.; Kuo, J.W.; Zhao, K.; Parikh, P.; Lu, Y.; et al. Abdominal, multi-organ, auto-contouring method for online adaptive magnetic resonance guided radiotherapy: An intelligent, multi-level fusion approach. Artif. Intell. Med. 2018, 90, 34–41. [Google Scholar] [CrossRef]
- Xia, X.; Wang, J.; Li, Y.; Peng, J.; Fan, J.; Zhang, J.; Wan, J.; Fang, Y.; Zhang, Z.; Hu, W. An Artificial Intelligence-Based Full-Process Solution for Radiotherapy: A Proof of Concept Study on Rectal Cancer. Front. Oncol. 2020, 10, 616721. [Google Scholar] [CrossRef]
- Savenije, M.H.F.; Maspero, M.; Sikkes, G.G.; van der Voort van Zyp, J.R.N.; Kotte, T.J.; Alexis, N.; Bol, G.H.; van den Berg, T.; Cornelis, A. Clinical implementation of MRI-based organs-at-risk auto-segmentation with convolutional networks for prostate radiotherapy. Radiat. Oncol. 2020, 15, 104. [Google Scholar] [CrossRef]
- Sartor, H.; Minarik, D.; Enqvist, O.; Ulén, J.; Wittrup, A.; Bjurberg, M.; Trägårdh, E. Auto-segmentations by convolutional neural network in cervical and anorectal cancer with clinical structure sets as the ground truth. Clin. Transl. Radiat. Oncol. 2020, 25, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Cha, E.; Elguindi, S.; Onochie, I.; Gorovets, D.; Deasy, J.O.; Zelefsky, M.; Gillespie, E.F. Clinical implementation of deep learning contour autosegmentation for prostate radiotherapy. Radiother. Oncol. 2021, 159, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.Y.; Zhou, J.Y.; Xu, X.T.; Guo, J.; Han, M.F.; Gao, Y.Z.; Du, H.; Stahl, J.N.; Maltz, J.S. Deep learning-based auto-segmentation of clinical target volumes for radiotherapy treatment of cervical cancer. J. Appl. Clin. Med. Phys. 2021, 23, e13470. [Google Scholar] [CrossRef] [PubMed]
- Byrne, M.; Archibald-Heeren, B.; Hu, Y.; Teh, A.; Beserminji, R.; Cai, E.; Liu, G.; Yates, A.; Rijken, J.; Collett, N.; et al. Varian ethos online adaptive radiotherapy for prostate cancer: Early results of contouring accuracy, treatment plan quality, and treatment time. J. Appl. Clin. Med. Phys. 2022, 23, e13479. [Google Scholar] [CrossRef]
- Brouwer, C.L.; Steenbakkers, R.J.H.M.; Bourhis, J.; Budach, W.; Grau, C.; Grégoire, V.; van Herk, M.; Lee, A.; Maingon, P.; Nutting, C.; et al. CT-based delineation of organs at risk in the head and neck region: DAHANCA, EORTC, GORTEC, HKNPCSG, NCIC CTG, NCRI, NRG Oncology and TROG consensus guidelines. Radiother. Oncol. 2015, 117, 83–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scoccianti, S.; Detti, B.; Gadda, D.; Greto, D.; Furfaro, I.; Meacci, F.; Simontacchi, G.; Di Brina, L.; Bonomo, P.; Giacomelli, I.; et al. Organs at risk in the brain and their dose-constraints in adults and in children: A radiation oncologist’s guide for delineation in everyday practice. Radiother. Oncol. 2015, 114, 230–238. [Google Scholar] [CrossRef]
- Offersen, B.V.; Boersma, L.J.; Kirkove, C.; Hol, S.; Aznar, M.C.; Biete Sola, A.; Kirova, Y.M.; Pignol, J.-P.; Remouchamps, V.; Verhoeven, K.; et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer. Radiother. Oncol. 2015, 114, 3–10. [Google Scholar] [CrossRef]
- Kong, F.M.; Ritter, T.; Quint, D.J.; Senan, S.; Gaspar, L.E.; Komaki, R.U.; Hurkmans, C.W.; Timmerman, R.; Bezjak, A.; Bradley, J.D.; et al. Consideration of dose limits for organs at risk of thoracic radiotherapy: Atlas for lung, proximal bronchial tree, esophagus, spinal cord, ribs, and brachial plexus. Int. J. Radiat. Oncol. Biol. Phys. 2011, 81, 1442–1457. [Google Scholar] [CrossRef] [Green Version]
