Guided Random Mask: Adaptively Regularizing Deep Neural Networks for Medical Image Analysis by Potential Lesions
Abstract
:1. Introduction
- (i)
- We found that the DNNs may bias toward the most prominent features and ignore the sub-clinical ones when the input image contains complicated lesions.
- (ii)
- A parameter-free data augmentation method called GRM is proposed, which utilizes visual interpretation of the prediction result to regularize the training of DNNs adaptively.
- (iii)
- Visual interpretation demonstrates that DNNs coupled with GRM can more effectively utilize the contextual information than the vanilla models.
- (iv)
- Ablation studies on multiple datasets, including OCT, X-ray, and ultrasound images, empirically show that the GRM substantially surpasses the benchmark method on various tasks.
2. Related Works
2.1. DNNs in Medical Image Analysis
2.2. Augmentation Methods for Training DCNNs
3. Data and Methodology
3.1. Data
3.2. Methodology
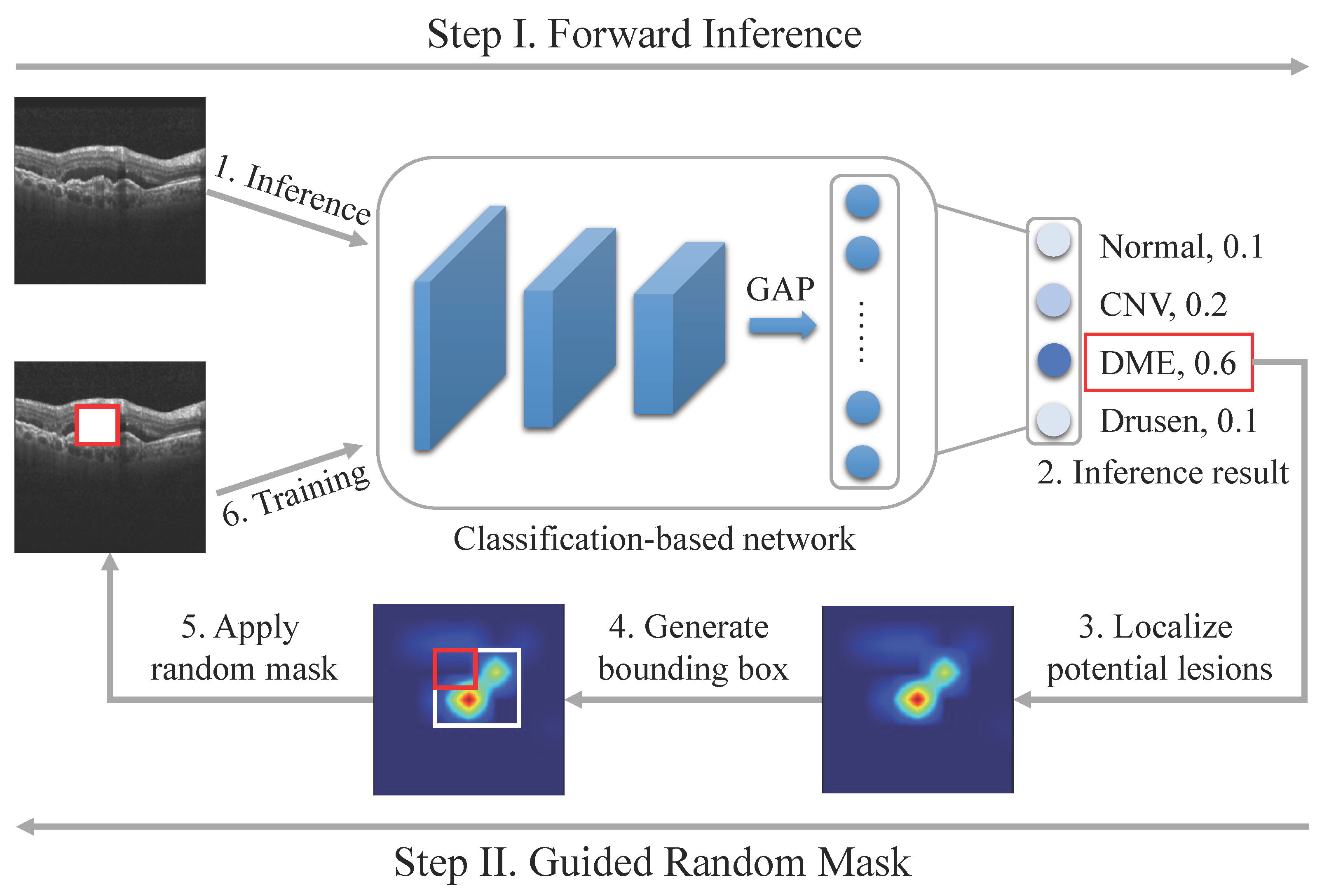
3.2.1. Localizing Potential Lesions
3.2.2. Guided Random Mask
| Algorithm 1: Algorithm of the proposed GRM method for the classification task. |
 |
4. Experimental Setup and Results
4.1. Experimental Setup
4.2. Ablation Studies of GRM
4.3. Comparison between GRM with Other Augmentation Methods
4.4. Visualization Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An Image is Worth 16 × 16 Words: Transformers for Image Recognition at Scale. In Proceedings of the International Conference on Learning Representations, Virtual, 26 April–1 May 2020. [Google Scholar]
- Hu, J.; Chen, Y.; Zhong, J.; Ju, R.; Yi, Z. Automated analysis for retinopathy of prematurity by deep neural networks. IEEE Trans. Med. Imaging 2018, 38, 269–279. [Google Scholar] [CrossRef] [PubMed]
- Kermany, D.S.; Goldbaum, M.; Cai, W.; Valentim, C.C.; Liang, H.; Baxter, S.L.; McKeown, A.; Yang, G.; Wu, X.; Yan, F.; et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell 2018, 172, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Chen, Y.; Yi, Z. Automated segmentation of macular edema in OCT using deep neural networks. Med. Image Anal. 2019, 55, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, L.; Shu, X.; Lv, Q.; Yi, Z. An end-to-end mammogram diagnosis: A new multi-instance and multiscale method based on single-image feature. IEEE Trans. Cogn. Dev. Syst. 2020, 13, 535–545. [Google Scholar] [CrossRef]
- Anthimopoulos, M.; Christodoulidis, S.; Ebner, L.; Christe, A.; Mougiakakou, S. Lung pattern classification for interstitial lung diseases using a deep convolutional neural network. IEEE Trans. Med. Imaging 2016, 35, 1207–1216. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, Y.; Zhao, D.; Liu, B.; Lai, Z.; Chen, J. CNN-G: Convolutional neural network combined with graph for image segmentation with theoretical analysis. IEEE Trans. Cogn. Dev. Syst. 2020, 13, 631–644. [Google Scholar] [CrossRef]
- DeVries, T.; Taylor, G.W. Learning confidence for out-of-distribution detection in neural networks. arXiv 2018, arXiv:1802.04865. [Google Scholar]
- Jiang, H.; Kim, B.; Guan, M.; Gupta, M. To trust or not to trust a classifier. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2018; pp. 5541–5552. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Object detectors emerge in deep scene cnns. arXiv 2014, arXiv:1412.6856. [Google Scholar]
