Abstract
AIDS, which is caused by the most widespread HIV-1 virus, attacks the immune system of the human body, and despite the incredible endeavors for finding proficient medication strategies, the continuing spread of AIDS and claiming subsequent infections has not yet been decreased. Consequently, the discovery of innovative medicinal methodologies is highly in demand. Some available therapies, based on peptides, proclaim the treatment for several deadly diseases such as AIDS and cancer. Since many experimental types of research are restricted by the analysis period and expenses, computational methods overcome the issues effectually. In computational technique, the peptide residues with anti-HIV-1 activity are predicted by classification method, and the learning process of the classification is improved with significant features. Rough set-based algorithms are capable of dealing with the gaps and imperfections present in real-time data. In this work, feature selection using Rough Set Improved Harmony Search Quick Reduct and Rough Set Improved Harmony Search Relative Reduct with Rough Set Classification framework is implemented to classify Anti-HIV-1 peptides. The primary objective of the proposed methodology is to predict the peptides with an anti-HIV-1 activity using effective feature selection and classification algorithms incorporated in the proposed framework. The results of the proposed algorithms are comparatively studied with existing rough set feature selection algorithms and benchmark classifiers, and the reliability of the algorithms implemented in the proposed framework is measured by validity measures, such as Precision, Recall, F-measure, Kulczynski Index, and Fowlkes–Mallows Index. The final results show that the proposed framework analyzed and classified the peptides with a high predictive accuracy of 96%. In this study, we have investigated the ability of a rough set-based framework with sequence-based numeric features to classify anti-HIV-1 peptides, and the experimentation results show that the proposed framework discloses the most satisfactory solutions, where it rapidly congregates in the problem space and finds the best reduct, which improves the prediction accuracy of the given dataset.
1. Introduction
AIDS (Acquired Immunodeficiency Syndrome) is a deadly and overwhelming disease caused by the HIV virus [1]. According to the World Health Organization (WHO) survey, more than 35 million people have died, and 36.9 million live with HIV globally. The HIV-1 type virus is the most widespread type and major stimulator of AIDS [2]. Many therapeutics have been discovered to save the infected person, but recovery is a challenging process; therefore, innovative and effective treatment for AIDS is needed. Long-term medication and toxicity may restrict the utilization of such treatments, and it should be frequently monitored and controlled by another supportive therapy [3]. Peptide molecules are created by the dehydration condensation reaction of amino acids joined by peptide bonds. They are used to treat a variety of diseases [4]. In recent years, peptides that have an anti-HIV-1 activity are being focused on and reveal promising results, which gradually reduce the effects of AIDS [5]. Investigating the enormous anti-HIV-1 peptides demands much time and effort, where simplified methodologies, such as machine learning, can be developed and implemented to predict the anti-HIV-1 peptides [6].
Feature Selection (FS) is one of the significant phases in the proposed framework, which eliminates redundant and irrelevant features to increase classification accuracy [7]. The prediction of anti-HIV-1 peptides (AHP) from a given sequence data would be well improvised, as it decreases the running time of the classification phase. The Harmony Search (HS) algorithm is based on a music enhancing process that searches for an optimal harmony [8,9,10,11]. The HS algorithm is modified by Mahdavi et al. [12,13] in terms of parameters. Rough Set theory is a tool to deal with the ambiguity and redundancy present in the dataset and reserves its originality [14]. The feature selection and classification techniques using rough set theory are effectively implemented in many studies [15,16]. An improved HS is embedded with RST for attribute selection to analyze protein sequences [17].
In the proposed framework, the essence of Rough Set Quick Reduct and Rough Set Relative Reduct with an improved HS algorithm is employed to select the significant features. A Rough Set Classification (RSC) is employed to form the rules for the selected features and predict anti-HIV-1 peptides. The RSC algorithm classifies the significant features, and the performance of the Rough Set Particle Swarm Optimization Quick Reduct (RSPSOQR) and Rough Set Particle Swarm Optimization Relative Reduct (RSPSORR) feature selection algorithms are comparatively analyzed with existing Particle Swarm Optimization (PSO) based algorithms. The accuracy of the RSC algorithm is evaluated with benchmark classifiers.
Research Motivation and Contribution
HIV is a deadly disease that should be treated with more effective therapy, and peptide-based treatment that fights against the virus has proved beneficial compared to other treatments. The conventional experiments on these peptides, as known, require much time and effort, where the peptide with AHP activity can be determined by computational intelligence techniques [6]. Generally, the dataset extracted from the biological sequences is large in dimension, with inconsistencies, where the dimensionality reduction method to handle the inconsistency in the peptide dataset is a requirement. The RSC algorithm is adopted in the proposed framework along with Rough Set Improved Harmony Search Quick Reduct (RSIHSQR) and Rough Set Improved Harmony Search Relative Reduct (RSIHSRR) feature selection algorithm. In Section 3 Figure 1 shows the proposed framework that depicts and predicts the anti-HIV-1 peptides with a good accuracy using finest selected features. As the RSIHSQR and RSIHSRR feature selection algorithm has revealed the best result in the previous studies [17], it is also employed for the peptide therapeutic field of this study.