- Jabbour, S.K.; Hashem, S.A.; Bosch, W.; Kim, T.K.; Finkelstein, S.E.; Anderson, B.M.; Ben-Josef, E.; Crane, C.H.; Goodman, K.A.; Haddock, M.G.; et al. Upper abdominal normal organ contouring guidelines and atlas: A Radiation Therapy Oncology Group consensus. Pract. Radiat. Oncol. 2014, 4, 82–89. [Google Scholar] [CrossRef] [Green Version]
- Gay, H.A.; Barthold, H.J.; O’Meara, E.; Bosch, W.R.; El Naqa, I.; Al-Lozi, R.; Rosenthal, S.A.; Lawton, C.; Lee, W.R.; Sandler, H.; et al. Pelvic normal tissue contouring guidelines for radiation therapy: A Radiation Therapy Oncology Group consensus panel atlas. Int. J. Radiat. Oncol. Biol. Phys. 2012, 83, e353–e362. [Google Scholar] [CrossRef] [Green Version]
- Salembier, C.; Villeirs, G.; De Bari, B.; Hoskin, P.; Pieters, B.R.; Van Vulpen, M.; Khoo, V.; Henry, A.; Bossi, A.; De Meerleer, G.; et al. ESTRO ACROP consensus guideline on CT- and MRI-based target volume delineation for primary radiation therapy of localized prostate cancer. Radiother. Oncol. 2018, 127, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Grégoire, V.; Ang, K.; Budach, W.; Grau, C.; Hamoir, M.; Langendijk, J.A.; Lee, A.; Le, Q.-T.; Maingon, P.; Nutting, C.; et al. Delineation of the neck node levels for head and neck tumors: A 2013 update. DAHANCA, EORTC, HKNPCSG, NCIC CTG, NCRI, RTOG, TROG consensus guidelines. Radiother. Oncol. 2014, 110, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Offersen, B.V.; Boersma, L.J.; Kirkove, C.; Hol, S.; Aznar, M.C.; Sola, A.B.; Kirova, Y.M.; Pignol, J.P.; Remouchamps, V.; Verhoeven, K.; et al. ESTRO consensus guideline on target volume delineation for elective radiation therapy of early stage breast cancer, version 1.1. Radiother. Oncol. 2016, 118, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Harris, V.A.; Staffurth, J.; Naismith, O.; Esmail, A.; Gulliford, S.; Khoo, V.; Lewis, R.; Littler, J.; McNair, H.; Sadoyze, A.; et al. Consensus Guidelines and Contouring Atlas for Pelvic Node Delineation in Prostate and Pelvic Node Intensity Modulated Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2015, 92, 874–883. [Google Scholar] [CrossRef]
- Lawton, C.A.F.; Michalski, J.; El-Naqa, I.; Buyyounouski, M.K.; Lee, W.R.; Menard, C.; O’Meara, E.; Rosenthal, S.A.; Ritter, M.; Seider, M. RTOG GU Radiation Oncology Specialists Reach Consensus on Pelvic Lymph Node Volumes for High-Risk Prostate Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 383–387. [Google Scholar] [CrossRef] [Green Version]
- Jeong, H.A.-O.; Ntolkeras, G.; Alhilani, M.A.-O.; Atefi, S.R.; Zöllei, L.; Fujimoto, K.; Pourvaziri, A.; Lev, M.H.; Grant, P.E.; Bonmassar, G. Development, validation, and pilot MRI safety study of a high-resolution, open source, whole body pediatric numerical simulation model. PLoS ONE 2021, 16, e0241682. [Google Scholar] [CrossRef]
- Cardenas, C.E.; Mohamed, A.S.R.; Yang, J.; Gooding, M.; Veeraraghavan, H.; Kalpathy-Cramer, J.; Ng, S.P.; Ding, Y.; Wang, J.; Lai, S.Y.; et al. Head and neck cancer patient images for determining auto-segmentation accuracy in T2-weighted magnetic resonance imaging through expert manual segmentations. Med. Phys. 2020, 47, 2317–2322. [Google Scholar] [CrossRef]
- Cardenas, C.E.; Yang, J.; Anderson, B.M.; Court, L.E.; Brock, K.B. Advances in Auto-Segmentation. Semin. Radiat. Oncol. 2019, 29, 185–197. [Google Scholar] [CrossRef]
