Abstract
The World Health Organization (WHO) predicted that 10 million people would have died of cancer by 2020. According to recent studies, liver cancer is the most prevalent cancer worldwide. Hepatocellular carcinoma (HCC) is the leading cause of early-stage liver cancer. However, HCC occurs most frequently in patients with chronic liver conditions (such as cirrhosis). Therefore, it is important to predict liver cancer more explicitly by using machine learning. This study examines the survival prediction of a dataset of HCC based on three strategies. Originally, missing values are estimated using mean, mode, and k-Nearest Neighbor (k-NN). We then compare the different select features using the wrapper and embedded methods. The embedded method employs Least Absolute Shrinkage and Selection Operator (LASSO) and ridge regression in conjunction with Logistic Regression (LR). In the wrapper method, gradient boosting and random forests eliminate features recursively. Classification algorithms for predicting results include k-NN, Random Forest (RF), and Logistic Regression. The experimental results indicate that Recursive Feature Elimination with Gradient Boosting (RFE-GB) produces better results, with a 96.66% accuracy rate and a 95.66% F1-score.
1. Introduction
The World Health Organization predicts that liver cancer will hit more than a million people in 2025, becoming a significant health issue worldwide. Most liver cancers are HCCs. Hospital admissions for HCC tripled between 1993 and 2005, leading to corresponding cost increases [1]. As a result of most HCC occurrences, hepatitis B and C have spread widely. Non-alcoholic steatohepatitis is associated with unique molecular pathogenesis [2].
HCC is found in patients who have cirrhosis or chronic liver disease [3]. A recent study revealed that liver cancer is the sixth-most-frequently diagnosed cancer worldwide, causing 600,000 deaths annually [4]. Therefore, early detection is essential to reduce the risk of death from HCC. In most cases, primary liver tumors develop into HCC [5]. Over 10% of HCC cases recur after five years [6]. Most deaths from cancer caused by HCC occur in the United States [7]. Early identification [8] is an effective strategy to avoid cancer, but it is difficult to detect due to the number of causes that cause cancer.
The use of machine learning regarding the HCC problem is motivated by two key factors. To begin with, enhancing a patient with HCC’s quality of life demands an accurate and quick diagnosis. The second reason is identifying relevant features to improve prediction-model accuracy.
The main aim of this work is structured as follows: The first step is to replace the missing values using the mean and mode method. The second step performs the following feature-selection methods on the data: logistic regression using LASSO and ridge regression; recursive feature elimination using a Gradient Boosting (GB) classifier and a RF classifier; and, finally, prediction utilizing DT, RF, LR, and k-NN algorithms. In addition, multiple feature-selection methods are utilized throughout this experiment to divide the data into two categories: alive and dead, depending on the dataset. Experimental work is carried out with missing-value replacements, data-preprocessing approaches, feature-selection methods, and classification algorithms. The major contributions of this work are:
- Examine the impact of missing-value replacement using mean and mode approaches.
- Utilize the feature-selection methods to select the relevant features causing HCC.
- Assess the impact of different machine-learning algorithms in HCC classification.
2. Related Works
The authors [9] proposed two preprocessing methods for missing and heterogeneous data and used k-means clustering. In [10], a dataset of 4000 chronic hepatitis C patients diagnosed at Cairo University’s multidisciplinary hospital is used with linear regression. The dataset is balanced using Synthetic Minority Over-sampling Technique (SMOTE) methods. The performance of LR and Neural Networks (NN) is 75.2% and 73%, respectively. CART, ADTree, and REP-Tree models give an excellent area under the receiver-operating-characteristic curve (AUROC), ranging between 95.5% and 99%. The high accuracy of HCC diagnosis ranges between 93.2% and 95.6%.
According to Książek and Gandor [11], LR is the best-known machine-learning model for binary classification. The authors proposed three experiments: genetically selecting logistic-regression parameters, selecting features, and training a logistic-regression model with genetically determined coefficients (weights) with an F1-score of 93.56%. Iterative optimization is employed to optimize the logistic-regression coefficients. First, Chronic Kidney Disease data is preprocessed by employing missing data-handling methods with mean and mode of statistical analysis [12]. Then, Recursive Feature Elimination (RFE) is applied to select the best relevant features. In that analysis, the classification algorithms used are SVM, k-NN, DT, and RF. Random Forest surpassed all competing algorithms with 100% accuracy based on all measures.
For prediction, the author [13] used fifteen different models for the HCC dataset. Several methods have evaluated the feature weights, including L-1 penalty or LASSO regression, L-2 penalty or ridge regression, Genetic Algorithm Optimization, and Regression Function. The RFGBEL model achieves 93.92% accuracy, 94.73% sensitivity, 93% F-1 score, a Log-Loss or cross-entropy score of 58.9%, and 72% Jaccard. Weighted Synthetic Minority Over-sampling Technique (WSMOTE) is an effective method for dealing with dataset imbalances for liver disease [14]. The Improved Fuzzy C Means clustering technique is applied to substitute missing information to increase the overall accuracy of the study. Combining Kernel Support Vector Machine and Fuzzy Convolutional Neural Network (FCNN) yielded a heterogeneous ensemble classifier that used Bootstrap aggregation to obtain accurate results. It has a classification rate of 99.12%, whereas other methods such as MCNN and FCNN have 90.75% and 92.48%, respectively.
The author [15] proposed an RFE-GB algorithm for heart disease. The proposed algorithm produced the greatest results (90.7%). Furthermore, the proposed RFE-GB algorithm has a significantly higher area under the curve than other algorithms. The robustness of HCC is assessed [16] with improved Fuzzy C Means clustering to impute missing values. The optimal feature subset is selected using multiple ensemble methods. Experimental results from the proposed scheme demonstrate that FCNN is more accurate in classifying than other techniques with precision, recall, F-measure, and accuracy scores of 90.78%, 97.36%, 93.96%, and 92.48%, respectively.
As a result of an inability to properly metabolize glucose, diabetes illnesses are becoming more prevalent [17]. The author in [18] proposed dynamic RFE (dRFE) for identifying the subset of features most closely associated with the class labels. The feature-selection scheme comprises RFE and L2 regularization in the wrapper-based method for diabetes, to overcome the overfitting problem. The algorithm predicts diabetes disease more accurately than other existing algorithms. In both of the datasets, GSE53045 and GSE66695, 100% accuracy is achieved. For the other three methylome datasets, GSE74845, GSE103186, and GSE80970, the best prediction accuracy obtained is 92.59%, 94.24%, and 86.01%, respectively.
Cervical cancer [19] prediction utilized SMOTE to balance classes and impute missing values in data preprocessing. The firefly algorithm is applied to identify the critical features and optimize the models. The Extreme GB is more accurate than the other two life-expectancy datasets in determining the target variables [20]. Additionally, visualizing, normalizing, data cleaning, reducing the number of features, etc., are applied. The accuracy of the three models is improved, with 95.32% for linear regression considering all attributes. Likewise, HCC [21] is diagnosed based on real-world data collected during medical practice. According to the most accurate hyperparameter, GB delivered an Area Under the Curve of 0.940, when used with the presence of HCC. The accuracy of the des-gamma-carboxyprothrombin, alpha-fetoprotein, and AFP-L3 used to predict disease progression is 74.91%, 70.67%, and 71.05%, respectively.