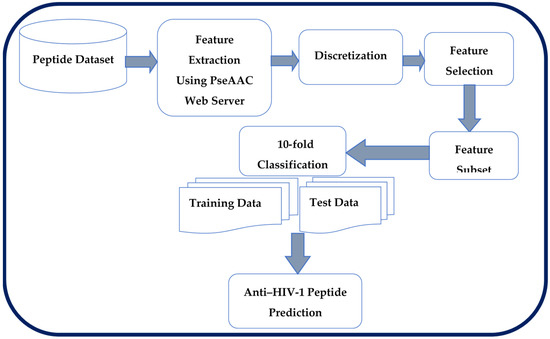
Figure 1.
The Proposed Framework.
The rest of the paper is organized as follows: Section 2 presents the related work of this study. Section 3 presents the proposed methods for Peptide Classification. Details about RSC are presented in Section 4. Experimental analysis and results are presented in Section 5. Discussion is presented in Section 6. Conclusions with future work are presented in Section 7.
2. Related Work
The implementation of the machine learning method in bioinformatics is affected by many factors. The peptide therapeutic field describes different types of frameworks using the computational method. It consumes enormous effort, time, and cost to analyze peptide information. The knowledge discovery for the bioinformatics applications will be a challenging process due to a large-sized database. The framework that embeds the feature selection before classification effectively handles the high-dimensional proteomic datasets. In the last two decades, significant research endeavors have been committed with data mining systems to acquire information in the field of disease diagnosis [15,18,19,20]. As accuracy is most important for diagnosing diseases and treating patients, the Rough Set is one such tool to deal with uncertain and imprecise data and yield accurate results. There are considerable numbers of studies on feature selection techniques, and classification methods are depicted in Table 1.

Table 1.
Related work of this study.
3. Proposed Method for Peptide Classification
The proposed methodology shown in Figure 1 collects the peptides with AHP from the HIV inhibitory peptides (HIPdb) database [34]. Physicochemical properties based on features are extracted from the peptides using Chou’s PseAAC method [20]. The extracted peptide dataset is discretized for the rough set-based analysis. It is subjected to a feature selection process to reveal the relevant features and classification algorithm used for peptides prediction. This section discusses the major components of the proposed framework.
3.1. Feature Extraction
Feature extraction is used in the proposed framework to convert sequences to numerical attributes, as shown in Figure 2. The peptides are converted to digital vector using PseAAC server for the computational process [20,35], where the peptide features are extracted from conventional AAC with six physicochemical properties with weight factor (0.05) and lambda given the value of 2, where the 6 physicochemical properties are: Hydrophobicity, Hydrophilicity, pK1 (Ca–COOH), pK2 (NH3), PI (25 _C), and molecular weight. The 63 combinations of physicochemical properties are considered, and 126 features are extracted (λ = 2) from the peptide dataset for sequence order information [36]. These features are generated from type 1 of the PseAAC web server (http://www.csbio.sjtu.edu.cn/bioinf/PseAAC, accessed on 10 January 2022).

Figure 2.
Features extracted from anti-HIV-1 peptide sequence.
3.2. Discretization of Peptide Features
The uncertain dataset with real-valued attributes is discretized for an effective classification [37,38]. Discretization is the process of transferring continuous values to discrete ones, and rough set theory works on discrete variables to deal with the ambiguity presented in the real dataset [39]. The peptide dataset experimented with within this study has real-valued attributes. Hence, it is discretized using the Max-Min discretization method [40,41], then it was experimented with by the feature selection algorithms in the proposed framework.
3.3. Rough Set Based Feature Selection
Feature selection (FS) is a dimension reduction method for datasets with many attributes, where the distinctiveness of the dataset is preserved [2]. The rough set selects the relevant features based on the FS algorithms, which improves the learning process of prediction methods [18,42]. In this work, RSIHSQR and RSIHSRR feature selection algorithms are implemented to diminish the redundant and immaterialized features of the available datasets and disclose the best features. Both algorithms are implemented for a large dataset and have shown likely results in protein analysis [22,43]. The spirit of the rough set quick reduct method, in the proposed algorithms, is inherited in the improved HS algorithm to converge towards the best solution, and the solution is achieved by the music improvisation technique [13,44], where both algorithms are briefly described in [17,26]. The results of Algorithm 1 and Algorithm 2 are relatively studied with RSPSOQR and RSPSORR algorithms [33,45], and the selected best features are used as an input to the classifier for the analysis and classification of anti-HIV-1 peptides.