- Liu, X.; Li, K.W.; Yang, R.; Geng, L.S. Review of Deep Learning Based Automatic Segmentation for Lung Cancer Radiotherapy. Front. Oncol. 2021, 11, 717039. [Google Scholar] [CrossRef]
- Maffei, N.; Manco, L.; Aluisio, G.; D’Angelo, E.; Ferrazza, P.; Vanoni, V.; Meduri, B.; Lohr, F.; Guidi, G. Radiomics classifier to quantify automatic segmentation quality of cardiac sub-structures for radiotherapy treatment planning. Phys. Med. 2021, 83, 278–286. [Google Scholar] [CrossRef]
- van Rooij, W.; Verbakel, W.F.; Slotman, B.J.; Dahele, M. Using Spatial Probability Maps to Highlight Potential Inaccuracies in Deep Learning-Based Contours: Facilitating Online Adaptive Radiation Therapy. Adv. Radiat. Oncol. 2021, 6, 100658. [Google Scholar] [CrossRef]
- Nijhuis, H.; van Rooij, W.; Gregoire, V.; Overgaard, J.; Slotman, B.J.; Verbakel, W.F.; Dahele, M. Investigating the potential of deep learning for patient-specific quality assurance of salivary gland contours using EORTC-1219-DAHANCA-29 clinical trial data. Acta Oncol. 2021, 60, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Maspero, M.; Savenije, M.H.F.; Dinkla, A.M.; Seevinck, P.R.; Intven, M.P.W.; Jurgenliemk-Schulz, I.M.; Kerkmeijer, L.G.W.; van den Berg, C.A.T. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy. Phys. Med. Biol. 2018, 63, 185001. [Google Scholar] [CrossRef] [PubMed]
- Barateau, A.; De Crevoisier, R.; Largent, A.; Mylona, E.; Perichon, N.; Castelli, J.; Chajon, E.; Acosta, O.; Simon, A.; Nunes, J.C.; et al. Comparison of CBCT-based dose calculation methods in head and neck cancer radiotherapy: From Hounsfield unit to density calibration curve to deep learning. Med. Phys. 2020, 47, 4683–4693. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.A.; Payne, G.S. Radiotherapy planning using MRI. Phys. Med. Biol. 2015, 60, R323–R361. [Google Scholar] [CrossRef] [PubMed]
- Devic, S. MRI simulation for radiotherapy treatment planning. Med. Phys. 2012, 39, 6701–6711. [Google Scholar] [CrossRef]
- Le, A.H.; Stojadinovic, S.; Timmerman, R.; Choy, H.; Duncan, R.L.; Jiang, S.B.; Pompos, A. Real-Time Whole-Brain Radiation Therapy: A Single-Institution Experience. Int. J. Radiat. Oncol. Biol. Phys. 2018, 100, 1280–1288. [Google Scholar] [CrossRef]
- Han, X. MR-based synthetic CT generation using a deep convolutional neural network method. Med. Phys. 2017, 44, 1408–1419. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Zhu, J.; Liu, Z.; Teng, J.; Xie, Q.; Zhang, L.; Liu, X.; Shi, J.; Chen, L. A preliminary study of using a deep convolution neural network to generate synthesized CT images based on CBCT for adaptive radiotherapy of nasopharyngeal carcinoma. Phys. Med. Biol. 2019, 64, 145010. [Google Scholar] [CrossRef]
- Dhont, J.; Verellen, D.; Mollaert, I.; Vanreusel, V.; Vandemeulebroucke, J. RealDRR—Rendering of realistic digitally reconstructed radiographs using locally trained image-to-image translation. Radiother. Oncol. 2020, 153, 213–219. [Google Scholar] [CrossRef]
- Bahrami, A.; Karimian, A.; Fatemizadeh, E.; Arabi, H.; Zaidi, H. A new deep convolutional neural network design with efficient learning capability: Application to CT image synthesis from MRI. Med. Phys. 2020, 47, 5158–5171. [Google Scholar] [CrossRef] [PubMed]
