Development of a Web Application for the Detection of Coronary Artery Calcium from Computed Tomography
Abstract
1. Introduction
2. Materials and Methods
2.1. Calculation of the Coronary Calcium Score
2.2. Image Dataset
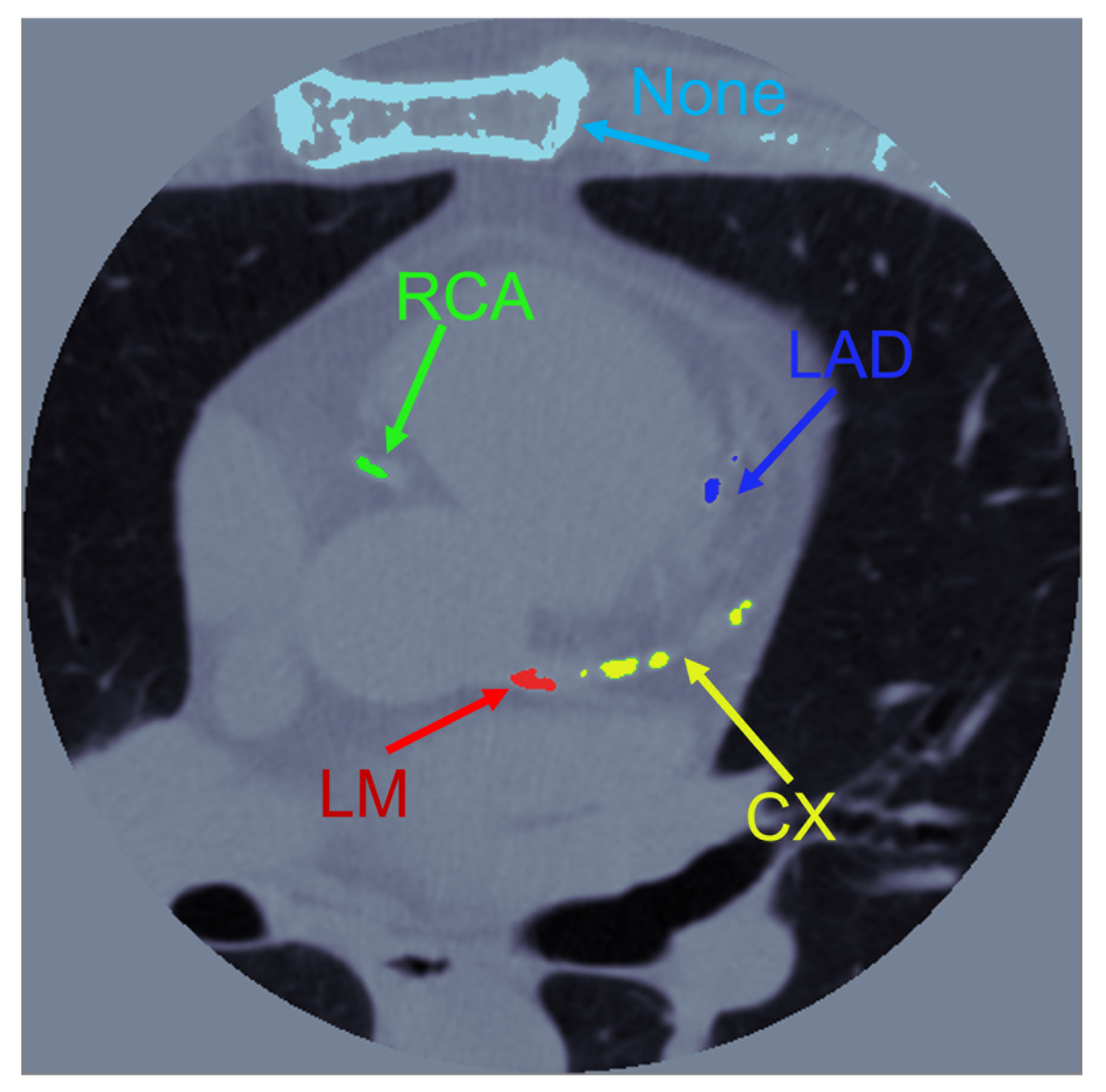
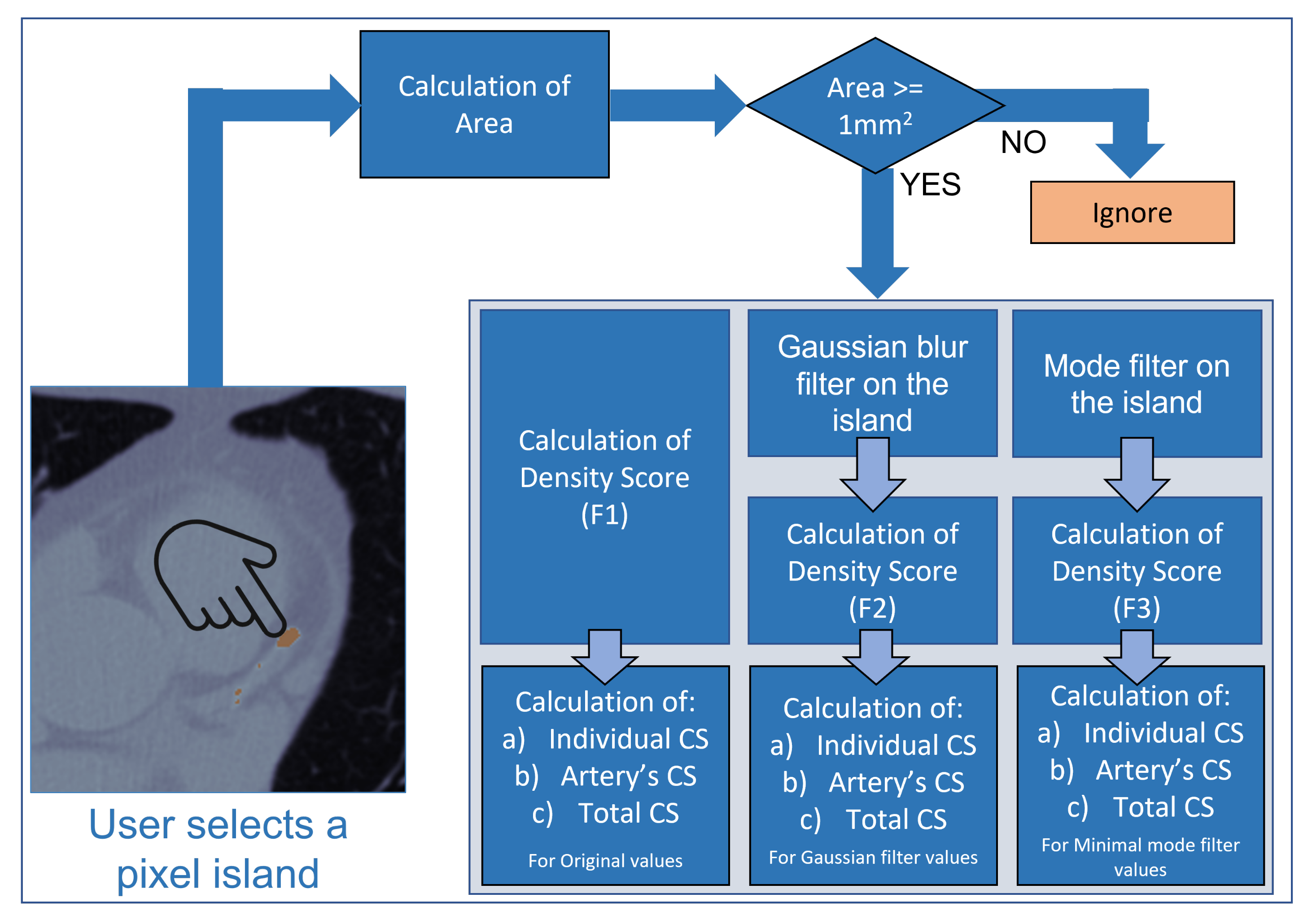
2.3. Image Processing
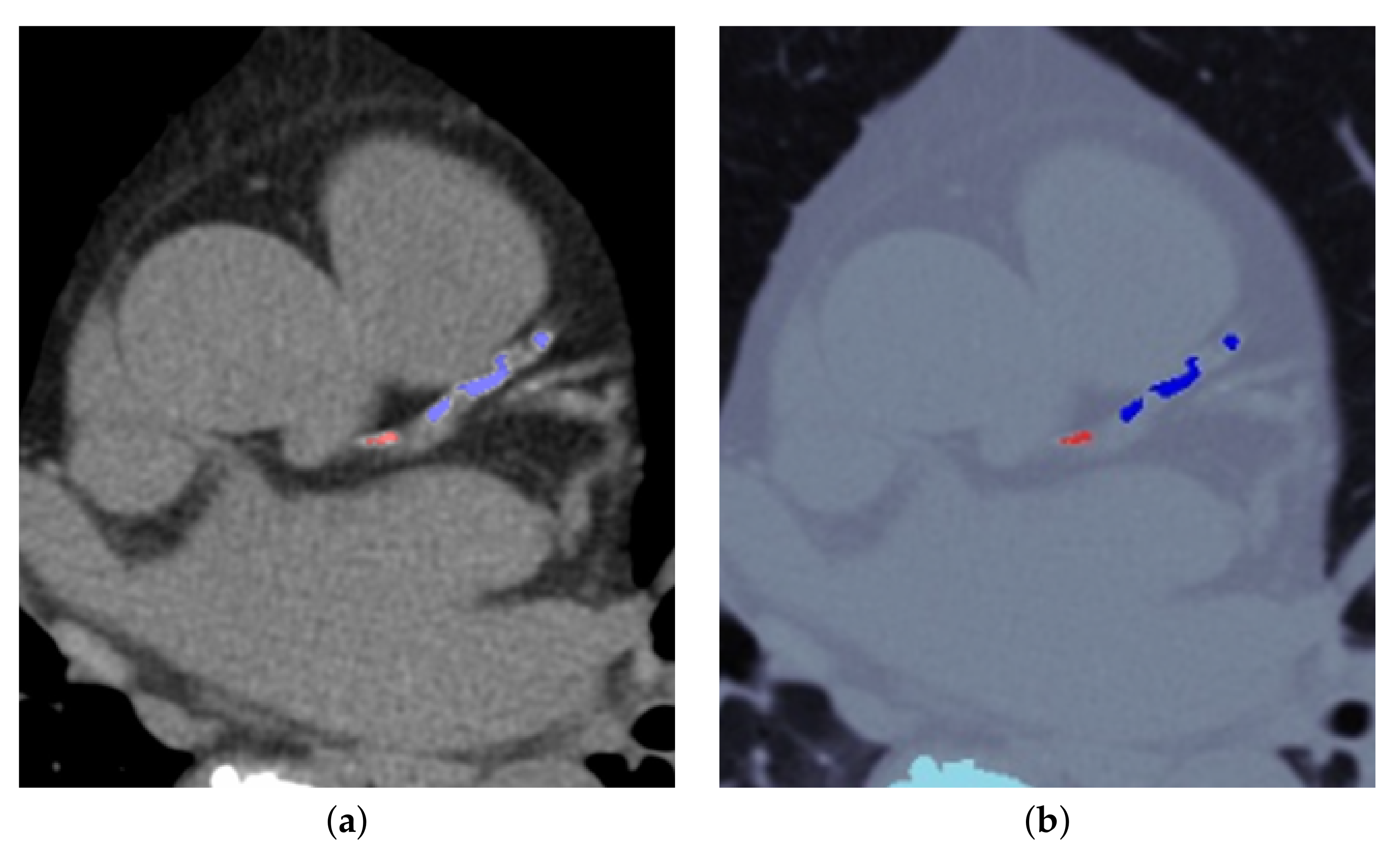