| Algorithm 1. RSIHSQR FS algorithm |
| Algorithm: RSIHSQR(C,De) |
| Input: C, the conditional attributes; |
| De, the class attribute; |
| Output: Optimal feature subset |
| Step I: The fitness function, f(X) to be defined. |
| Initialize the parameters HMS = 30 |
| HMCR = 0.9 // HM constraint |
| MaxIt = 100 // iteration count |
| PVB//feasible value limit |
| PARmin,PARmax, bwmin, bwmax, // Pitch Adjusting Rate & bandwidth ∈ (0 to 1) |
| fit = 0; |
| Xold = X1; bestfit = X1; bestreduct = {}; |
| Step II: The Harmony Memory to be set as, HM = (X1, X2, …, XHMS) |
| For i = 1:HMS |
| ∀: Xi // Xi is the ith harmony of HM |
| R ← Xi (1’s of Xi) |
| ∀x ∈ (C − R) |
| R∪{x}(De) = |
| f(Xi) = R∪{x}(De) for all X ⊂ R, X(De) ≠ C(De) |
| if f(Xi) > fit |
| fit ← f(Xi) |
| Xold ← Xi |
| End if |
| End for |
| Step III: The new HM to be improvised. |
| While itr ≤ MaxIt | fit == 1 |
| for j = 1:NVAR |
| ∀:Xold (j) |
| Update PAR(); |
| Update bw(); |
| if random ( ) ≤ HMCR //random numberbetween 0 and 1 |
| Xnew ← Xold; |
| if random ( ) ≤ PAR |
| Xnew(j) = Xnew(j) ± random() * bw |
| end if |
| else |
| // select Xnew |
| Xnew(j) = PVBlower + random( ) * (PVBupper – PVBlower) |
| end if |
| end for |
| Step IV: The new HM to be updated |
| Calculate fitness for Xnew---(Step II) |
| if f(Xnew) f(Xold) |
| // Substitute the older harmony with new harmony, if it is best |
| Xold ← Xnew; |
| if f(Xnew) > fit |
| fit ← f(Xnew); |
| bestfit ← Xnew; |
| End if |
| Exit |
| end if |
| end while |
| bestreduct ← selected attributes of bestfit |
| Algorithm 2. RSIHSRR FS algorithm |
| Algorithm: RSIHSRR(C,De) |
| Input: C, the conditional attribute set; |
| De, the decision attribute |
| Output: Optimal feature subset |
| Step I: The fitness function, f(X) to be defined. |
| Initialize the parameters HMS = 30 |
| HMCR = 0.9 // HM constraint |
| MaxIt = 100 // iteration count |
| PVB//feasible value limit |
| PARmin, PARmax, bwmin, bwmax,// Pitch Adjusting Rate & bandwidth ∈ (0 to 1) |
| fit = 0; |
| Xold = X1; bestfit = X1; bestreduct = {}; |
| Step II: The Harmony Memory to be set as, HM = (X1,X2,….XHMS) |
| For i = 1:HMS |
| ∀: Xi // Xi is the ith harmony of HM |
| R ← Xi (1′s of Xi) |
| ∀x ∈ R |
| ƘR-{x}(De) = |
| f(Xi) = ƘR-{x}(De) for all X⊂R, ƘX(De) ≠ ƘC(De) |
| if f(Xi) > fit |
| fit ← f(Xi) |
| Xold ← Xi |
| End if |
| End for |
| if fit == 1 |
| bestfit = fit; |
| Return R; |
| Endif |
| Step III: The new HM to be improvised. |
| While itr ≤ MaxIt | fit != 1 |
| for j = 1:NVAR |
| ∀:Xold (j) |
| Update PAR(); |
| Update bw(); |
| if random ( ) ≤ HMCR |
| Xnew ← Xold; |
| if random ( ) ≤ PAR |
| Xnew(j) = Xnew(j) ± random() * bw |
| end if |
| else |
| //selectXnew |
| Xnew(j)=PVBlower + random( ) * (PVBupper—PVBlower) |
| end if |
| end for |
| Step IV: The new HM to be updated |
| Calculate fitness for Xnew ---(Step II). |
| if f(Xnew) == 1 |
| bestfit = Xnew; |
| Return R; |
| End if |
| if f(Xnew) f(Xold) |
| // Substitute the older harmony, if new harmony is acceptable. |
| Xold ← Xnew; |
| if f(Xnew) > fit |
| fit ← f(Xnew); |
| bestfit ← Xnew; |
| End if |
| Exit |
| end if |
| end while |
| bestreduct ← selected attributes of bestfit |
4. Rough Set Classification (RSC)
Classification is another vital method for discovering knowledge in bioinformatics applications [7]. In the proposed framework, the peptide dataset is classified using a Rough RSC algorithm based on the following objectives [46,47]: (a) RST is simple to implement, and it can be extended with any algorithms; (b) it does not require any input parameter except dataset; (c) it delivers better analysis result for the knowledge-making process; (d) it withstands uncertainties and imperfections present in the dataset. It works on a 10-fold cross-validation method to produce the decision rules and the lower and upper approximation of the RST. The equivalence relation of conditional and decision attributes classifies the peptides into two disjoint classes: anti-HIV-1 activity and low/nil anti-HIV-1 activity. The peptides are classified to forecast the anti-HIV-1 activity based on the constructed rules. The functioning of the RSC algorithm is comparatively assessed with five benchmark classifiers [48] in terms of Precision, Recall, F-Measure, Kulczynski Index, and Fowlkes–Mallows Index [49]. The proposed RSC algorithm is represented in Algorithm 3 in the following table.