3. Proposed Framework
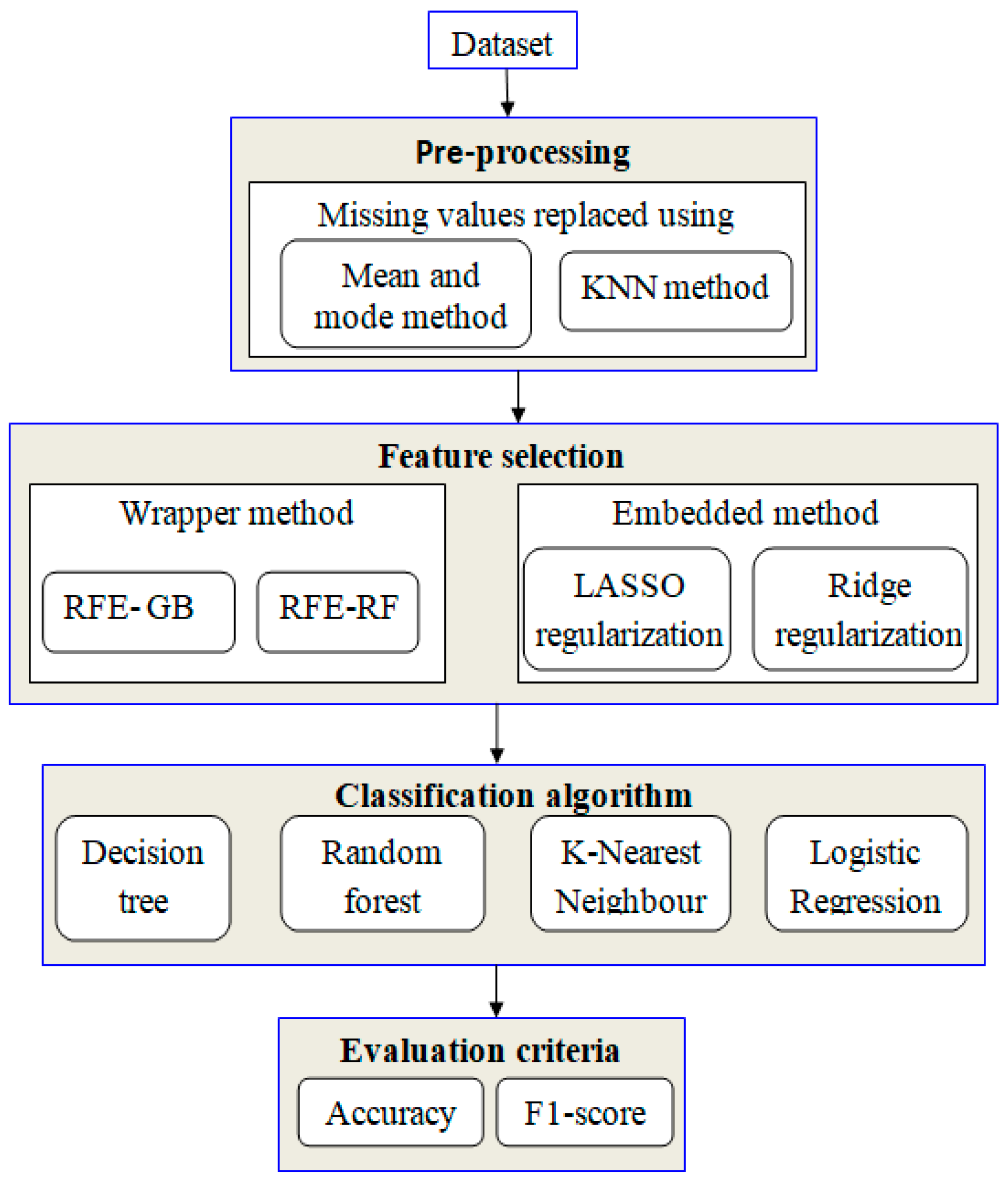
The proposed study uses machine-learning techniques to perform the classification of HCC. For data classification, four algorithms are applied: RF, k-NN, DT, and LR. When dealing with unbalanced data, a random oversampling method is applied. Finally, the embedded and wrapper methods are employed to identify the essential features. Figure 1 illustrates the conceptual framework for the research.
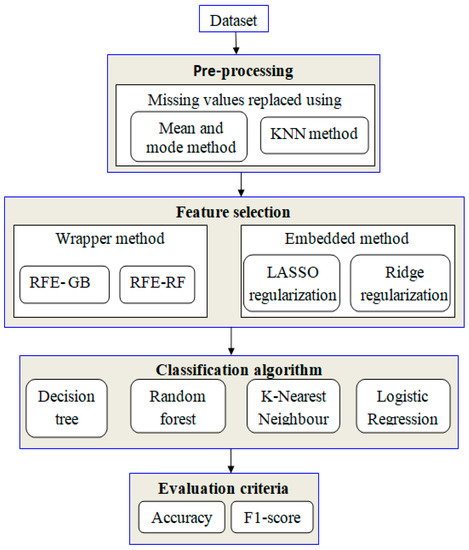
Figure 1.
Proposed framework.
3.1. Data Preprocessing
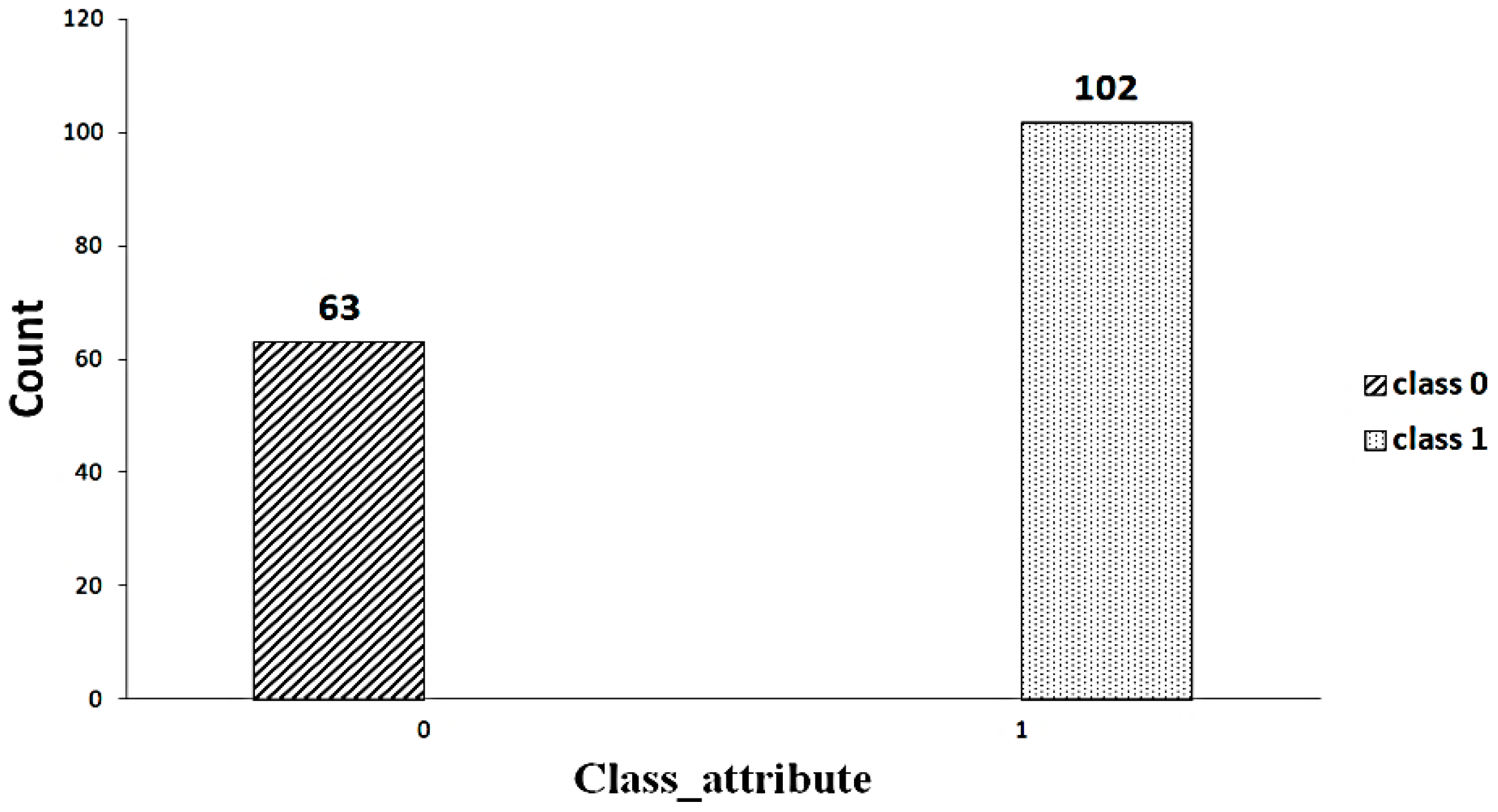
For this study, the database is taken from UCI’s repository. The dataset contained information about 165 patients with 49 features. Twenty-six qualitative and twenty-three quantitative variables are considered [9]. The data contain a lot of missing values. Furthermore, only four patients have complete information in the data. The dataset has an unbalanced distribution of classes (102 versus 63). The detail of the dataset is given in Table 1.

Table 1.
HCC dataset details.
The dataset is cleaned up in the preprocessing stage because it contains missing values. The dataset contains four instances that are filled, while the rest are with missing data. The dataset consists of binary-, ordinal-, and scalar-type features. Two methods are used for the missing values: mean and mode and the k-NN method.
3.2. Handling Missing Values
Table 1 also shows the number of missing values for each feature. The values vary between 0 and 80 for specific features. The missing values for binary and ordinal features are substituted with the mode and mean method. Mode refers to a number or the value appearing most often in a dataset. A mean is the arithmetic average of all data points in a collection for scalar-type features. It is computed by adding the values in a dataset and dividing it by the total number of records, as represented in Equation (1).
where x represents values and N denotes the number of records.