- Maspero, M.; Bentvelzen, L.G.; Savenije, M.H.F.; Guerreiro, F.; Seravalli, E.; Janssens, G.O.; van den Berg, C.A.T.; Philippens, M.E.P. Deep learning-based synthetic CT generation for paediatric brain MR-only photon and proton radiotherapy. Radiother. Oncol. 2020, 153, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Lei, Y.; Tian, Z.; Wang, T.; Liu, T.; Curran, W.J.; Yang, X. Deep learning-based volumetric image generation from projection imaging for prostate radiotherapy. In Proceedings of the Medical Imaging 2021: Image-Guided Procedures, Robotic Interventions, and Modeling, Online, 15–19 February 2021. [Google Scholar]
- Tong, F.; Nakao, M.; Wu, S.; Nakamura, M.; Matsuda, T. X-ray2Shape: Reconstruction of 3D Liver Shape from a Single 2D Projection Image. In Proceedings of the 2020 42nd Annual International Conference of the IEEE Engineering in Medicine & Biology Society (EMBC), Montréal, QC, Canada, 20–24 July 2020; pp. 1608–1611. [Google Scholar]
- Cusumano, D.; Lenkowicz, J.; Votta, C.; Boldrini, L.; Placidi, L.; Catucci, F.; Dinapoli, N.; Antonelli, M.V.; Romano, A.; De Luca, V.; et al. A deep learning approach to generate synthetic CT in low field MR-guided adaptive radiotherapy for abdominal and pelvic cases. Radiother. Oncol. 2020, 153, 205–212. [Google Scholar] [CrossRef]
- Wang, M.; Zhang, Q.; Lam, S.; Cai, J.; Yang, R. A Review on Application of Deep Learning Algorithms in External Beam Radiotherapy Automated Treatment Planning. Front. Oncol. 2020, 10, 580919. [Google Scholar] [CrossRef]
- Cilla, S.; Deodato, F.; Romano, C.; Ianiro, A.; Macchia, G.; Re, A.; Buwenge, M.; Boldrini, L.; Indovina, L.; Valentini, V.; et al. Personalized automation of treatment planning in head-neck cancer: A step forward for quality in radiation therapy? Phys. Med. 2021, 82, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Hrinivich, W.T.; Lee, J. Artificial intelligence-based radiotherapy machine parameter optimization using reinforcement learning. Med. Phys. 2020, 47, 6140–6150. [Google Scholar] [CrossRef]
- Cilla, S.; Romano, C.; Morabito, V.E.; Macchia, G.; Buwenge, M.; Dinapoli, N.; Indovina, L.; Strigari, L.; Morganti, A.G.; Valentini, V.; et al. Personalized Treatment Planning Automation in Prostate Cancer Radiation Oncology: A Comprehensive Dosimetric Study. Front. Oncol. 2021, 11, 636529. [Google Scholar] [CrossRef]
- Cilla, S.; Macchia, G.; Romano, C.; Morabito, V.E.; Boccardi, M.; Picardi, V.; Valentini, V.; Morganti, A.G.; Deodato, F. Challenges in lung and heart avoidance for postmastectomy breast cancer radiotherapy: Is automated planning the answer? Med. Dosim. 2021, 46, 295–303. [Google Scholar] [CrossRef]
- Kida, S.; Nakamoto, T.; Nakano, M.; Nawa, K.; Haga, A.; Kotoku, J.; Yamashita, H.; Nakagawa, K. Cone Beam Computed Tomography Image Quality Improvement Using a Deep Convolutional Neural Network. Cureus 2018, 10, e2548. [Google Scholar] [CrossRef] [Green Version]
- Kurosawa, T.; Nishio, T.; Moriya, S.; Tsuneda, M.; Karasawa, K. Feasibility of image quality improvement for high-speed CBCT imaging using deep convolutional neural network for image-guided radiotherapy in prostate cancer. Phys. Med. 2020, 80, 84–91. [Google Scholar] [CrossRef]
- Rostampour, N.; Jabbari, K.; Esmaeili, M.; Mohammadi, M.; Nabavi, S. Markerless Respiratory Tumor Motion Prediction Using an Adaptive Neuro-fuzzy Approach. J. Med. Signals Sens. 2018, 8, 25–30. [Google Scholar] [PubMed]