| Algorithm 3. Rough Set Classification Algorithm |
| Algorithm: RSC (C,D) |
| Input: Conditional attributes 1, 2, …, n − 1 and the Decision attribute D. |
| Output: Generated Decision Rules |
| Step 1: Generate training data set and test data in 10:1 ratio, respectively. |
| Step 2: The equivalence relation for the conditional attributes is to be constructed in the training set. |
| Step 3: The equivalence relation for the decision attribute is constructed in the training set. |
| Step 4: The rough set lower approximation for indiscernibility relation for conditional attributes and decision attribute to be built. |
| Step 5: The rough set upper approximation for indiscernibility relation for conditional attributes and the decision attribute to be constructed. |
| Step 6: Generate the specific rules from the lower approximation of the rough set. |
| Step 7: Generate the possible rules from rough set upper approximation. |
| Step 8: Remove the redundant rules from the rough approximation space. |
5. Experimental Analysis
In the proposed framework, the rough set algorithms analyze the peptides to predict anti-HIV-1 activity. Many features are reduced by selecting the significant feature subset by RSIHSQR and RSIHSRR algorithms. The feature subset is used as an input to the RSC algorithm, which classifies the peptides into two distinct classes as mentioned earlier, and the performance of the FS and the classification algorithms were analyzed with existing RSPSOQR and RSPSORR algorithms using Naïve Bayes, IBK, J48, Random Forest, and JRIP classifiers.
5.1. Results
The results of the feature selection algorithms are given in Table 2, and the outcome of classification algorithms are given in Table 3.

Table 2.
Features Selected by FS algorithms.

Table 3.
Results of the proposed framework and other FS and classification algorithms.
5.1.1. Performance Evaluation of Proposed FS Algorithms
This study proposes using the rough set rapid reduct and relative reduct with better HS FS to reduce the dimensions of a given dataset. The proposed algorithms investigated the peptides collected from the HIPdb database. There are six physicochemical properties of the peptides, and 126 numerical features were generated, and the extracted features are subjected to the feature selection process to extract the best features. The proposed RSIHSQR and RSIHSRR algorithms are evaluated with RSPSOQR and RSPSORR algorithms. Table 2 explicitly illustrates the result of the proposed FS algorithms, which selects the minimum features of the existing algorithms. The classification algorithms evaluate the reliability of the feature subsets.
5.1.2. Assessment of FS with a Classification Algorithm
Six classification algorithms assess the feature subsets selected by the four FS algorithms. The reliability of the FS algorithms is measured by Precision, Recall, F-measure, Kulczynski Index, and Fowlkes–Mallows Index [49,50]. These measures depend on True Positive (TP), False Positive (FP), and False Negative (FN).
Precision = TP/(TP + FP)
Recall = TP/(TP + FN)
F-Measure = 2 * ((Precision X Recall)/(Precision + Recall))
Kulczynski Index = ½ (Precision + Recall)
Fowlkes–Mallows Index=√(Precision * Recall)
The results of the proposed framework of this study are evaluated using the performance measures (Equations (1)–(5)) of the 10-fold classification methods [51,52]. Table 3 shows the results of performance measures, in which many classifiers have revealed good results (>0.8) with feature subset of proposed RSIHSQR and RSIHSRR feature selection algorithms. The validity measure greater than 0.8 represents the accuracy of the prediction rate, particularly with the peptide dataset [6].
5.1.3. Assessment of RSC with Other Classification Algorithms
Table 3 shows the performance of the RSC algorithm in the proposed framework compared with benchmark classifiers. It is believed that the rough set classifier has discovered better classification results of the anti-HIV-1 peptides and the RSC algorithm exposed high predictive accuracies with the proposed FS algorithms.