A point in a multidimensional space matched with its k-NN is beneficial [22] to associate with an instance. This method is especially useful in dealing with missing data that is continuous, discrete, ordinal, or categorical. First, a Euclidean distance metric with ten nearest neighbors is used to impute missing values. For each feature, the mean value is replaced against the missing values.
3.3. Random Oversampling/Undersampling
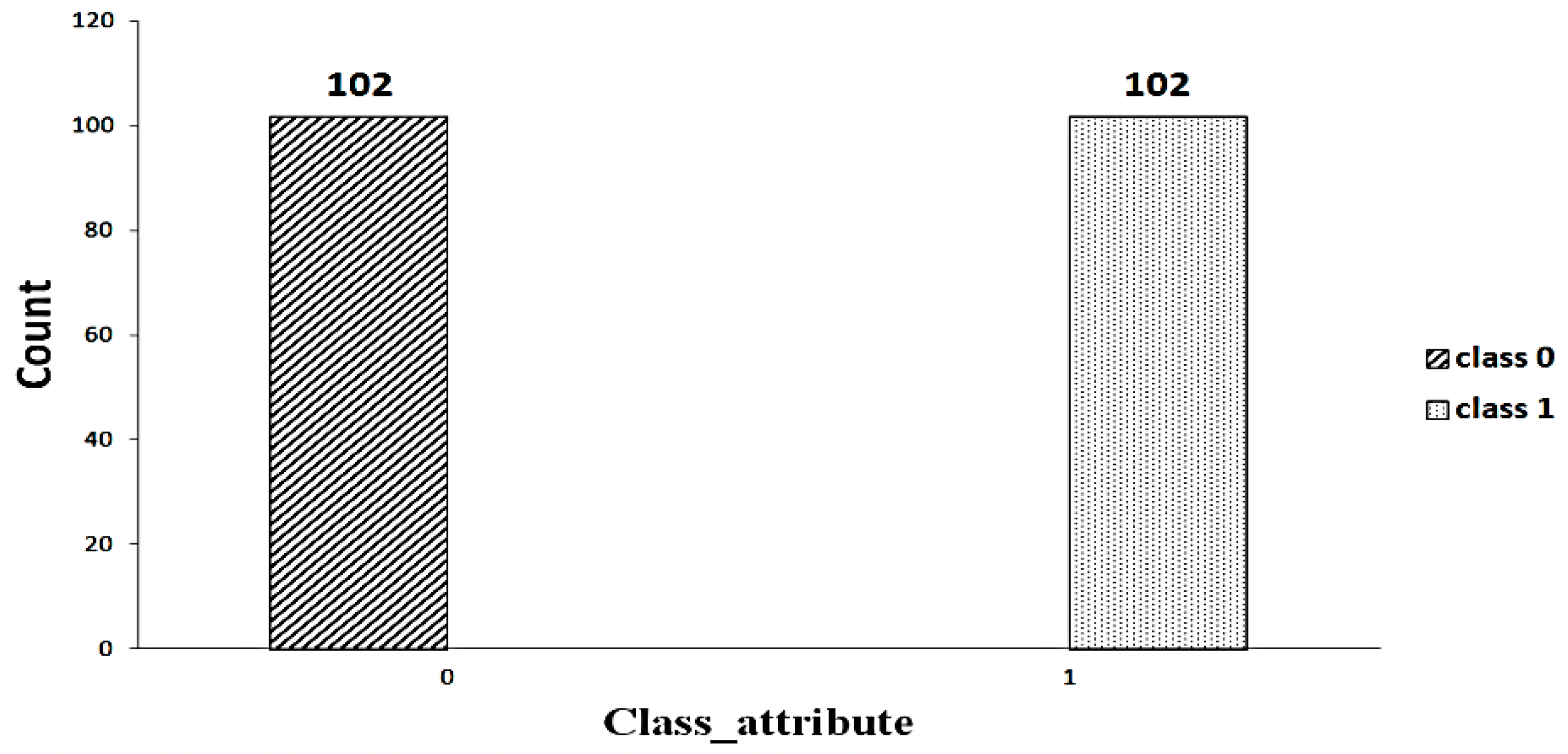
A significant component of machine learning lies with the distribution of data. An imbalanced dataset [23] indicates that some classes have a higher number of instances than others. As shown in Figure 2, the dataset includes an imbalanced set of data (63 versus 102). Random oversampling/undersampling is a method used to duplicate the data from the minority class, add it to the training set, and remove the data from the majority class as well as from the training set to make the data balanced. This process is repeated until the minority class is balanced with the majority class. Figure 3 shows the balanced data after applying the random oversampling/undersampling method.
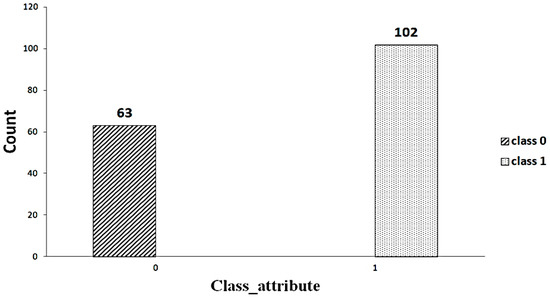
Figure 2.
Imbalanced data.
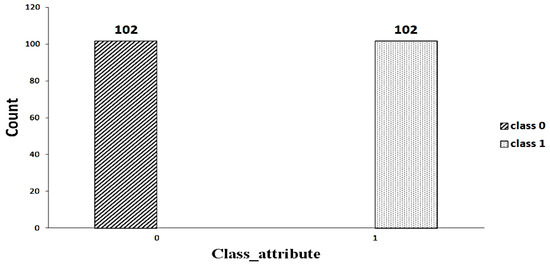
Figure 3.
Balanced data.
3.4. Feature Selection
Feature selection has become a key component for attaining the most desirable subset of features. An algorithm for feature selection proposes a solution based on the distinct feature subsets and ranks those subsets based on three criteria: embedded, wrappers, and filters. This work examines two methods, wrapper and embedded.
3.4.1. Wrapper Method
The wrapper method identifies the most suitable feature for machine-learning algorithms, and the main goal is to enhance the learning process. A machine-learning algorithm is needed in a wrapper method, and its performance is used as an evaluation criterion. Unfortunately, wrapper methods [24] are over-complicated, though they produce the most reliable outcomes based on the explicit-learning technique [25].
RFE is a common approach that combines other machine-learning techniques. Lately, RFE has emerged as a method employed in numerous biomedical fields such as protein classification, selection of genes [26,27], areas of expression analysis, cancer diagnosis, and many others [28]. These methods are created iteratively and determine each feature’s best or worst traits. Once all elements are explored, these methods rank the models according to how features have been eliminated. The importance of specific attributes increases until it reaches an appropriate level [26].
The GB algorithm is used in the GB-RFE method to train the classifier. Boosting applies a sequential-training approach, where every learner tries to change the previous one. Loss functions are calculated as gradients, indicating how well the model coefficients fit the data. This research estimates feature importance with a frequency-based measure of algorithm performance called the Gini index, per Equation (2). The Gini-index value is high for an important feature. Class variable c denotes the number of observations in the class; it indicates the ratio of both instances where each node has its particular type.
An ensemble method such as RF bagging becomes an excellent method in prediction accuracy, performing a great representative for bagging algorithms. RF algorithms use two methods to calculate feature importance from a training model and measure variable significance. Equation (3) computes the relevant ranking of the features. The mean reduction in accuracy shows that the model accuracy reduces when the variable values are adjusted [29,30]. This metric quantifies how observation lowers node impurity over time, weighted by the proportion of data reaching that node, with a mean drop in the Gini coefficient.
In this work, we have used n_estimators = 100, random_state = 10, and n_features_to_select = 1–30, which are chosen as the optimal hyperparameters of RFE-RF that help in selecting the most important features.
3.4.2. Embedded Method
The author of [31] represented a geometrical approach for analyzing high-dimensional data. An embedded method performs feature selection during the model creation process itself. A regularization method is one of the most used methods for penalizing a feature based on a coefficient threshold [32]. If the objective function connects with the absolute values concerning the model parameters, LASSO (L1) regularization allows a penalty term by shrinking some coefficients to zero [33]. L1 provides the regression coefficients to be overcome to zero, collecting aggregated important features concurrently. Equation (4) illustrates how to determine the outcome. A regularization parameter lambda provides the model input. A higher lambda reduces overfitting. We have used C = 0.3, penalty = ‘l1’, solver = ‘liblinear’, and max_iter = 300 as the optimal hyperparameters of GB-RFE that help in selecting the most important features.