2.4. Validation of Image Processing Calculation Results
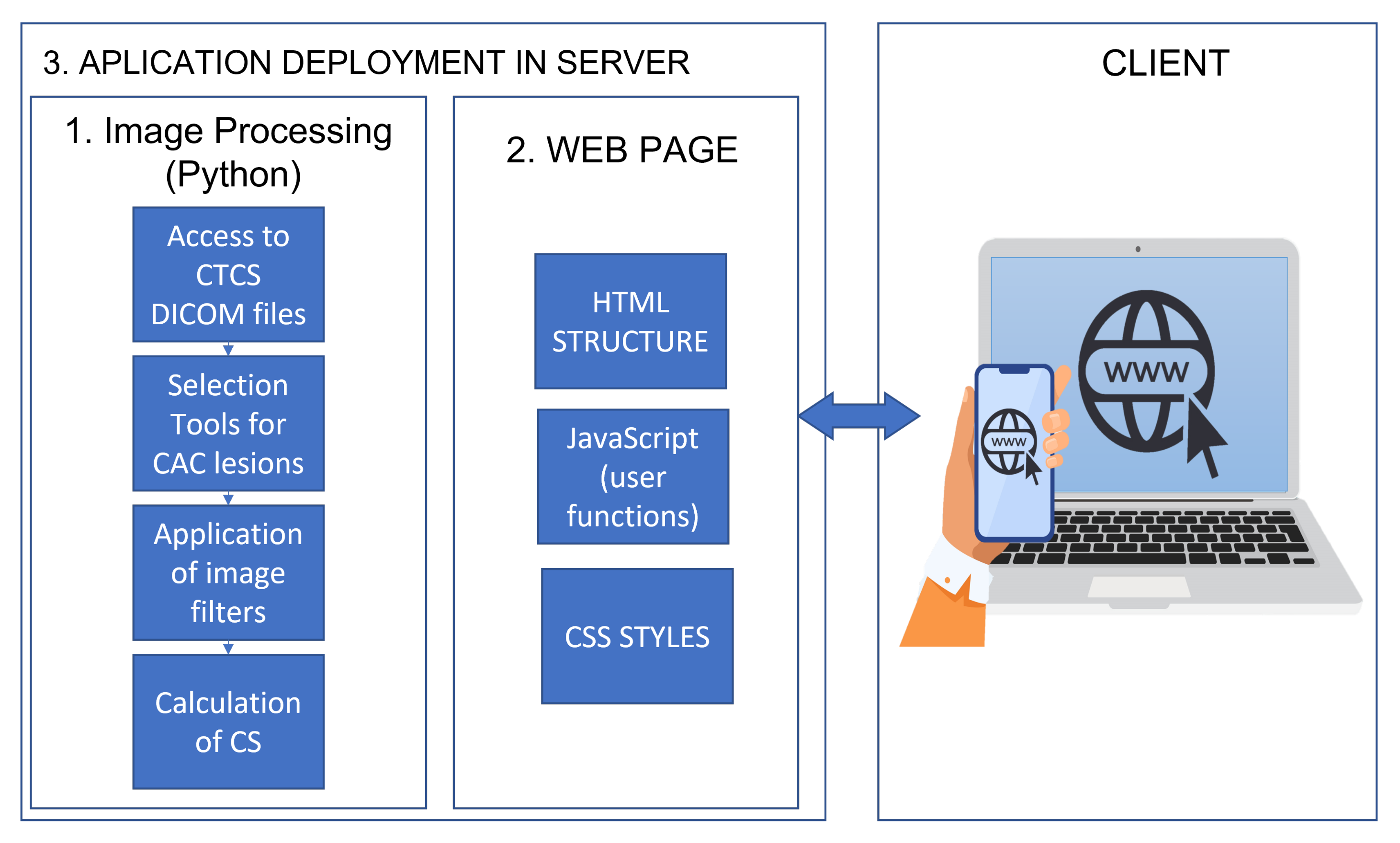
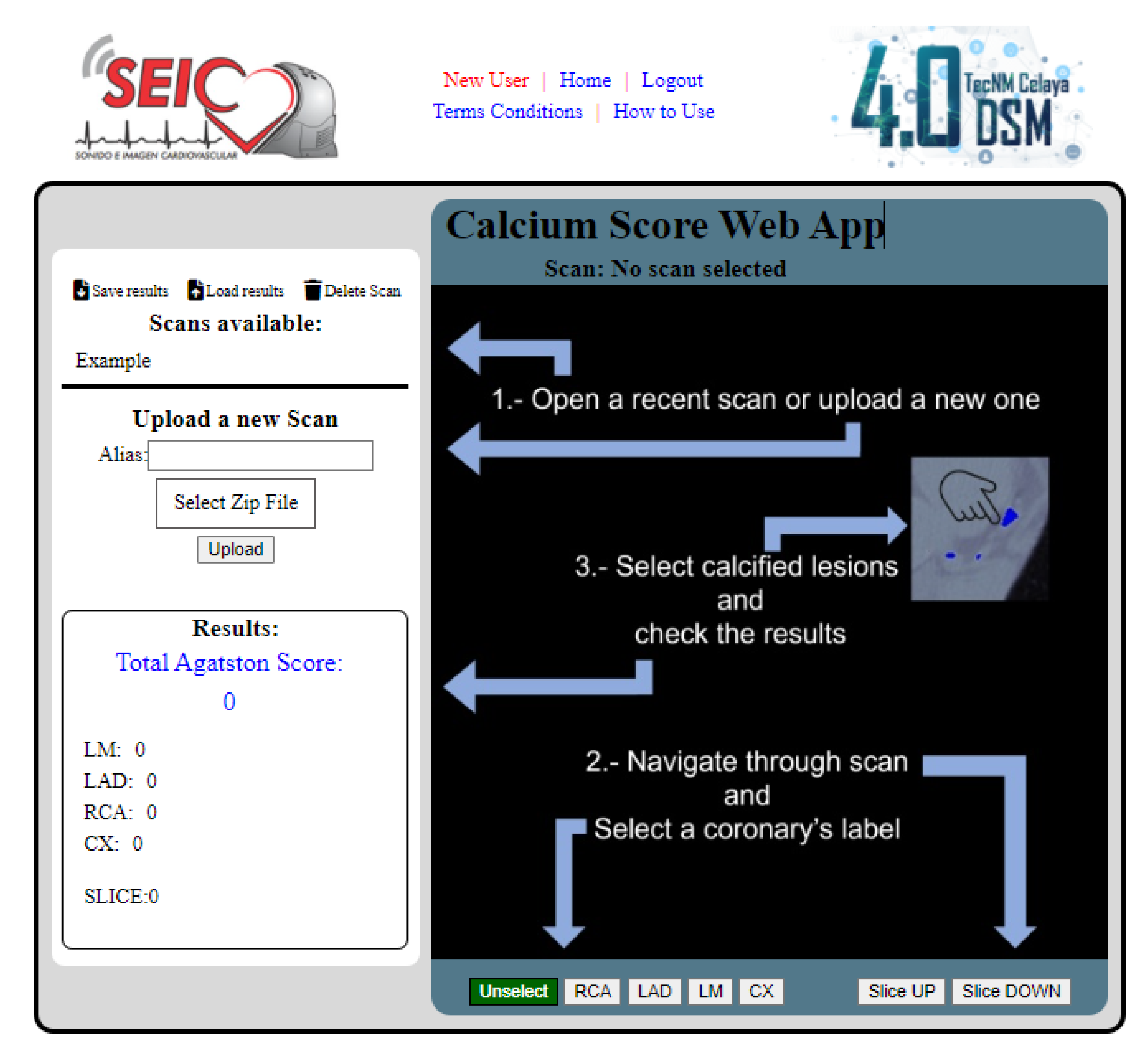
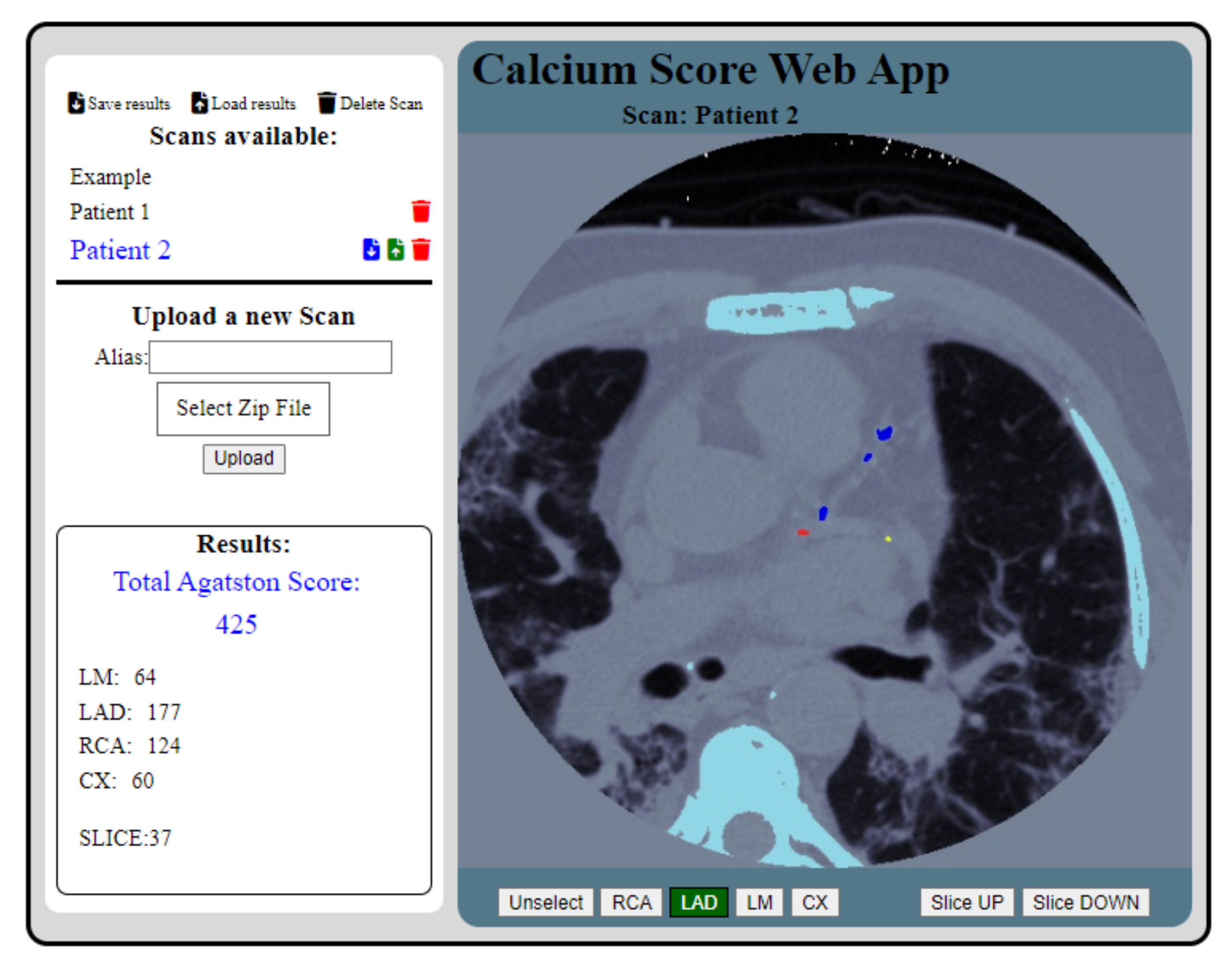