6. Discussion
Twenty-two different amino acids constitute numerous body proteins present in the human body. Of them, two are particular amino acids—selenocysteine and pyrrolysine, which were recently discovered. Each amino acid is unique in its own way and has specific characters that are responsible for its function. For example, histidine has a pK value of 6.1, which is close to the physiological pH of the blood, which makes it a critical blood buffer. It shields against changes in blood pH and maintains it within a narrow range of 7.38 to 7.42. The primary structure of proteins is formed by amino acids linked together by peptide bonds linearly. Some proteins also contain a circular primary structure, whereas some show branching. Hence, the functioning of the protein depends on its primary structure made of amino acids. Thus, in the same way, the functioning of an AHP is also, in turn, dependent on its amino acid composition. The amino acid may be linear or cyclic (aromatic), branched or unbranched, hydrophobic or hydrophilic, acidic or basic, and may also contain special groups with them.
This work on AHP focuses on understanding what essential attributes of an amino acid sequence determine if the AHP is effective against the virus. It is further focused on improving the efficiency in identifying the AHP and concurrently reduces the number of features needed to classify a peptide sequence to be AHP or not. This, in turn, makes the process easier in identifying AHP. The attributes chosen include:
Hydrophobicity: It is the property of an amino acid to present it as non-polar (chargeless) and, having no charges, they do not have any charge-to-charge communications that enable them to interface with water, and they dissolve in water very rarely; additionally, their substances can easily transverse the cell membrane made of the lipid bilayer. Almost all of the hydrophobic substances are essentially lipophilic.
Hydrophilicity: It is the property of the amino acid to present it as polar (charged); they are lipophobic and cannot transverse the cell membrane.
Molecular Weight: It is calculated as the sum of the atomic weights of each element. For macromolecules such as proteins, methods based on viscosity and light-scattering can find molecular mass when crystallographic data are unavailable. Peptides of high molecular weight are less likely to be taken up by the cell compared to low molecular weight peptides.
pI (Isoelectric pH): Amino acids have the unique property to exist as zwitterions. The pH at which the amino acid has no net charge (or an equal amount of positive and negative charges) is called pK. The amino acid will have nil mobility in an electric field at this pH. The solubility and buffering capacity will be minimum, whereas the perceptibility will be maximum.
pK1: When a particular amino acid is titrated with hydrochloric acid, the pH at which 50% of it exists as zwitterions and the other 50% as cations is called pK1.
pK2: The taken amino acid is titrated with Sodium Hydroxide. The pH at which 50% of the molecules exist as zwitterions and the other 50% as anions is called pK2.
The proposed framework utilized these six attributes and proved maximum accuracy over existing FS and classification algorithms. The proposed RSIHSRR and RSC algorithms are observed as an appropriate framework for the accurate classification system with 96% in predicting anti-HIV-1 peptides.
7. Conclusions
HIV-1-fighting peptides have gained much attention in HIV/AIDS therapeutics. Researchers have adopted computational methods and algorithms in recent years to provide a cost-effective method in screening anti-HIV1 peptides, and feature selection is the vital pre-processing tool in peptide therapy. This study proposed a rough set-based FS technique, hybridized with population-based meta-heuristic algorithms, to classify the peptide sequences and solve dimensionality problems. Experimentation results show that the proposed framework discloses the most satisfactory solutions. It rapidly congregates in the problem space and finds the best reduct, which improves the predictive accuracy of the given dataset. The performance analysis report proved the effectiveness of RSIHSQR and RSIHSRR feature selection and Rough Set Classifier in analyzing peptide sequences to predict its activities.
As a future work, the proposed framework is recommended to any disease prediction analysis on peptide-related studies, such as viruses, bacteria, and microbes. The application of proposed framework can be extended by incorporating time complexity and deep learning algorithms.
Author Contributions
Conceptualization, A.T.A., B.M. and H.H.I.; methodology, B.M., H.H.I., A.T.A., N.A.K. and K.M.F.; software, B.M. and H.H.I.; validation, A.T.A., N.A.K., Y.J. and K.M.F.; formal analysis, B.M., A.T.A., H.H.I., N.A.K. and K.M.F.; investigation, J.L., H.H.I., A.T.A., N.A.K., K.M.F. and Y.J.; resources, J.L., B.M., N.A.K., K.M.F. and Y.J.; data curation, J.L., B.M. and Y.J.; writing—original draft preparation B.M., H.H.I., A.T.A., N.A.K., K.M.F. and J.L.; writing—review and editing B.M., H.H.I., A.T.A., N.A.K., K.M.F., Y.J. and J.L.; visualization, N.A.K., K.M.F. and Y.J.; supervision, H.H.I. and A.T.A. All authors have read and agreed to the published version of the manuscript.
Funding
The work is funded by Prince Sultan, Riyadh, Saudi Arabia.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets underlying this article are accessible through the National Practitioner Data Bank portal [http://npdb-hipdb.com] accessed on 8 February 2022, antiviral peptides (AVPs) prediction web server [http://crdd.osdd.net/servers/avppred]. Accessed on 8 February 2022.