In Ridge (L2) regularization, the function is penalized for adding the sum of the squares from the model parameters. The author of [3] described that L2 regularization is a method that accounts for nearly all the features and attempts to withdraw many coefficient measures towards zero in Equation (5).
when a lambda is too large, it adds too much weight and causes underfitting. Meanwhile, overfitting is avoided effectively with the L2-regularization technique. The optimal hyperparameter of this classifier for the data used in the present study are C = 0.5, penalty = ‘l2’, and max_iter = 300.
3.5. Classification Algorithms
This work utilized LR, k-NN, DT, and RF to assess the classification accuracy of HCC. Learning technologies create predictive structures based on training and validation data [34].
3.5.1. Decision Tree
Decision trees are used in supervised learning techniques to identify relevant attributes for classification [12]. A root node represents the entire dataset, a node represents a feature, and a branch represents a decision rule. The root node compares the characteristics with the records (real dataset) and makes the appropriate decisions, per the decision tree. The next step compares the second node’s features with the sub node’s features, and this procedure is repetitive when it has reached a leaf node.
3.5.2. Random Forest
Classification and regression can be performed using the ensemble-classification algorithm. Combining the output from individual decision trees, the RF creates a class of results that are then classified. Random Forests are generally suitable for massive datasets with many input variables [35]. The optimal hyperparameter used in this classifier is n_estimators = 100.
3.5.3. k-Nearest Neighbors
Data is classified based on their distance from each other and their distance from the data location [36]. In addition, all the different data groups are called neighbors. The user chooses the number of neighbors, which is very important when analyzing the dataset. A Euclidean distance (Di) is calculated in two-dimensional space based on the features vector, per Equation (6).
where xk and yk are the kth attributes of x and y respectively.
3.5.4. Logistic Regression
For linear regression to become LR, sigmoid functions are applied [37]. A logistic function reduces the range of y values from 0 to 1 using a large scale. In logistic regression, maximum-likelihood estimators are reliable for calculating linear-regression coefficients, a technique that is generic to machine learning, despite making assumptions about the data distribution.
3.6. Cross-Validation Method
Generally, the performance of a classifier is evaluated by constructing a model with the given data. However, using a single-fold data split reduces the model’s generalization capability. Hence the k-fold cross-validation method is employed to split the data into k-folds. While building the model, k-1 folds are used for training, and one fold is used for testing. Likewise, the different combinations of folds are used for constructing the model, and the average classification accuracy is used to assess the performance. This study used a 10-fold cross-validation method to evaluate the performance of the classifiers.