2.5. Programming the Web Page and HTTP Requests
2.6. Setting Up the Application on the Internet
3. Results
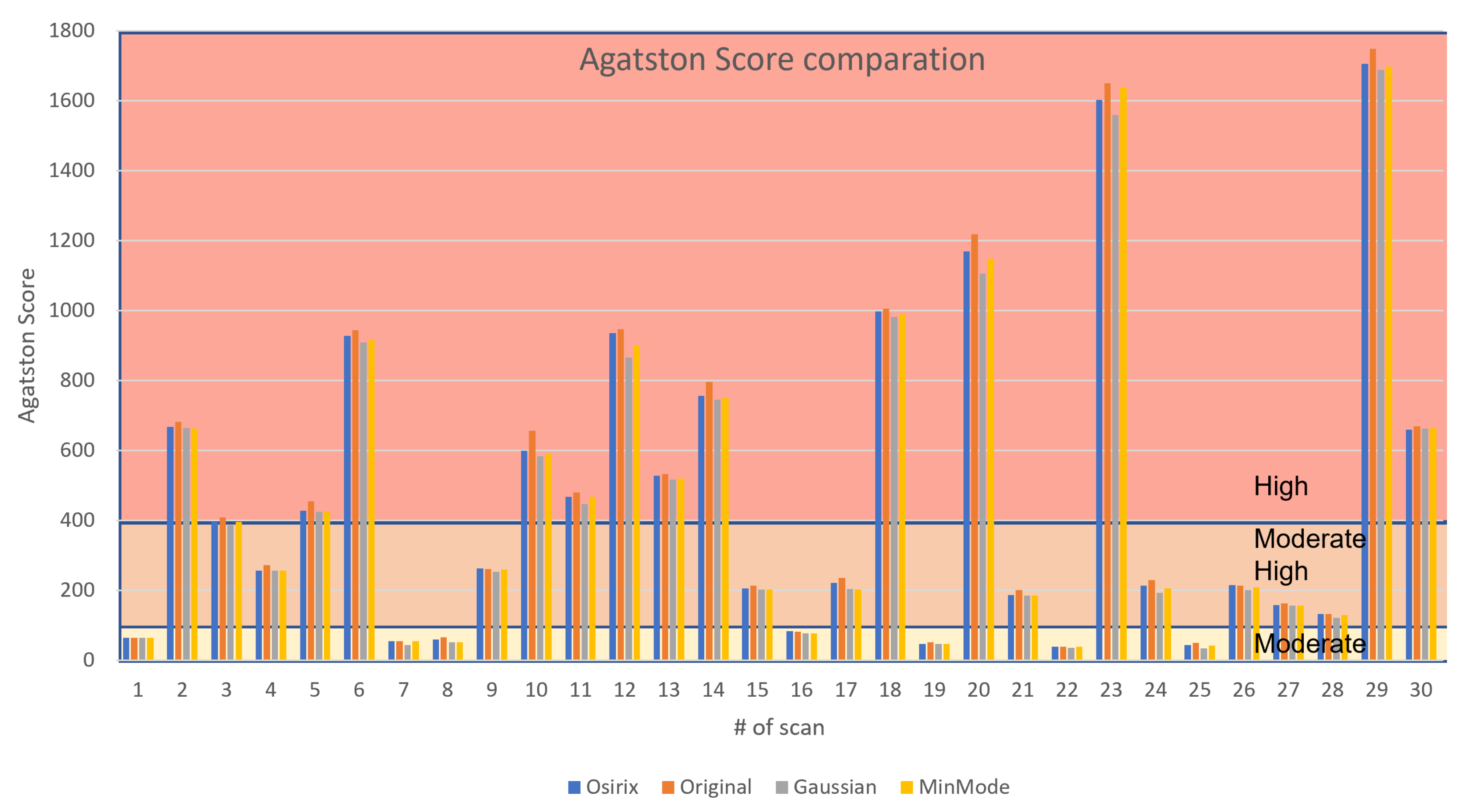
3.1. CS Calculation and Validation
3.2. Access to the Web Application
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Cardiovascular Diseases. Fact Sheet. 2021. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 10 July 2022).