- Springenberg, J.T.; Dosovitskiy, A.; Brox, T.; Riedmiller, M. Striving for simplicity: The all convolutional net. arXiv 2014, arXiv:1412.6806. [Google Scholar]
- Zhou, B.; Khosla, A.; Lapedriza, A.; Oliva, A.; Torralba, A. Learning deep features for discriminative localization. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2921–2929. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 27–29 October 2017; pp. 618–626. [Google Scholar]
- Yang, H.; Kim, J.Y.; Kim, H.; Adhikari, S.P. Guided soft attention network for classification of breast cancer histopathology images. IEEE Trans. Med. Imaging 2019, 39, 1306–1315. [Google Scholar] [CrossRef]
- DeVries, T.; Taylor, G.W. Improved regularization of convolutional neural networks with cutout. arXiv 2017, arXiv:1708.04552. [Google Scholar]
- Krizhevsky, A.; Hinton, G. Learning Multiple Layers of Features from Tiny Images; Citeseer: State College, PA, USA, 2009. [Google Scholar]
- Netzer, Y.; Wang, T.; Coates, A.; Bissacco, A.; Wu, B.; Ng, A.Y. Reading digits in natural images with unsupervised feature learning. In Proceedings of the NIPS Workshop on Deep Learning and Unsupervised Feature Learning, Granada, Spain, 16 December 2011. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2012; pp. 1097–1105. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.F. ImageNet: A large-scale hierarchical image database. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Szegedy, C.; Vanhoucke, V.; Ioffe, S.; Shlens, J.; Wojna, Z. Rethinking the inception architecture for computer vision. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 2818–2826. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Munich, Germany, 5–9 October 2015; Springer: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Oktay, O.; Schlemper, J.; Folgoc, L.L.; Lee, M.; Heinrich, M.; Misawa, K.; Mori, K.; McDonagh, S.; Hammerla, N.Y.; Kainz, B.; et al. Attention u-net: Learning where to look for the pancreas. arXiv 2018, arXiv:1804.03999. [Google Scholar]
- Roy, A.G.; Navab, N.; Wachinger, C. Concurrent spatial and channel ‘squeeze & excitation’ in fully convolutional networks. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Granada, Spain, 16–20 September 2018; Springer: Berlin/Heidelberg, Germany, 2018; pp. 421–429. [Google Scholar]
- Alom, M.Z.; Hasan, M.; Yakopcic, C.; Taha, T.M.; Asari, V.K. Recurrent residual convolutional neural network based on u-net (r2u-net) for medical image segmentation. arXiv 2018, arXiv:1802.06955. [Google Scholar]
- Shan, H.; Padole, A.; Homayounieh, F.; Kruger, U.; Khera, R.D.; Nitiwarangkul, C.; Kalra, M.K.; Wang, G. Competitive performance of a modularized deep neural network compared to commercial algorithms for low-dose CT image reconstruction. Nat. Mach. Intell. 2019, 1, 269–276. [Google Scholar] [CrossRef]
- Yang, Q.; Yan, P.; Zhang, Y.; Yu, H.; Shi, Y.; Mou, X.; Kalra, M.K.; Zhang, Y.; Sun, L.; Wang, G. Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans. Med. Imaging 2018, 37, 1348–1357. [Google Scholar] [CrossRef]