3.7. Evaluation Metrics
A confusion matrix is an N X N matrix used to evaluate model performance. The machine-learning models are compared with the actual target values [38,39].
The accuracy of the classification is measured by the percentage of instances that are classified correctly, per Equation (7)
Accuracy = (TP + TN)/(TP + TN+ FP + FN)
In order to calculate precision, divide the true-positive results with the sum of true positives and false positives, per Equation (8).
Precision = TP/(TP + FP)
Similarly, Equation (9) is used to calculate the recall by dividing the true positives with true positives and false negatives.
Recall = TP/(TP + FN)
The F1-score combines the precision and recall into a single metric that captures both properties, as shown in Equation (10).
F1-score = (2 × Precision × Recall)/(Precision + Recall)
4. Result and Discussion
The information collected from Coimbra Hospital (at the university center) in Portugal is taken from the UCI repository. The missing values are replaced with the mean and mode method in the first phase. Here, a mean value is used for the quantitative attributes, and the mode method is applied for the qualitative attributes. Accuracy and F1-score are chosen as basic metrics. This proposed work consists of three stages: First, it selects the most significant representative features based on the embedded and wrapper methods. Following that, four machine-learning algorithms, namely, LR, k-NN, DT, and RF, are employed for classifying HCC. Finally, a comparative study is done based on the smaller number of selected features with the highest accuracy.
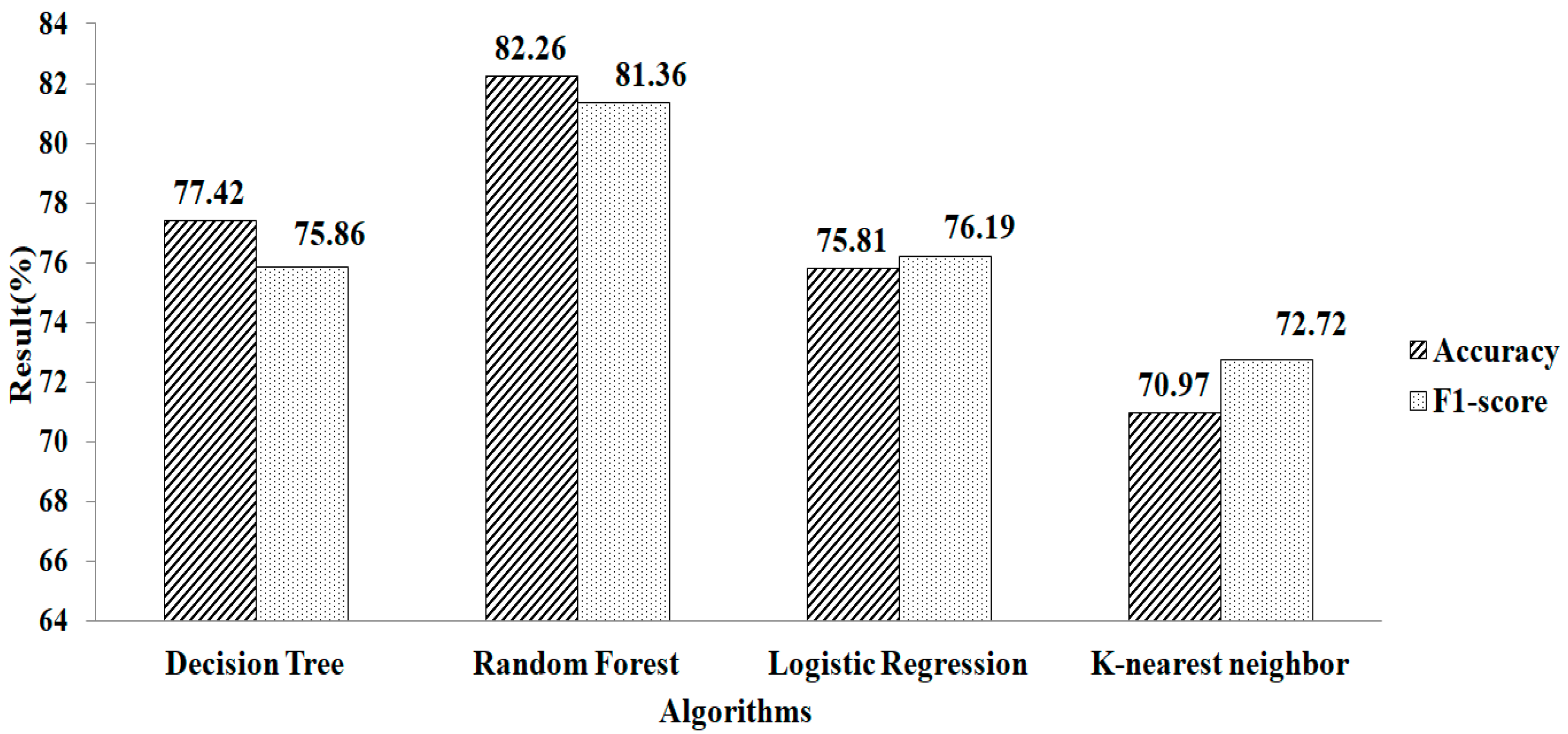
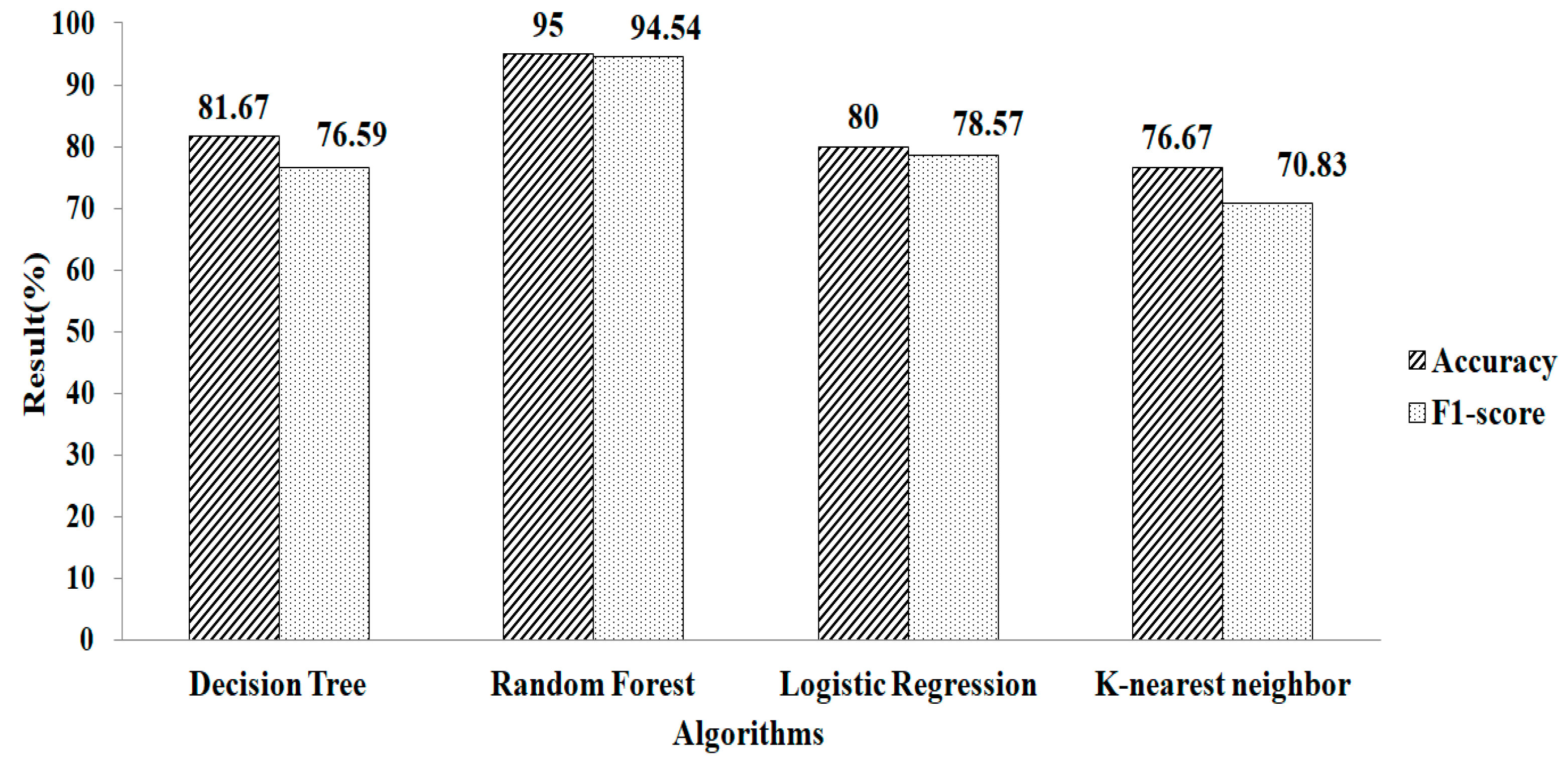
Figure 4 shows the accuracy and F1-score obtained after filling in the missing values using the mean and mode method. The result showed that compared with other classification algorithms, RF gives a greater accuracy value and a higher F1-score value, of 82.26% and 81.36%, respectively, using the parameter values of n_estimators = 100, criterion = “entropy”, and random_state = 35.
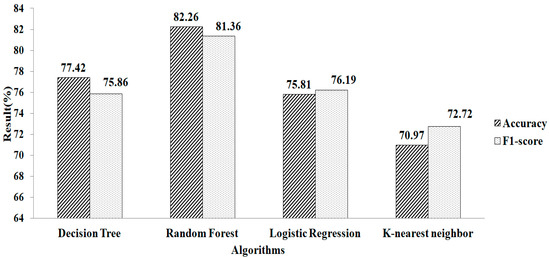
Figure 4.
Mean and mode methods without using feature selection.
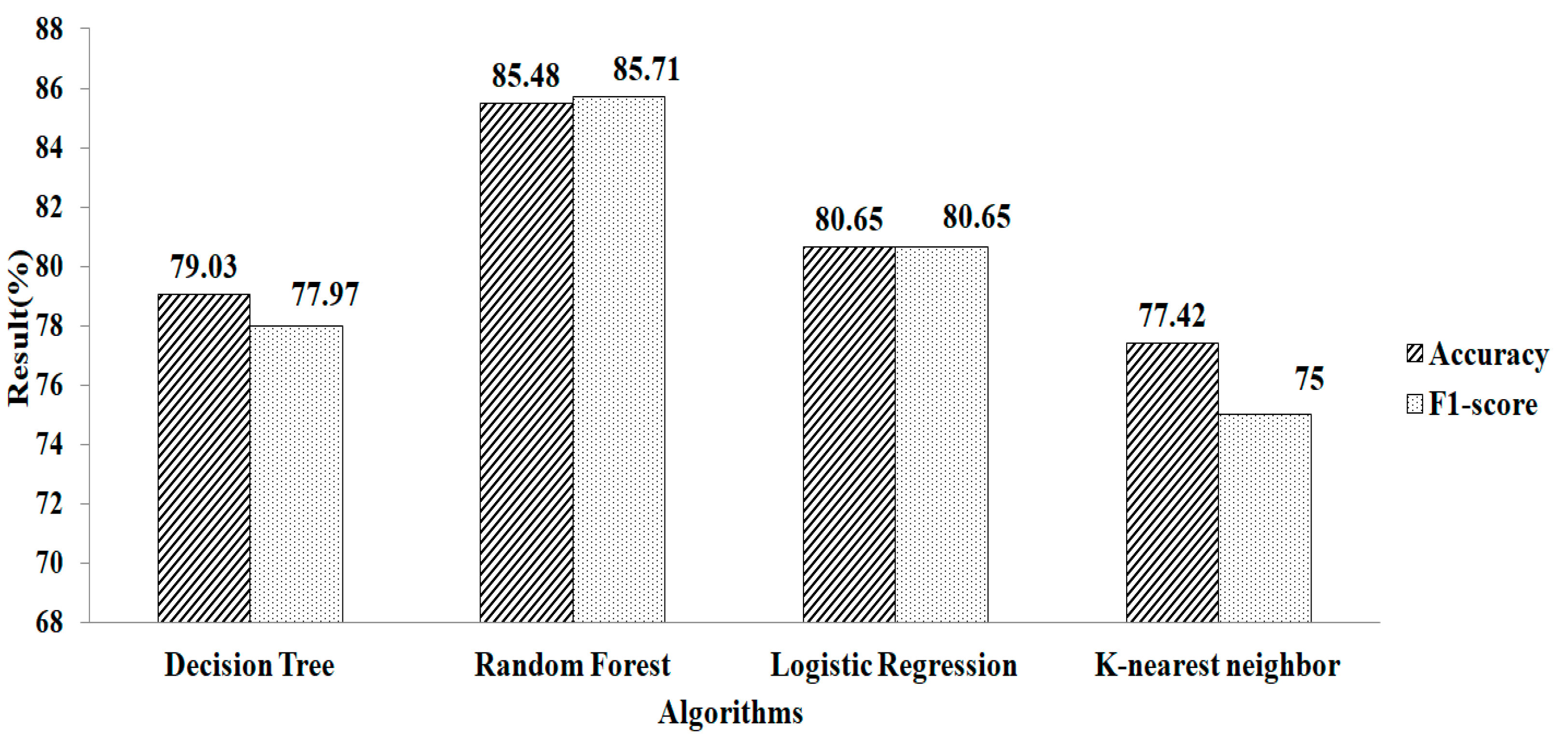
Figure 5 depicts the result obtained after replacing the missing values using k-NN. The experimental results indicate that RF gives a greater accuracy value and a higher F1-score value, of 85.48% and 85.71%, respectively, using the parameters of n_estimators = 100, criterion = “entropy”, and random_state = 35. Compared with the two missing-value-replacement methods, the k-NN method provides a more accurate result without using the feature-selection technique. Similarly, the RF method gives a higher accuracy.
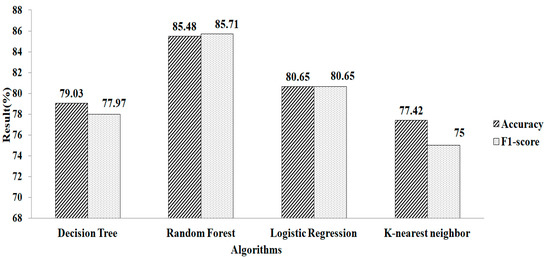
Figure 5.
k-NN method without using feature selection.