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Patel, J.; Blaha, M.J.; McEvoy, J.W.; Qadir, S.; Tota-Maharaj, R.; Shaw, L.J.; Rumberger, J.A.; Callister, T.Q.; Berman, D.S.; Min, J.K.; et al. All-cause mortality in asymptomatic persons with extensive Agatston scores above 1000. J. Cardiovasc. Comput. Tomogr. 2014, 8, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Hecht, H.S. Coronary Artery Calcium and Prevention Guidelines. JACC Cardiovasc. Imaging 2020, 13, 1187–1190. [Google Scholar] [CrossRef] [PubMed]
- Greenland, P.; LaBree, L.; Azen, S.P.; Doherty, T.M.; Detrano, R.C. Coronary Artery Calcium Score Combined With Framingham Score for Risk Prediction in Asymptomatic Individuals. JAMA 2004, 291, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.J.; Bindeman, J.; Feuerstein, I.; Cao, F.; Brazaitis, M.; O’Malley, P.G. Coronary Calcium Independently Predicts Incident Premature Coronary Heart Disease Over Measured Cardiovascular Risk Factors: Mean Three-Year Outcomes in the Prospective Army Coronary Calcium (PACC) Project. J. Am. Coll. Cardiol. 2005, 46, 807–814. [Google Scholar] [CrossRef]
- Shaw, L.J.; Raggi, P.; Schisterman, E.; Berman, D.S.; Callister, T.Q. Prognostic Value of Cardiac Risk Factors and Coronary Artery Calcium Screening for All-Cause Mortality. Radiology 2003, 228, 826–833. [Google Scholar] [CrossRef]
- Isgum, I.; Bartels-Rutten, A.; Prokop, M.; Ginneken, B. Detection of coronary calcifications from computed tomography scans for automated risk assessment of coronary artery disease. Med. Phys. 2007, 34, 1450–1461. [Google Scholar] [CrossRef]
- Agatston, A.S.; Janowitz, W.R.; Hildner, F.J.; Zusmer, N.R.; Viamonte, M.; Detrano, R. Quantification of coronary artery calcium using ultrafast computed tomography. J. Am. Coll. Cardiol. 1990, 15, 827–832. [Google Scholar] [CrossRef]
- Palomares, J.F.; Evangelista, A. Cuantificación del calcio aórtico y arteriosclerosis vascular en individuos asintomáticos: más allá de las arterias coronarias. Rev. Esp. Cardiol. 2016, 69, 813–816. [Google Scholar] [CrossRef]
- Yoon, W.J.; Crisostomo, P.; Halandras, P.; Bechara, C.F.; Aulivola, B. The use of the Agatston calcium score in predicting carotid plaque vulnerability. Ann. Vasc. Surg. 2019, 54, 22–26. [Google Scholar] [CrossRef]
- 3D Slicer Image Computing Platform. Available online: https://www.slicer.org/ (accessed on 10 July 2022).
- Kay, F.U.; Abbara, S.; Joshi, P.H.; Garg, S.; Khera, A.; Peshock, R.M. Identification of high-risk left ventricular hypertrophy on calcium scoring cardiac computed tomography scans: validation in the DHS. Circ.Cardiovasc. Imaging 2020, 13, e009678. [Google Scholar] [CrossRef] [PubMed]
- Foldyna, B.; Eslami, P.; Scholtz, J.E.; Baltrusaitis, K.; Lu, M.T.; Massaro, J.M.; D’Agostino, R.B.; Ferencik, M.; Aerts, H.J.; O’Donnell, C.J.; et al. Density and morphology of coronary artery calcium for the prediction of cardiovascular events: insights from the Framingham Heart Study. Eur. Radiol. 2019, 29, 6140–6148. [Google Scholar] [CrossRef] [PubMed]
- Dransfield, M.T.; Huang, F.; Nath, H.; Singh, S.P.; Bailey, W.C.; Washko, G.R. CT emphysema predicts thoracic aortic calcification in smokers with and without COPD. COPD. 2010, 7, 404–410. [Google Scholar] [CrossRef]
- Image Processing and Analysis in Java. Available online: https://imagej.nih.gov/ij/index.html (accessed on 10 July 2022).
- Sun, Z.; Ng, C.K. High calcium scores in coronary CT angiography: effects of image post-processing on visualization and measurement of coronary lumen diameter. J. Med. Imaging. Health. Inform. 2015, 5, 110–116. [Google Scholar] [CrossRef]
- Phillips-Eakley, A.K.; McKenney-Drake, M.L.; Bahls, M.; Newcomer, S.C.; Radcliffe, J.S.; Wastney, M.E.; Van Alstine, W.G.; Jackson, G.; Alloosh, M.; Martin, B.R.; et al. Effect of high-calcium diet on coronary artery disease in Ossabaw miniature swine with metabolic syndrome. J. Am. Heart. Assoc. 2015, 4, e001620. [Google Scholar] [CrossRef]
- Cahalane, R.M.; Broderick, S.P.; Kavanagh, E.G.; Moloney, M.A.; Mongrain, R.; Purtill, H.; Walsh, M.T.; O’Brien, J.M. Comparative analysis of calcification parameters with Agatston Score approximations for ex vivo atherosclerotic lesions. J. Cardiovasc. Comput. Tomogr. 2020, 14, 20–26. [Google Scholar] [CrossRef]
- Bos, D.; Ikram, M.A.; Elias-Smale, S.E.; Krestin, G.P.; Hofman, A.; Witteman, J.C.; van der Lugt, A.; Vernooij, M.W. Calcification in major vessel beds relates to vascular brain disease. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2331–2337. [Google Scholar] [CrossRef]
- OpenCV. Available online: https://opencv.org/ (accessed on 10 July 2022).