- Shan, H.; Zhang, Y.; Yang, Q.; Kruger, U.; Kalra, M.K.; Sun, L.; Cong, W.; Wang, G. 3-D convolutional encoder-decoder network for low-dose CT via transfer learning from a 2-D trained network. IEEE Trans. Med. Imaging 2018, 37, 1522–1534. [Google Scholar] [CrossRef]
- Vaswani, A.; Shazeer, N.; Parmar, N.; Uszkoreit, J.; Jones, L.; Gomez, A.N.; Kaiser, Ł.; Polosukhin, I. Attention is all you need. In Advances in Neural Information Processing Systems; The MIT Press: Cambridge, MA, USA, 2017; Volume 30. [Google Scholar]
- Hatamizadeh, A.; Tang, Y.; Nath, V.; Yang, D.; Myronenko, A.; Landman, B.; Roth, H.R.; Xu, D. UNETR: Transformers for 3D Medical Image Segmentation. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision, Waikoloa, HI, USA, 4–8 January 2022; pp. 574–584. [Google Scholar]
- Landman, B.; Xu, Z.; Igelsias, J.; Styner, M.; Langerak, T.; Klein, A. MICCAI multi-atlas labeling beyond the cranial vault–workshop and challenge. In Proceedings of the MICCAI Multi-Atlas Labeling Beyond Cranial Vaul—Workshop Challenge, Munich, Germany, 5–9 October 2015; Volume 5, p. 12. [Google Scholar]
- Simpson, A.L.; Antonelli, M.; Bakas, S.; Bilello, M.; Farahani, K.; Van Ginneken, B.; Kopp-Schneider, A.; Landman, B.A.; Litjens, G.; Menze, B.; et al. A large annotated medical image dataset for the development and evaluation of segmentation algorithms. arXiv 2019, arXiv:1902.09063. [Google Scholar]
- Huang, S.; Li, J.; Xiao, Y.; Shen, N.; Xu, T. RTNet: Relation Transformer Network for Diabetic Retinopathy Multi-lesion Segmentation. IEEE Trans. Med. Imaging 2022, 41, 1596–1607. [Google Scholar] [CrossRef]
- Dai, Y.; Gao, Y.; Liu, F. Transmed: Transformers advance multi-modal medical image classification. Diagnostics 2021, 11, 1384. [Google Scholar] [CrossRef]
- Liu, Z.; Mao, H.; Wu, C.Y.; Feichtenhofer, C.; Darrell, T.; Xie, S. A ConvNet for the 2020s. arXiv 2022, arXiv:2201.03545. [Google Scholar]
- Dong, H.; Yang, G.; Liu, F.; Mo, Y.; Guo, Y. Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In Proceedings of the Annual Conference on Medical Image Understanding and Analysis, Edinburgh, UK, 11–13 July 2017; Springer: Berlin/Heidelberg, Germany, 2017; pp. 506–517. [Google Scholar]
- Liskowski, P.; Krawiec, K. Segmenting retinal blood vessels with deep neural networks. IEEE Trans. Med. Imaging 2016, 35, 2369–2380. [Google Scholar] [CrossRef] [PubMed]
- Christ, P.F.; Elshaer, M.E.A.; Ettlinger, F.; Tatavarty, S.; Bickel, M.; Bilic, P.; Rempfler, M.; Armbruster, M.; Hofmann, F.; D’Anastasi, M.; et al. Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention, Athens, Greece, 17–21 October 2016; Springer: Berlin/Heidelberg, Germany, 2016; pp. 415–423. [Google Scholar]
- Sirinukunwattana, K.; Pluim, J.P.; Chen, H.; Qi, X.; Heng, P.A.; Guo, Y.B.; Wang, L.Y.; Matuszewski, B.J.; Bruni, E.; Sanchez, U.; et al. Gland segmentation in colon histology images: The glas challenge contest. Med. Image Anal. 2017, 35, 489–502. [Google Scholar] [CrossRef]
- Zhang, H.; Cisse, M.; Dauphin, Y.N.; Lopez-Paz, D. mixup: Beyond empirical risk minimization. arXiv 2017, arXiv:1710.09412. [Google Scholar]