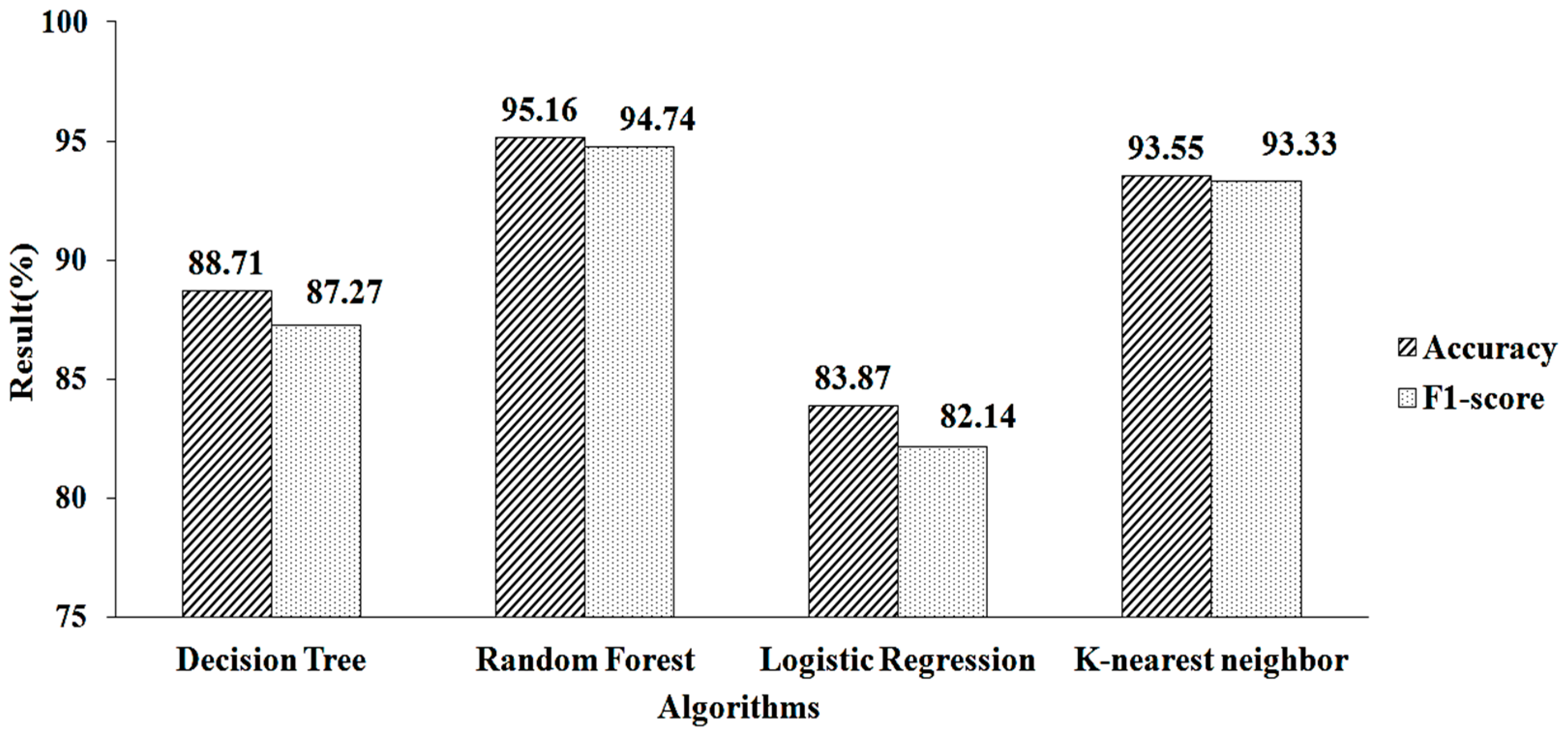
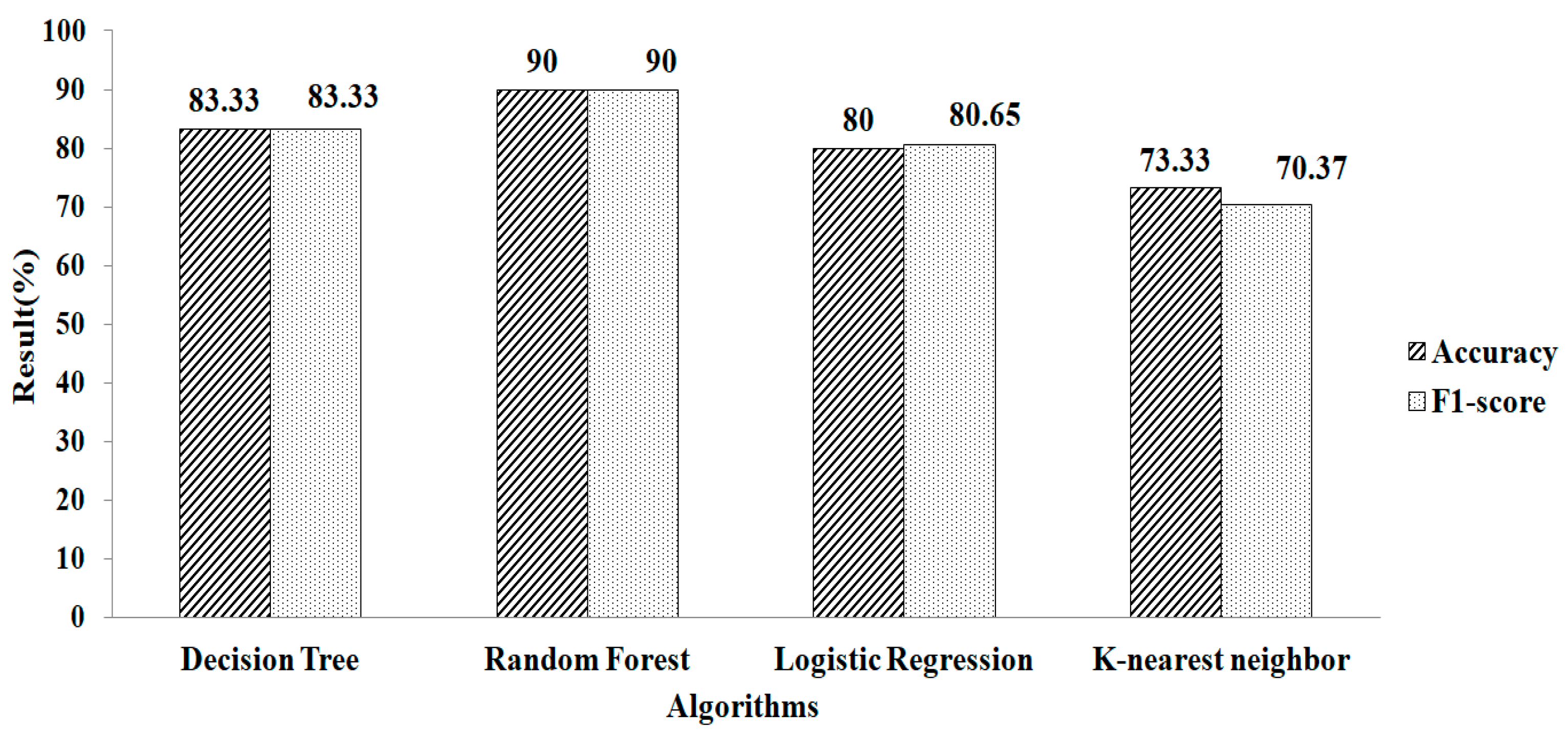
The feature-selection method is used to increase accuracy. Table 2 shows the features chosen by employing mean- and model-based missing-value replacement with LASSO regularization. When employing this strategy, 28 features are chosen. The machine-learning algorithm receives these selected features as input. RF produces a greater accuracy value and a higher F1-score value, of 95.16% and 94.74%, respectively, when employing parameter values of n estimators = 100, criterion = “entropy,” and random state = 35, as shown in Figure 6.

Table 2.
Features selected based on different feature selections.
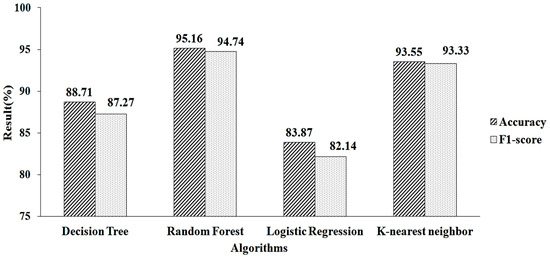
Figure 6.
LASSO regressions for mean and mode.