- Durlak, F.; Wels, M.; Schwemmer, C.; Sühling, M.; Steidl, S.; Maier, A. Growing a random forest with fuzzy spatial features for fully automatic artery-specific coronary calcium scoring. In International Workshop on Machine Learning in Medical Imaging; Springer: Berlin/Heidelberg, Germany, 2017; pp. 27–35. [Google Scholar] [CrossRef]
- Toji, B.; Ohmiya, J.; Kondo, S.; Ishikawa, K.; Yamamoto, M. Fully Automatic Extraction of Carotid Artery Contours from Ultrasound Images. IEICE Trans. Inf. Syst. 2014, 97, 2493–2500. [Google Scholar] [CrossRef]
- Mirunalini, P.; Aravindan, C.; Nambi, A.T.; Poorvaja, S.; Priya, V.P. Segmentation of Coronary Arteries from CTA axial slices using Deep Learning techniques. In Proceedings of the TENCON 2019—2019 IEEE Region 10 Conference (TENCON), Kochi, India, 17–20 October 2019; pp. 2074–2080. [Google Scholar] [CrossRef]
- De Vos, B.D.; Wolterink, J.M.; Leiner, T.; de Jong, P.A.; Lessmann, N.; Išgum, I. Direct automatic coronary calcium scoring in cardiac and chest CT. IEEE Trans. Med. Imaging 2019, 38, 2127–2138. [Google Scholar] [CrossRef]
- Garousi, V.; Mesbah, A.; Betin-Can, A.; Mirshokraie, S. A systematic mapping study of web application testing. Inf. Softw. Technol. 2013, 55, 1374–1396. [Google Scholar] [CrossRef]
- Verber, D.; Novak, D.; Borovič, M.; Dugonik, J.; Flisar, D. EQUIDopa: A responsive web application for the levodopa equivalent dose calculator. Comput. Methods Programs Biomed. 2020, 196, 105633. [Google Scholar] [CrossRef]
- Al-Waisy, A.S.; Alruban, A.; Al-Fahdawi, S.; Qahwaji, R.; Ponirakis, G.; Malik, R.A.; Mohammed, M.A.; Kadry, S. CellsDeepNet: A Novel Deep Learning-Based Web Application for the Automated Morphometric Analysis of Corneal Endothelial Cells. Mathematics 2022, 10, 320. [Google Scholar] [CrossRef]
- Pemmaraju, R.; Minahan, R.; Wang, E.; Schadl, K.; Daldrup-Link, H.; Habte, F. Web-Based Application for Biomedical Image Registry, Analysis, and Translation (BiRAT). Tomography 2022, 8, 1453–1462. [Google Scholar] [CrossRef]
- Mora-Aguilera, G.; Martínez-Bustamante, V.; Acevedo-Sánchez, G.; Coria-Contreras, J.J.; Guzmán-Hernández, E.; Flores-Colorado, O.E.; Mendoza-Ramos, C.; Hernández-Nava, G.; Álvarez-Maya, I.; Gutiérrez-Espinosa, M.A.; et al. Surveillance web system and mouthwash-saliva qPCR for labor ambulatory SARS-CoV-2 detection and prevention. Int. J. Environ. Res. Public Health 2022, 19, 1271. [Google Scholar] [CrossRef]
- Villavicencio, C.N.; Macrohon, J.J.; Inbaraj, X.A.; Jeng, J.H.; Hsieh, J.G. Development of a Machine Learning Based Web Application for Early Diagnosis of COVID-19 Based on Symptoms. Diagnostics 2022, 12, 821. [Google Scholar] [CrossRef]
- Lanera, C.; Azzolina, D.; Pirotti, F.; Prosepe, I.; Lorenzoni, G.; Berchialla, P.; Gregori, D. A Web-Based Application to Monitor and Inform about the COVID-19 Outbreak in Italy: The {COVID-19ita} Initiative. Healthcare 2022, 10, 473. [Google Scholar] [CrossRef]
- Nawi, N.A.M.M.; Sapiai, N.S.; Ghazali, S.A.M.; Rusok, N.H.M.; Zulkifli, F.Z.; Mazlan, F.M. Developing an E-College Monitoring System as a Web-Based Monitoring Tool Application. Proceedings 2022, 82, 25. [Google Scholar] [CrossRef]
- Mejía, S.; Muñoz, I.C.; Serna, L.Y.; Sarmiento, C.A.; Bravo, C.L.; Hernández, A.M. Web Applications for Teaching the Respiratory System: Content Validation. Appl. Sci. 2022, 12, 4289. [Google Scholar] [CrossRef]
- Udias, A.; Pistocchi, A.; Vigiak, O.; Grizzetti, B.; Bouraoui, F.; Alfaro, C. ESPRES: A web application for interactive analysis of multiple pressures in aquatic ecosystems. Sci. Total Environ. 2020, 744, 140792. [Google Scholar] [CrossRef] [PubMed]
- Andreo-Martínez, P.; Ortiz-Martínez, V.M.; Muñoz, A.; Menchón-Sánchez, P.; Quesada-Medina, J. A web application to estimate the carbon footprint of constructed wetlands. Environ. Model Softw. 2021, 135, 104898. [Google Scholar] [CrossRef]
- Cochard, T.; Branger, M.; Supply, P.; Sreevatsan, S.; Biet, F. MAC-INMV-SSR: a web application dedicated to genotyping members of Mycobacterium avium complex (MAC) including Mycobacterium avium subsp. paratuberculosis strains. Infect. Genet. Evol. 2020, 77, 104075. [Google Scholar] [CrossRef] [PubMed]