- Gustafsson, C.J.; Sward, J.; Adalbjornsson, S.I.; Jakobsson, A.; Olsson, L.E. Development and evaluation of a deep learning based artificial intelligence for automatic identification of gold fiducial markers in an MRI-only prostate radiotherapy workflow. Phys. Med. Biol. 2020, 65, 225011. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, W.; Oshikawa, S.; Mori, S. Real-time markerless tumour tracking with patient-specific deep learning using a personalised data generation strategy: Proof of concept by phantom study. Br. J. Radiol. 2020, 93, 20190420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maspero, M.; Houweling, A.C.; Savenije, M.H.F.; van Heijst, T.C.F.; Verhoeff, J.J.C.; Kotte, A.; van den Berg, C.A.T. A single neural network for cone-beam computed tomography-based radiotherapy of head-and-neck, lung and breast cancer. Phys. Imaging Radiat. Oncol. 2020, 14, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhu, X.; Hong, J.C.; Zheng, D. Artificial Intelligence in Radiotherapy Treatment Planning: Present and Future. Technol. Cancer Res. Treat. 2019, 18, 1533033819873922. [Google Scholar] [CrossRef]
- Ford, E.; Conroy, L.; Dong, L.; de Los Santos, L.F.; Greener, A.; Gwe-Ya Kim, G.; Johnson, J.; Johnson, P.; Mechalakos, J.G.; Napolitano, B.; et al. Strategies for effective physics plan and chart review in radiation therapy: Report of AAPM Task Group 275. Med. Phys. 2020, 47, e236–e272. [Google Scholar] [CrossRef] [Green Version]
- El Naqa, I.; Ruan, D.; Valdes, G.; Dekker, A.; McNutt, T.; Ge, Y.; Wu, Q.J.; Oh, J.H.; Thor, M.; Smith, W.; et al. Machine learning and modeling: Data, validation, communication challenges. Med. Phys. 2018, 45, e834–e840. [Google Scholar] [CrossRef] [Green Version]
- Xia, P.; Sintay, B.J.; Colussi, V.C.; Chuang, C.; Lo, Y.-C.; Schofield, D.; Wells, M.; Zhou, S. Medical Physics Practice Guideline (MPPG) 11.a: Plan and chart review in external beam radiotherapy and brachytherapy. J. Appl. Clin. Med. Phys. 2021, 22, 4–19. [Google Scholar] [CrossRef]
- Xu, H.; Zhang, B.; Guerrero, M.; Lee, S.W.; Lamichhane, N.; Chen, S.; Yi, B. Toward automation of initial chart check for photon/electron EBRT: The clinical implementation of new AAPM task group reports and automation techniques. J. Appl. Clin. Med. Phys. 2021, 22, 234–245. [Google Scholar] [CrossRef]
- Osman, A.A.-O.X.; Maalej, N.M. Applications of machine and deep learning to patient-specific IMRT/VMAT quality assurance. J. Appl. Clin. Med Phys. 2021, 22, 20–36. [Google Scholar] [CrossRef]
- Cho, Y.B.; Farrokhkish, M.; Norrlinger, B.; Heaton, R.; Jaffray, D.; Islam, M. An artificial neural network to model response of a radiotherapy beam monitoring system. Med. Phys. 2020, 47, 1983–1994. [Google Scholar] [CrossRef] [PubMed]
- Luk, S.M.H.; Meyer, J.; Young, L.A.; Cao, N.; Ford, E.C.; Phillips, M.H.; Kalet, A.M. Characterization of a Bayesian network-based radiotherapy plan verification model. Med. Phys. 2019, 46, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Hu, C.; Chi, C.; Jiang, Z.; Tong, Y.; Zhao, C. An Artificial Intelligence Model for Predicting 1-Year Survival of Bone Metastases in Non-Small-Cell Lung Cancer Patients Based on XGBoost Algorithm. Biomed. Res. Int. 2020, 2020, 3462363. [Google Scholar] [CrossRef]
- Du, R.; Lee, V.H.; Yuan, H.; Lam, K.O.; Pang, H.H.; Chen, Y.; Lam, E.Y.; Khong, P.L.; Lee, A.W.; Kwong, D.L.; et al. Radiomics Model to Predict Early Progression of Nonmetastatic Nasopharyngeal Carcinoma after Intensity Modulation Radiation Therapy: A Multicenter Study. Radiol. Artif. Intell. 2019, 1, e180075. [Google Scholar] [CrossRef] [PubMed]