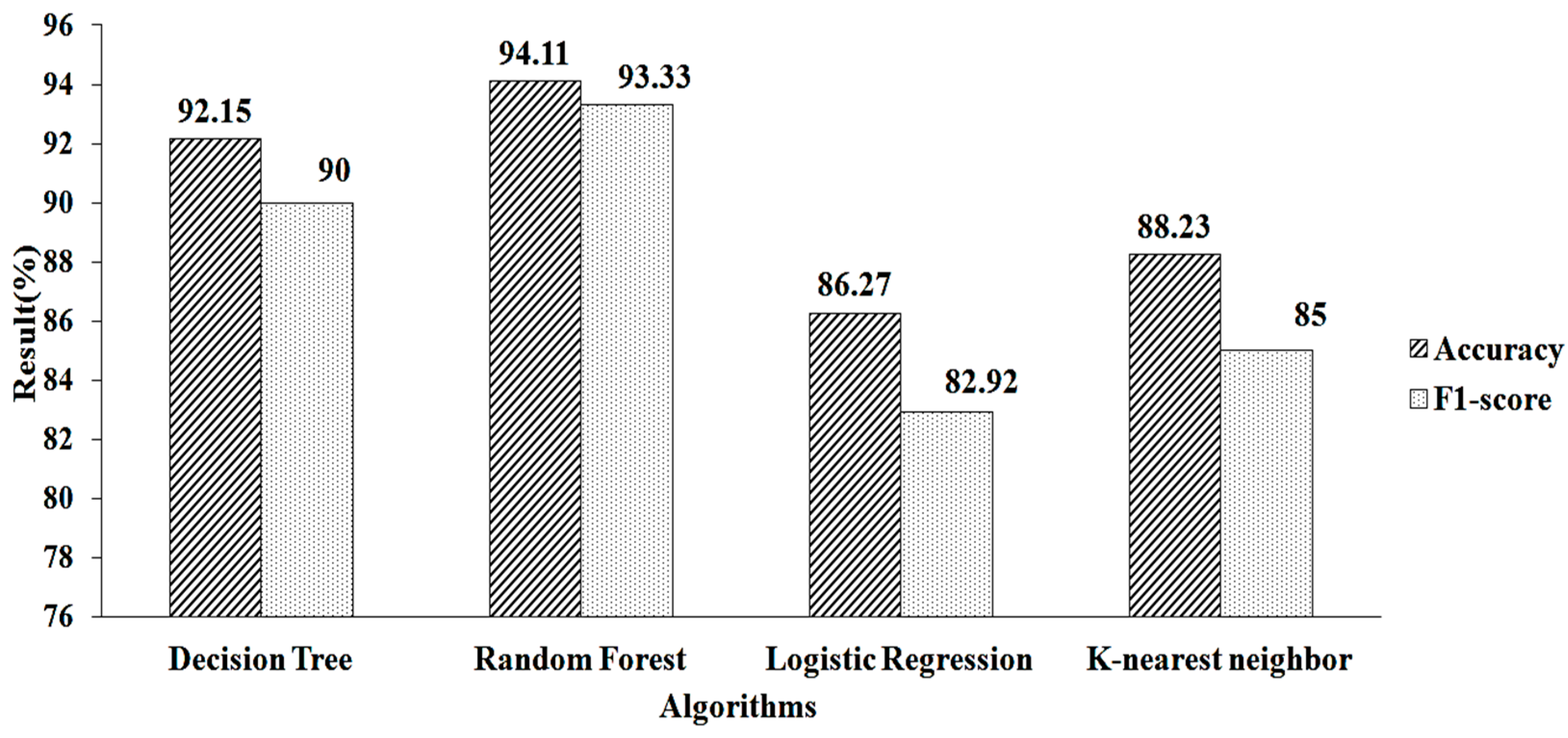
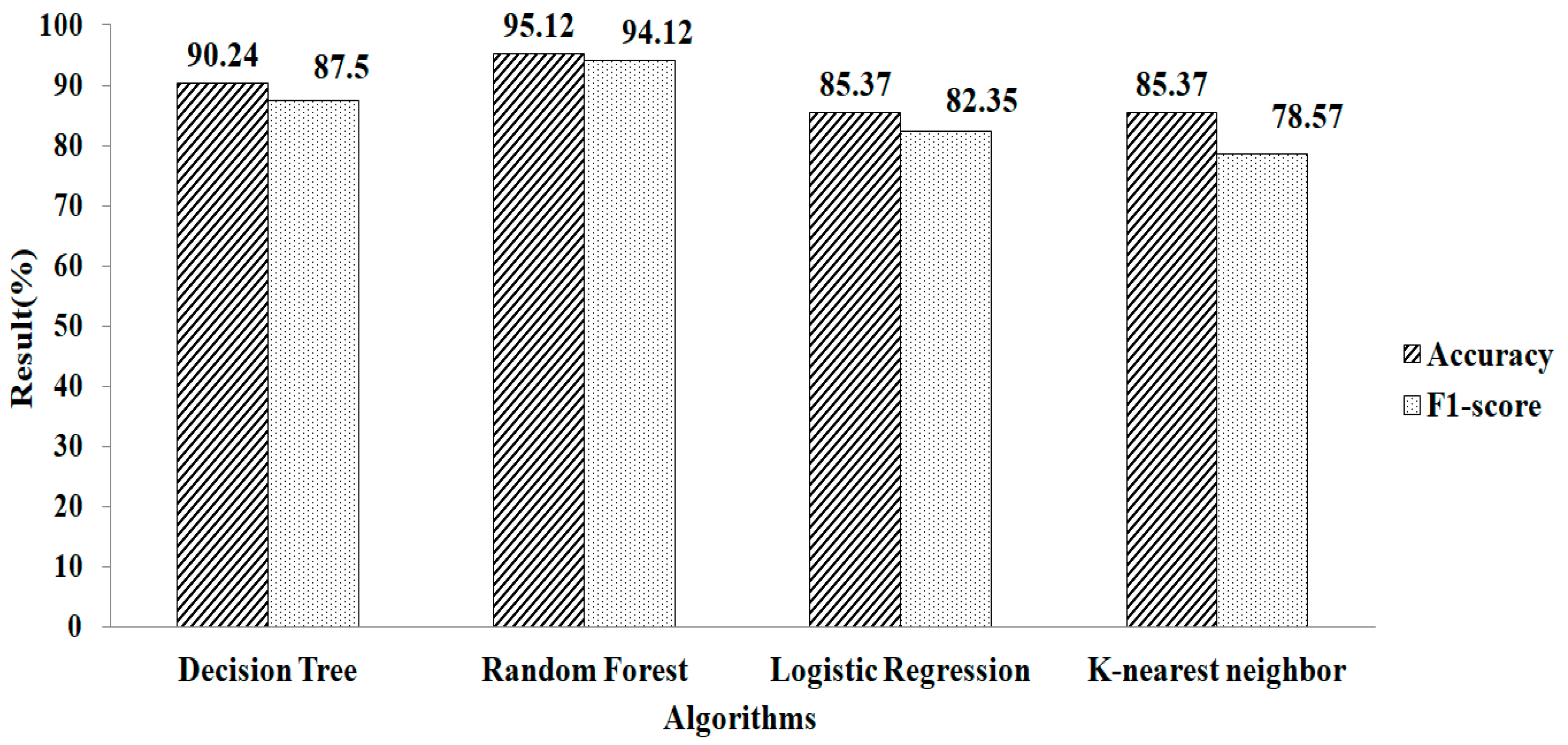
The features obtained for the k-NN using LASSO regularization are shown in Table 2. When this approach is used, 25 features are obtained. These pre-selected features are fed into the machine-learning algorithms. When using parameter values (n estimators = 50, criterion = “entropy”, random state = 60), RF algorithm provides a greater accuracy value and a higher F1-score value, of 94.11% and 93.33%, respectively, as shown in Figure 7.
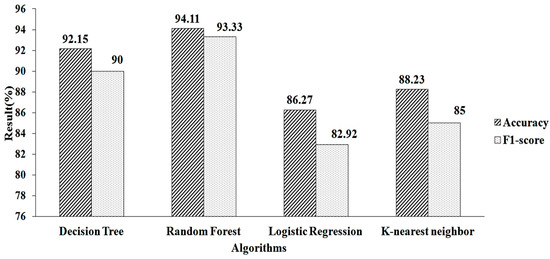
Figure 7.
LASSO regression for k-NN.