Acknowledgments
The authors would like to acknowledge the support of Prince Sultan University for paying the Article Processing Charges (APC) of this publication. Special acknowledgement to Automated Systems & Soft Computing Lab (ASSCL), Prince Sultan University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Niinomi, M. Titanium Alloys. In Encyclopedia of Biomedical Engineering; Elsevier: Amsterdam, The Netherlands, 2019; Volume 5, pp. 213–224. ISBN 9780128012383. [Google Scholar]
- Tinguely, C.; Schild-Spycher, T.; Bahador, Z.; Gowland, P.; Stolz, M.; Niederhauser, C. Comparison of a conventional HIV 1/2 line immunoassay with a rapid confirmatory HIV 1/2 assay. J. Virol. Methods 2014, 206, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Mehellou, Y.; Clercq, E.D. Twenty-six years of anti-HIV drug discovery: Where do we stand and where do we go? J. Med. Chem. 2021, 53, 521–538. [Google Scholar] [CrossRef]
- Xiao, Y.F.; Jie, M.M.; Li, B.S.; Hu, C.J.; Xie, R.; Tang, B.; Yang, S.M. Peptide-Based Treatment: A Promising Cancer Therapy. J. Immunol. Res. 2015, 2015, 761820. [Google Scholar] [CrossRef] [Green Version]
- Chertov, O.; Zhang, N.; Chen, X.; Oppenheim, J.J.; Lubkowski, J.; McGrath, C.; Ii, R.C.S.; Crise, B.J.; Malyguine, A.; Kutzler, M.A.; et al. Novel Peptides Based on HIV-1 gp120 Sequence with Homology to Chemokines Inhibit HIV Infection in Cell Culture. PLoS ONE 2011, 6, e14474. [Google Scholar] [CrossRef] [PubMed]
- Poorinmohammad, N.; Mohabatkar, H. A Comparison of Different Machine Learning Algorithms for the Prediction of Anti-HIV-1 Peptides Based on Their Sequence-Related Properties. Int. J. Pept. Res. Ther. 2015, 21, 57–62. [Google Scholar] [CrossRef]
- Iqbal, M.J.; Faye, I.; Samir, B.B.; Said, A.M. Efficient Feature Selection and Classification of Protein Sequence Data in Bioinformatics. Sci. World J. 2014, 2014, 173869. [Google Scholar] [CrossRef] [Green Version]
- Al-Betar, M.; Khader, A.; Liao, I. A harmony search with multi-pitch adjusting rate for the university course timetabling. In Recent Advances in Harmony Search Algorithm; Geem, Z.W., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 147–161. [Google Scholar]
- Alia, O.M.; Mandava, R. The variants of the harmony search algorithm: An Overview. Artif. Intell. Rev. 2011, 36, 49–68. [Google Scholar] [CrossRef]
- Zhu, Q.; Tang, X.; Elahi, A. Application of the novel harmony search optimization algorithm for DBSCAN clustering. Expert Syst. Appl. 2021, 178, 115054. [Google Scholar] [CrossRef]
- Manjarres, D.; Landa-Torres, I.; Gil-Lopez, S.; Del Ser, J.; Bilbao, M.; Salcedo-Sanz, S.; Geem, Z.W. A survey on applications of the harmony search algorithm. Eng. Appl. Artif. Intell. 2013, 26, 1818–1831. [Google Scholar] [CrossRef]
- Hasan, B.H.F.; Abu Doush, I.; Al Maghayreh, E.; Alkhateeb, F.; Hamdan, M. Hybridizing Harmony Search algorithm with different mutation operators for continuous problems. Appl. Math. Comput. 2014, 232, 1166–1182. [Google Scholar] [CrossRef]
- Poursalehi, N.; Zolfaghari, A.; Minuchehr, A. Differential harmony search algorithm to optimize PWRs loading pattern. Nucl. Eng. Des. 2013, 257, 161–174. [Google Scholar] [CrossRef]
- Yao, N.; Miao, D.; Pedrycz, W.; Zhang, H.; Zhang, Z. Causality measures and analysis: A rough set framework. Expert Syst. Appl. 2019, 136, 187–200. [Google Scholar] [CrossRef]
- Inbarani, H.H.; Bagyamathi, M.; Azar, A.T. A Novel Hybrid Feature Selection Method Based on Rough Set and Improved Harmony Search. Neural Computing and Applications; Springer: Berlin/Heidelberg, Germany, 2015; Volume 26, pp. 1–22. [Google Scholar]
- Mac Parthalain, N.; Jensen, R. Unsupervised fuzzy-rough set-based dimensionality reduction. Inf. Sci. 2013, 229, 106–121. [Google Scholar] [CrossRef]