- De Felice, F.; Valentini, V.; De Vincentiis, M.; Di Gioia, C.R.T.; Musio, D.; Tummulo, A.A.; Ricci, L.I.; Converti, V.; Mezi, S.; Messineo, D.; et al. Prediction of Recurrence by Machine Learning in Salivary Gland Cancer Patients After Adjuvant (Chemo)Radiotherapy. In Vivo 2021, 35, 3355–3360. [Google Scholar] [CrossRef]
- Lee, S.; Kerns, S.; Ostrer, H.; Rosenstein, B.; Deasy, J.O.; Oh, J.H. Machine Learning on a Genome-wide Association Study to Predict Late Genitourinary Toxicity After Prostate Radiation Therapy. Int. J. Radiat. Oncol. 2018, 101, 128–135. [Google Scholar] [CrossRef]
- Tian, Z.; Yen, A.; Zhou, Z.; Shen, C.; Albuquerque, K.; Hrycushko, B. A machine-learning-based prediction model of fistula formation after interstitial brachytherapy for locally advanced gynecological malignancies. Brachytherapy 2019, 18, 530–538. [Google Scholar] [CrossRef]
- van Velzen, S.G.M.; Gal, R.; Teske, A.J.; van der Leij, F.; van den Bongard, D.; Viergever, M.A.; Verkooijen, H.M.; Išgum, I. AI-Based Radiation Dose Quantification for Estimation of Heart Disease Risk in Breast Cancer Survivors After Radiation Therapy. Int. J. Radiat. Oncol. Biol. Phys. 2021, 112, 621–632. [Google Scholar] [CrossRef]
- Tabl, A.A.; Alkhateeb, A.; ElMaraghy, W.; Rueda, L.; Ngom, A. A machine learning approach for identifying gene biomarkers guiding the treatment of breast cancer. Front. Genet. 2019, 10, 256. [Google Scholar] [CrossRef] [Green Version]
- Ubaldi, L.; Valenti, V.; Borgese, R.F.; Collura, G.; Fantacci, M.E.; Ferrera, G.; Iacoviello, G.; Abbate, B.F.; Laruina, F.; Tripoli, A.; et al. Strategies to develop radiomics and machine learning models for lung cancer stage and histology prediction using small data samples. Phys. Med. 2021, 90, 13–22. [Google Scholar] [CrossRef]
- Kawahara, D.; Murakami, Y.; Tani, S.; Nagata, Y. A prediction model for degree of differentiation for resectable locally advanced esophageal squamous cell carcinoma based on CT images using radiomics and machine-learning. Br. J. Radiol. 2021, 94, 20210525. [Google Scholar] [CrossRef] [PubMed]
- Lou, B.; Doken, S.; Zhuang, T.; Wingerter, D.; Gidwani, M.; Mistry, N.; Ladic, L.; Kamen, A.; Abazeed, M.E. An image-based deep learning framework for individualizing radiotherapy dose. Lancet Digit. Health 2019, 1, e136–e147. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Jiao, Y.; Zhang, Y.; Ren, X.; Li, P.; Yu, Q.; Zhang, Q.; Wang, Q.; Fu, S. Imaging-Based Individualized Response Prediction Of Carbon Ion Radiotherapy For Prostate Cancer Patients. Cancer Manag. Res. 2019, 11, 9121–9131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haak, H.E.; Gao, X.; Maas, M.; Waktola, S.; Benson, S.; Beets-Tan, R.G.H.; Beets, G.L.; van Leerdam, M.; Melenhorst, J. The use of deep learning on endoscopic images to assess the response of rectal cancer after chemoradiation. Surg. Endosc. 2021, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Osman, S.O.S.; Leijenaar, R.T.H.; Cole, A.J.; Lyons, C.A.; Hounsell, A.R.; Prise, K.M.; O’Sullivan, J.M.; Lambin, P.; McGarry, C.K.; Jain, S. Computed Tomography-based Radiomics for Risk Stratification in Prostate Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2019, 105, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Olszewski, D.; He, C.; Pintea, G.; Lian, J.; Chou, T.; Chen, R.C.; Shtylla, B. Machine learning and statistical prediction of patient quality-of-life after prostate radiation therapy. Comput. Biol. Med. 2021, 129, 104127. [Google Scholar] [CrossRef]