- Karatzas, E.; Baltoumas, F.A.; Kasionis, I.; Sanoudou, D.; Eliopoulos, A.G.; Theodosiou, T.; Iliopoulos, I.; Pavlopoulos, G.A. Darling: A Web Application for Detecting Disease-Related Biomedical Entity Associations with Literature Mining. Biomolecules 2022, 12, 520. [Google Scholar] [CrossRef] [PubMed]
- Bianchetti, G.; Abeltino, A.; Serantoni, C.; Ardito, F.; Malta, D.; De Spirito, M.; Maulucci, G. Personalized Self-Monitoring of Energy Balance through Integration in a Web-Application of Dietary, Anthropometric, and Physical Activity Data. J. Pers. Med. 2022, 12, 568. [Google Scholar] [CrossRef]
- Cubillas, J.J.; Ramos, M.I.; Jurado, J.M.; Feito, F.R. A Machine Learning Model for Early Prediction of Crop Yield, Nested in a Web Application in the Cloud: A Case Study in an Olive Grove in Southern Spain. Agriculture 2022, 12, 1345. [Google Scholar] [CrossRef]
- Paraschiv, L.S.; Acomi, N.; Serban, A.; Paraschiv, S. A web application for analysis of heat transfer through building walls and calculation of optimal insulation thickness. Energy Rep. 2020, 6, 343–353. [Google Scholar] [CrossRef]
- Mason, D. SU-E-T-33: pydicom: An open source DICOM library. Med. Phys. 2011, 38, 3493. [Google Scholar] [CrossRef]
- Pulli, K.; Baksheev, A.; Kornyakov, K.; Eruhimov, V. Real-time computer vision with OpenCV. Commun. ACM 2012, 55, 61–69. [Google Scholar] [CrossRef]
- Nelson, J.C.; Kronmal, R.A.; Carr, J.J.; McNitt-Gray, M.F.; Wong, N.D.; Loria, C.M.; Goldin, J.G.; Williams, O.D.; Detrano, R. Measuring coronary calcium on CT images adjusted for attenuation differences. Radiology 2005, 235, 403–414. [Google Scholar] [CrossRef]
- McRoy, C.; Patel, L.; Gaddam, D.S.; Rothenberg, S.; Herring, A.; Hamm, J.; Chelala, L.; Weinstein, J.; Smith, E.; Awan, O. Radiology education in the time of COVID-19: a novel distance learning workstation experience for residents. Acad. Radiol. 2020, 27, 1467–1474. [Google Scholar] [CrossRef]
- Awan, O.A.; Klein, J.S. Stepping Up to the Challenge: Overcoming Barriers to Radiology Training in the United States During COVID-19. Can. Assoc. Radiol. J. 2021, 72, 11–12. [Google Scholar] [CrossRef]
- Sugi, M.D.; Kennedy, T.A.; Shah, V.; Hartung, M.P. Bridging the gap: interactive, case-based learning in radiology education. Abdom. Radiol. 2021, 46, 5503–5508. [Google Scholar] [CrossRef] [PubMed]
- Aiello, M.; Esposito, G.; Pagliari, G.; Borrelli, P.; Brancato, V.; Salvatore, M. How does DICOM support big data management? Investigating its use in medical imaging community. Insights Imaging 2021, 12, 1–21. [Google Scholar] [CrossRef] [PubMed]
- De Ronde, M.W.; Khoshiwal, A.; Planken, R.N.; Boekholdt, S.M.; Biemond, M.; Budoff, M.J.; Cooil, B.; Lotufo, P.A.; Bensenor, I.M.; Ohmoto-Sekine, Y.; et al. A pooled-analysis of age and sex based coronary artery calcium scores percentiles. J. Cardiovasc. Comput. Tomogr. 2020, 14, 414–420. [Google Scholar] [CrossRef] [PubMed]
- McClelland, R.L.; Chung, H.; Detrano, R.; Post, W.; Kronmal, R.A. Distribution of coronary artery calcium by race, gender, and age: results from the Multi-Ethnic Study of Atherosclerosis (MESA). Circulation 2006, 113, 30–37. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aguilera-Alvarez, J.; Martínez-Nolasco, J.; Olmos-Temois, S.; Padilla-Medina, J.; Sámano-Ortega, V.; Bravo-Sanchez, M. Development of a Web Application for the Detection of Coronary Artery Calcium from Computed Tomography. Appl. Sci. 2022, 12, 12281. https://doi.org/10.3390/app122312281
Aguilera-Alvarez J, Martínez-Nolasco J, Olmos-Temois S, Padilla-Medina J, Sámano-Ortega V, Bravo-Sanchez M. Development of a Web Application for the Detection of Coronary Artery Calcium from Computed Tomography. Applied Sciences. 2022; 12(23):12281. https://doi.org/10.3390/app122312281
Chicago/Turabian StyleAguilera-Alvarez, Juan, Juan Martínez-Nolasco, Sergio Olmos-Temois, José Padilla-Medina, Víctor Sámano-Ortega, and Micael Bravo-Sanchez. 2022. "Development of a Web Application for the Detection of Coronary Artery Calcium from Computed Tomography" Applied Sciences 12, no. 23: 12281. https://doi.org/10.3390/app122312281
APA StyleAguilera-Alvarez, J., Martínez-Nolasco, J., Olmos-Temois, S., Padilla-Medina, J., Sámano-Ortega, V., & Bravo-Sanchez, M. (2022). Development of a Web Application for the Detection of Coronary Artery Calcium from Computed Tomography. Applied Sciences, 12(23), 12281. https://doi.org/10.3390/app122312281