- Bagyamathi, M.; Inbarani, H.H. A Novel Hybridized Rough Set and Improved Harmony Search Based Feature Selection for Protein Sequence Classification. Big Data in Complex Systems: Challenges and Opportunities, Studies in Big Data; Springer: Berlin/Heidelberg, Germany, 2015; Volume 9, pp. 173–204. [Google Scholar]
- Cao, Y.; Liu, S.; Zhang, L.; Qin, J.; Wang, J.; Tang, K. Prediction of protein structural class with Rough Sets. BMC Bioinform. 2006, 7, 20. [Google Scholar] [CrossRef] [Green Version]
- Anand, A.; Pugalenthi, G.; Suganthan, P. Predicting protein structural class by SVM with class-wise optimized features and decision probabilities. J. Theor. Biol. 2008, 253, 375–380. [Google Scholar] [CrossRef]
- Velayutham, C.; Thangavel, K. Unsupervised quick reduct algorithm using rough set theory. J. Electron. Sci. Technol. 2011, 9, 193–201. [Google Scholar]
- Bagyamathi, M.; Azar, A.T.; Inbarani, H.H. Hybrid Rough Set with Black Hole Optimization Based Feature Selection Algorithm for Protein Structure Prediction. Int. J. Adv. Intell. Paradig. 2018, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Bagyamathi, M.; Inbarani, H.H. Prediction of Protein Structural Classes by Pseudo Amino Acid Composition Using Improved Harmony Search Relative Reduct Feature Selection and Rough Set Classification Algorithms. Int. J. Invent. Comput. Sci. Eng. 2017, 4, 55–65. [Google Scholar]
- Meher, P.K.; Sahu, T.K.; Saini V and Roa, A.R. Predicting anti-microbial peptides with improved accuracy by incorporating the compositional, physico-chemical and structural features into Chou’s general PseAAC. Sci. Rep. 2017, 7, 42362. [Google Scholar] [CrossRef]
- Azar, A.T.; Kumar, S.S.; Inbarani, H.H.; Hassanien, A.E. Pessimistic multi-granulation rough set-based classification for heart valve disease diagnosis. Int. J. Modeling Identif. Control. 2016, 26, 42–51. [Google Scholar] [CrossRef]
- Zare, M.; Mohabatkar, H.; Faramarzi, F.K.; Beigi, M.M.; Behbahani, M. Using Chou’s Pseudo Amino Acid Composition and Machine Learning Method to predict the Antiviral Peptides. Open Bioinform. J. 2015, 9, 13–19. [Google Scholar] [CrossRef]
- Bagyamathi, M.; Inbarani, H.H. Feature Selection using Improved Harmony Search Hybridized with Relative Reduct for Medical Data Classification. Int. J. Appl. Eng. Res. (IJAER) 2015, 10, 19476–19480. [Google Scholar]
- Bagyamathi, M.; Inbarani, H.H. Feature Selection using Relative Reduct hybridized with Improved Harmony Search for Protein Sequence Classification. Int. J. Trend Res. Dev. 2015. Available online: http://www.ijtrd.com/papers/IJTRD1328.pdf (accessed on 10 January 2022).
- Barrett, R.; Jiang, S.; White, A.D. Classifying antimicrobial and multifunctional peptides with Bayesian network models. Pept. Sci. 2018, 110, e24079. [Google Scholar] [CrossRef] [Green Version]
- Tantisatirapong, S.; Davies, N.P.; Rodriguez, D.; Abernethy, L.; Auer, D.P.; Clark, C.A.; Arvanitis, T.N. Magnetic Resonance Texture Analysis: Optimal Feature Selection in Classifying Child Brain Tumors. In Proceedings of the XIII Mediterranean Conference on Medical and Biological Engineering and Computing, Seville, Spain, 25–28 September 2013; pp. 309–312. [Google Scholar]
- Hajisharifi, Z.; Piryaiee, M.; Beigi, M.M.; Behbahani, M.; Mohabatkar, H. Predicting anticancer peptides with Chou’s pseudo amino acid composition and investigating their mutagenicity via Ames test. J. Theor. Biol. 2014, 341, 34–40. [Google Scholar] [CrossRef]
- Inbarani, H.H.; Azar, A.T.; Jothi, G. Supervised hybrid feature selection based on PSO and rough sets for medical diagnosis. Comput. Methods Programs Biomed. 2014, 113, 175–185. [Google Scholar] [CrossRef]
- Azar, A.T. Neuro-fuzzy feature selection approach based on linguistic hedges for medical diagnosis. Int. J. Model. Identif Control. (IJMIC) 2014, 22, 195–206. [Google Scholar] [CrossRef]
- Jothi, G.; Inbarani, H.H.; Azar, A.T. Hybrid tolerance-PSO based supervised feature selection for digital mammogram images. Int. J. Fuzzy Syst Appl (IJFSA) 2013, 3, 15–30. [Google Scholar]