- Jochems, A.; El-Naqa, I.; Kessler, M.; Mayo, C.S.; Jolly, S.; Matuszak, M.; Faivre-Finn, C.; Price, G.; Holloway, L.; Vinod, S.; et al. A prediction model for early death in non-small cell lung cancer patients following curative-intent chemoradiotherapy. Acta Oncol. 2018, 57, 226–230. [Google Scholar] [CrossRef] [Green Version]
- Blackledge, M.D.; Winfield, J.M.; Miah, A.; Strauss, D.; Thway, K.; Morgan, V.A.; Collins, D.J.; Koh, D.M.; Leach, M.O.; Messiou, C. Supervised Machine-Learning Enables Segmentation and Evaluation of Heterogeneous Post-treatment Changes in Multi-Parametric MRI of Soft-Tissue Sarcoma. Front. Oncol. 2019, 9, 941. [Google Scholar] [CrossRef] [Green Version]
- Stenhouse, K.; Roumeliotis, M.; Ciunkiewicz, P.; Banerjee, R.; Yanushkevich, S.; McGeachy, P. Development of a Machine Learning Model for Optimal Applicator Selection in High-Dose-Rate Cervical Brachytherapy. Front. Oncol. 2021, 11, 611437. [Google Scholar] [CrossRef]
- Li, H.; Galperin-Aizenberg, M.; Pryma, D.; Simone, C.B., II; Fan, Y. Unsupervised machine learning of radiomic features for predicting treatment response and overall survival of early stage non-small cell lung cancer patients treated with stereotactic body radiation therapy. Radiother. Oncol. 2018, 129, 218–226. [Google Scholar] [CrossRef]
- Sleeman Iv, W.C.; Nalluri, J.; Syed, K.; Ghosh, P.; Krawczyk, B.; Hagan, M.; Palta, J.; Kapoor, R. A Machine Learning method for relabeling arbitrary DICOM structure sets to TG-263 defined labels. J. Biomed. Inform. 2020, 109, 103527. [Google Scholar] [CrossRef] [PubMed]
- Syed, K.; Iv, W.S.; Ivey, K.; Hagan, M.; Palta, J.; Kapoor, R.; Ghosh, P. Integrated Natural Language Processing and Machine Learning Models for Standardizing Radiotherapy Structure Names. Healthcare 2020, 8, 120. [Google Scholar] [CrossRef]
- Haga, A.; Takahashi, W.; Aoki, S.; Nawa, K.; Yamashita, H.; Abe, O.; Nakagawa, K. Standardization of imaging features for radiomics analysis. J. Med. Investig. 2019, 66, 35–37. [Google Scholar] [CrossRef] [PubMed]
- Peltola, M.K.; Lehikoinen, J.S.; Sippola, L.T.; Saarilahti, K.; Mäkitie, A.A. A Novel Digital Patient-Reported Outcome Platform for Head and Neck Oncology Patients—A Pilot Study. Clin. Med. Insights Ear Nose Throat 2016, 9, 1–6. [Google Scholar] [CrossRef]
- Batumalai, V.; Jameson, M.G.; King, O.; Walker, R.; Slater, C.; Dundas, K.; Dinsdale, G.; Wallis, A.; Ochoa, C.; Gray, R.; et al. Cautiously optimistic: A survey of radiation oncology professionals’ perceptions of automation in radiotherapy planning. Tech. Innov. Patient Support Radiat. Oncol. 2020, 16, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Luna, D.; Almerares, A.; Mayan, J.C., 3rd; González Bernaldo de Quirós, F.; Otero, C. Health Informatics in Developing Countries: Going beyond Pilot Practices to Sustainable Implementations: A Review of the Current Challenges. Healthc. Inform. Res. 2014, 20, 3–10. [Google Scholar] [CrossRef]
- Abernethy, A.P.; Etheredge, L.M.; Ganz, P.A.; Wallace, P.; German, R.R.; Neti, C.; Bach, P.B.; Murphy, S.B. Rapid-learning system for cancer care. J. Clin. Oncol. 2010, 28, 4268–4274. [Google Scholar] [CrossRef] [Green Version]
- Halford, G.S.; Baker, R.; McCredden, J.E.; Bain, J.D. How many variables can humans process? Psychol. Sci. 2005, 16, 70–76. [Google Scholar] [CrossRef]