- Bdair, T.; Navab, N.; Albarqouni, S. ROAM: Random Layer Mixup for Semi-Supervised Learning in Medical Imaging. arXiv 2020, arXiv:2003.09439. [Google Scholar] [CrossRef]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Lin, M.; Chen, Q.; Yan, S. Network in network. In Proceedings of the International Conference on Learning Representations, Scottsdale, AZ, USA, 2–4 May 2013. [Google Scholar]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Kingma, D.; Ba, J. ADADELTA: An Adaptive Learning Rate Method. arXiv 2012, arXiv:1212.5701. [Google Scholar]
- Loshchilov, I.; Hutter, F. Decoupled Weight Decay Regularization. In Proceedings of the International Conference on Learning Representations, Vancouver, BC, Canada, 30 April–3 May 2018. [Google Scholar]
- Paszke, A.; Gross, S.; Chintala, S.; Chanan, G.; Yang, E.; DeVito, Z.; Lin, Z.; Desmaison, A.; Antiga, L.; Lerer, A. Automatic differentiation in PyTorch. In Proceedings of the 31st Conference of Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; Available online: ttps://openreview.net/forum?id=BJJsrmfCZ (accessed on 30 July 2022).




| Part | Modality | Task | Classes | Training Samples | Validation Samples | Test Samples | |
|---|---|---|---|---|---|---|---|
| Retinal diseases | Eyes | OCT | Classification | 4 | 4000 | 1000 | 1000 |
| Pneumonia | Chest | X-ray | Classification | 2 | 4632 | 600 | 624 |
| Glaucoma | Eyes | Color fundus image | Classification | 2 | 5232 | 744 | 744 |
| Task | Network | Vanilla | GRM |
|---|---|---|---|
| Retinal diseases | Inception-V3 | 94.9 | 96.7 |
| ResNet-50 | 93.7 | 96.3 | |
| DenseNet-121 | 93.6 | 96.0 | |
| ViT | 89.9 | 92.6 | |
| Pneumonia | Inception-V3 | 90.4 | 92.8 |
| ResNet-50 | 90.0 | 94.2 | |
| DenseNet-121 | 88.8 | 92.1 | |
| ViT | 90.2 | 91.8 | |
| Glaucoma | Inception-V3 | 89.2 | 91.6 |
| ResNet-50 | 87.5 | 90.6 | |
| DenseNet-121 | 88.9 | 90.0 | |
| ViT | 87.5 | 88.5 |
| Task | Network | GRM | Cutout | Mixup |
|---|---|---|---|---|
| Retinal diseases | Inception-V3 | 96.7 | 94.3 | 95.8 |
| ResNet-50 | 96.3 | 94.1 | 92.8 | |
| DenseNet-121 | 96.0 | 95.1 | 95.6 | |
| ViT | 92.6 | 92.0 | 92.8 | |
| Pneumonia | Inception-V3 | 92.8 | 89.2 | 91.2 |
| ResNet-50 | 94.2 | 89.7 | 91.5 | |
| DenseNet-121 | 92.1 | 92.0 | 91.0 | |
| ViT | 91.8 | 91.1 | 89.1 | |
| Glaucoma | Inception-V3 | 91.6 | 89.0 | 91.4 |
| ResNet-50 | 90.6 | 87.3 | 89.4 | |
| DenseNet-121 | 90.0 | 89.6 | 88.8 | |
| ViT | 88.5 | 86.2 | 86.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, X.; Wang, S.; Hu, J. Guided Random Mask: Adaptively Regularizing Deep Neural Networks for Medical Image Analysis by Potential Lesions. Appl. Sci. 2022, 12, 9099. https://doi.org/10.3390/app12189099
Yu X, Wang S, Hu J. Guided Random Mask: Adaptively Regularizing Deep Neural Networks for Medical Image Analysis by Potential Lesions. Applied Sciences. 2022; 12(18):9099. https://doi.org/10.3390/app12189099
Chicago/Turabian StyleYu, Xiaorui, Shuqi Wang, and Junjie Hu. 2022. "Guided Random Mask: Adaptively Regularizing Deep Neural Networks for Medical Image Analysis by Potential Lesions" Applied Sciences 12, no. 18: 9099. https://doi.org/10.3390/app12189099
APA StyleYu, X., Wang, S., & Hu, J. (2022). Guided Random Mask: Adaptively Regularizing Deep Neural Networks for Medical Image Analysis by Potential Lesions. Applied Sciences, 12(18), 9099. https://doi.org/10.3390/app12189099