- Qureshi, A.; Thakur, N.; Kumar, M. HIPdb: A database of experimentally validated HIV inhibiting peptides. PLoS ONE 2013, 8, e54908. [Google Scholar] [CrossRef] [Green Version]
- Du, P.; Wang, X.; Xu, C.; Gao, Y. PseAAC-Builder: A Cross-Platform Stand-Alone Program for Generating Various Special Chou’s Pseudo-Amino Acid Compositions. Anal. Biochem. 2012, 425, 117–119. [Google Scholar] [CrossRef]
- Khosravian, M.; Faramarzi, F.K.; Beigi, M.M.; Behbahani, M.; Mohabatkar, H. Predicting Antibacterial Peptides by the Concept of Chou’s Pseudo-Amino Acid Composition and Machine Learning Methods. Protein Pept. Lett. 2013, 20, 180–186. [Google Scholar] [CrossRef]
- Beniwal, S.; Arora, J. Classification and feature selection techniques in data Mining. Int. J. Eng. Research Technol. 2012, 1, 2278–2284. [Google Scholar]
- Kotsiantis, S.; Kanellopoulos, D. Discretization Techniques: A recent survey. GESTS Int. Trans. Comput. Sci. Eng. 2006, 32, 47–58. [Google Scholar]
- Ali, R.; Siddiqi, M.H.; Lee, S. Rough set-based approaches for discretization: A compact review. Artif. Intell. Rev. 2015, 44, 235–263. [Google Scholar] [CrossRef]
- Tsoukalas, A.; Parpas, P.; Rustem, B. A smoothing algorithm for finite min–max–min problems. Optim. Lett. 2008, 3, 49–62. [Google Scholar] [CrossRef]
- Sathishkumar, E.N.; Thangavel, K.; Nishama, A. Comparative analysis of discretization methods for gene selection of breast cancer gene expression data. In Computational Intelligence, Cyber Security and Computational Models; Springer: Coimbatore, India, 2014; pp. 373–378. [Google Scholar]
- Anaraki, J.R.; Eftekhari, M. Rough set based feature selection: A Review Fifth conference on information and knowledge technology (IKT). IEEE 2013, 2013, 301–306. [Google Scholar] [CrossRef]
- Bagyamathi, M.; Inbarani, H.H. Prediction of Protein Structural Classes using Rough Set based Feature Selection and Classification Framework. J. Recent Res. Eng. Technol. 2017, 4, 1–9. [Google Scholar]
- Geem, Z.W. Particle-Swarm Harmony Search for Water Network Design. Eng. Optim. 2009, 41, 297–311. [Google Scholar] [CrossRef]
- Inbarani, H.H.; Banu, P.K.N.; Azar, A.T. Feature selection using swarm based relative reduct technique for fetal heart rate. Neural Comput. Appl. 2014, 25, 793–806. [Google Scholar] [CrossRef]
- Kumar, S.U.; Inbarani, H.H.; Kumar, S.S. Improved Bijective-Soft-Set-Based Classification for Gene Expression Data. Computational Intelligence, Cyber Security and Computational Models. In Advances in Intelligent Systems and Computing; Springer: Berlin/Heidelberg, Germany, 2014; Volume 246, pp. 127–132. [Google Scholar]
- Kumar, S.U.; Inbarani, H.H. PSO-based feature selection and neighborhood rough set-based classification for BCI multiclass motor imagery task. Neural Comput. Appl. 2016, 28, 3239–3258. [Google Scholar] [CrossRef]
- Hall, M.; Frank, E.; Holmes, G.; Pfahringer, G.; Reutemann, P.; Witten, I.H. The WEKA data mining software: An update. ACM SIGKDD Explor. Newsl. 2009, 11, 10–18. [Google Scholar] [CrossRef]
- Witten, I.H.; Frank, E.; Hall, M.A.; Pal, C.J. Data Mining: Practical Machine Learning Tools and Techniques; Morgan Kaufmann Publishers: Amsterdam, The Netherlands, 2016; ISBN 978-0-12-804291-5. [Google Scholar]
- Thakur, N.; Qureshi, A.; Kumar, M. AVPpred: Collection and prediction of highly effective antiviral peptides. Nucleic Acids Res. 2012, 40, W199–W204. [Google Scholar] [CrossRef] [Green Version]
- Salam, M.A.; Azar, A.T.; Elgendy, M.S.; Fouad, K.M. The Effect of Different Dimensionality Reduction Techniques on Machine Learning Overfitting Problem. Int. J. Adv. Comput. Sci. Appl. 2021, 12, 641–655. [Google Scholar] [CrossRef]
- Azar, A.T.; Anter, A.M.; Fouad, K.M. Intelligent system for feature selection based on rough set and chaotic binary grey wolf optimization. Int. J. Comput. Appl. Technol. 2020, 63, 4–24. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).