- Vayena, E.A.-O.; Blasimme, A.A.-O.; Cohen, I.G. Machine learning in medicine: Addressing ethical challenges. PLoS Med. 2018, 15, e1002689. [Google Scholar] [CrossRef]
- Scheetz, J.; Rothschild, P.; McGuinness, M.; Hadoux, X.; Soyer, H.P.; Janda, M.; Condon, J.J.J.; Oakden-Rayner, L.; Palmer, L.J.; Keel, S.; et al. A survey of clinicians on the use of artificial intelligence in ophthalmology, dermatology, radiology and radiation oncology. Sci. Rep. 2021, 11, 5193. [Google Scholar] [CrossRef]
- Victor Mugabe, K. Barriers and facilitators to the adoption of artificial intelligence in radiation oncology: A New Zealand study. Tech. Innov. Patient Support Radiat. Oncol. 2021, 18, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Zanca, F.; Hernandez-Giron, I.; Avanzo, M.; Guidi, G.; Crijns, W.; Diaz, O.; Kagadis, G.C.; Rampado, O.; Lønne, P.I.; Ken, S.; et al. Expanding the medical physicist curricular and professional programme to include Artificial Intelligence. Phys. Med. 2021, 83, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Atwood, T.F.; Brown, D.W.; Murphy, J.D.; Moore, K.L.; Mundt, A.J.; Pawlicki, T. Establishing a New Clinical Role for Medical Physicists: A Prospective Phase II Trial. Int. J. Radiat. Oncol. 2018, 102, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Netherton, T.J.; Cardenas, C.E.; Rhee, D.J.; Court, L.E.; Beadle, B.M. The Emergence of Artificial Intelligence within Radiation Oncology Treatment Planning. Oncology 2020, 99, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, M.A.-O.; Sollini, M.A.-O.; Ninatti, G.A.-O.; Loiacono, D.A.-O.; Giacomello, E.A.-O.; Gozzi, N.A.-O.; Amigoni, F.A.-O.; Mainardi, L.A.-O.; Lanzi, P.A.-O.; Chiti, A.A.-O. Distributed learning: A reliable privacy-preserving strategy to change multicenter collaborations using AI. Eur. J. Pediatr. 2021, 48, 3791–3804. [Google Scholar] [CrossRef]
- Korreman, S.; Eriksen, J.G.; Grau, C. The changing role of radiation oncology professionals in a world of AI—Just jobs lost—Or a solution to the under-provision of radiotherapy? Clin. Transl. Radiat. Oncol. 2021, 26, 104–107. [Google Scholar] [CrossRef]
- McBee, M.P.; Awan, O.A.; Colucci, A.T.; Ghobadi, C.W.; Kadom, N.; Kansagra, A.P.; Tridandapani, S.; Auffermann, W.F. Deep Learning in Radiology. Acad. Radiol. 2018, 25, 1472–1480. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santoro, M.; Strolin, S.; Paolani, G.; Della Gala, G.; Bartoloni, A.; Giacometti, C.; Ammendolia, I.; Morganti, A.G.; Strigari, L. Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond. Appl. Sci. 2022, 12, 3223. https://doi.org/10.3390/app12073223
Santoro M, Strolin S, Paolani G, Della Gala G, Bartoloni A, Giacometti C, Ammendolia I, Morganti AG, Strigari L. Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond. Applied Sciences. 2022; 12(7):3223. https://doi.org/10.3390/app12073223
Chicago/Turabian StyleSantoro, Miriam, Silvia Strolin, Giulia Paolani, Giuseppe Della Gala, Alessandro Bartoloni, Cinzia Giacometti, Ilario Ammendolia, Alessio Giuseppe Morganti, and Lidia Strigari. 2022. "Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond" Applied Sciences 12, no. 7: 3223. https://doi.org/10.3390/app12073223
APA StyleSantoro, M., Strolin, S., Paolani, G., Della Gala, G., Bartoloni, A., Giacometti, C., Ammendolia, I., Morganti, A. G., & Strigari, L. (2022). Recent Applications of Artificial Intelligence in Radiotherapy: Where We Are and Beyond. Applied Sciences, 12(7), 3223. https://doi.org/10.3390/app12073223