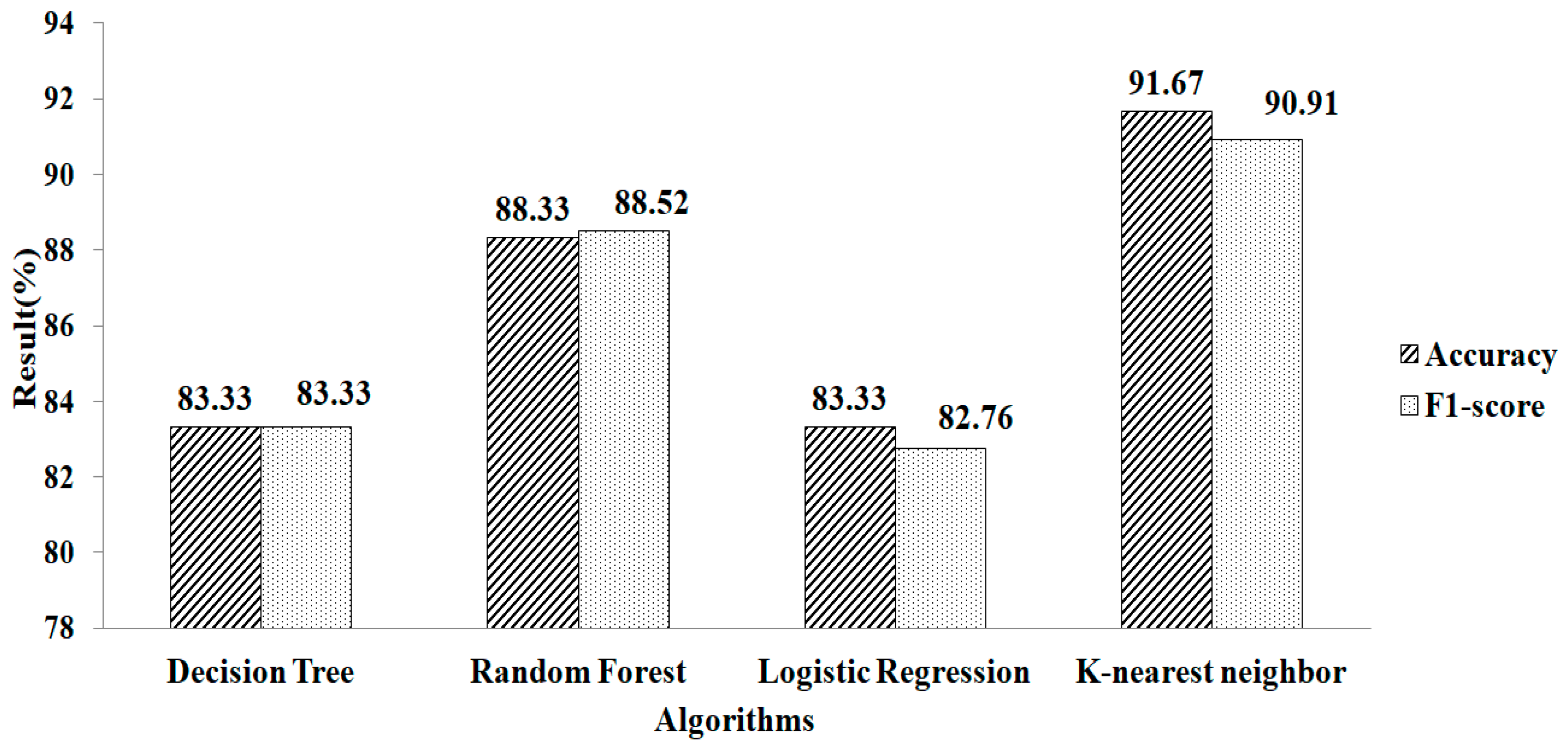
The features chosen for mean and mode based on l2 regularization are shown in Table 2. Here, 17 attributes are chosen. These selected features are fed into the machine-learning algorithm. When parameter values (metric = ‘manhattan’, n neighbors = 1, weights = ‘distance’) are used, k-NN provides a greater accuracy value and a higher F1-score value, of 91.67% and 90.91%, respectively, as shown in Figure 8.
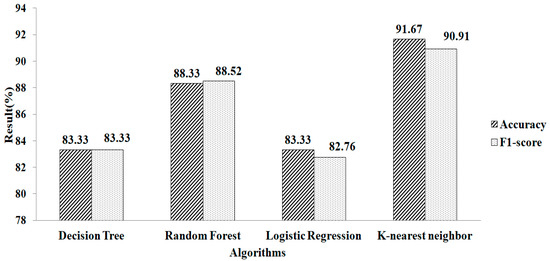
Figure 8.
Ridge regressions for mean and mode.
Table 2 shows the l2- regularization-based feature selection employing k-NN to handle missing values. This approach identifies eighteen features from the dataset. Classification using the selected features is performed with the different machine learning algorithms. Figure 9 indicates that RF produces a greater accuracy value and a higher F1-score value, both of 90%, when employing the parameter values of n estimators = 200, criterion = “entropy”, random_state = 60.
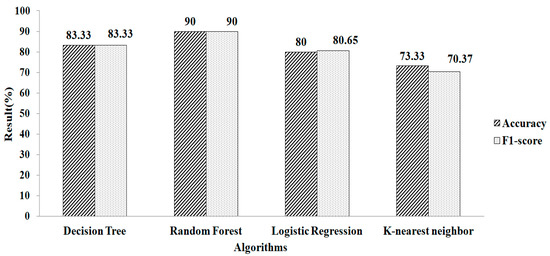
Figure 9.
Ridge regressions for k-NN.
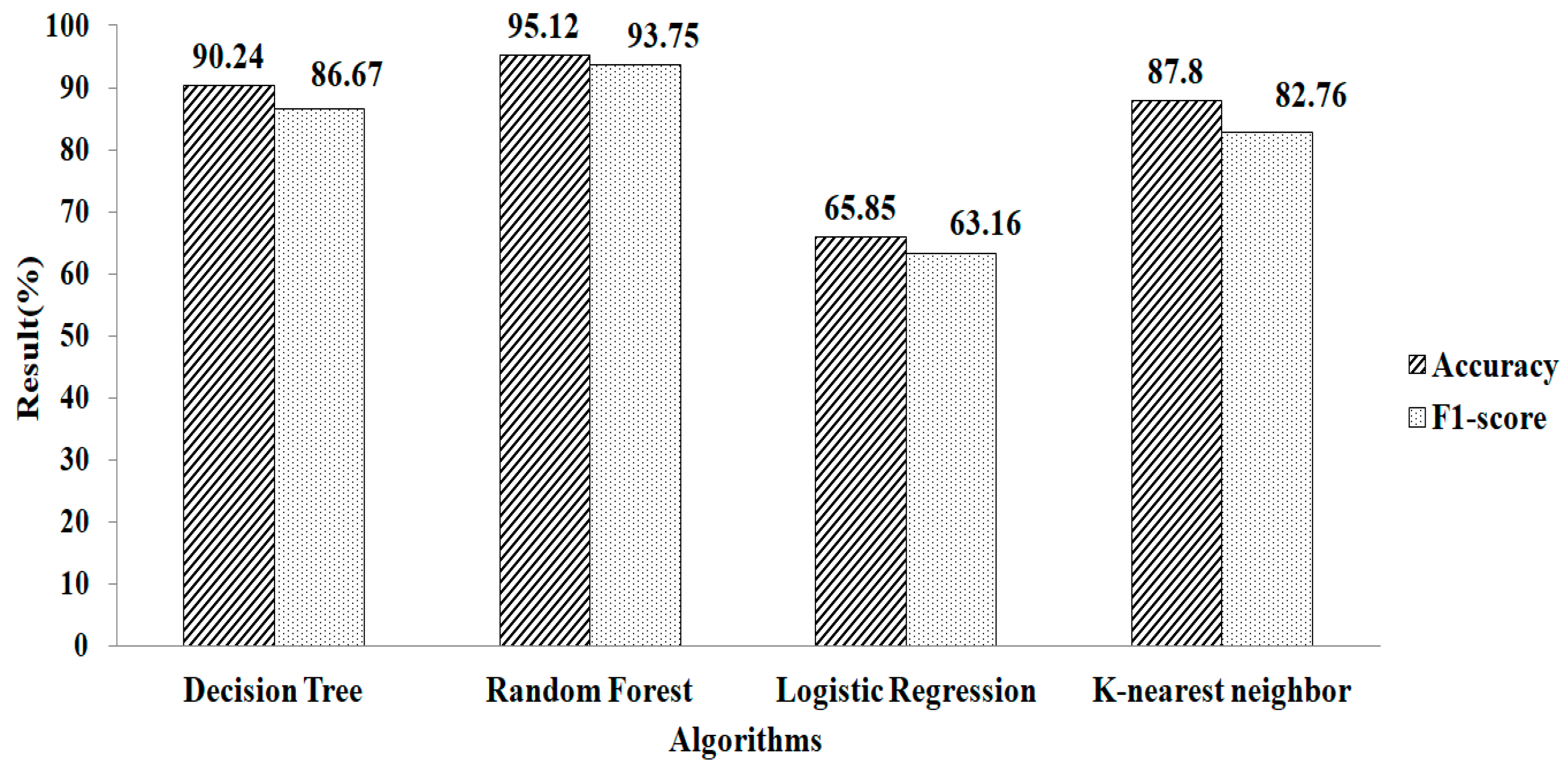
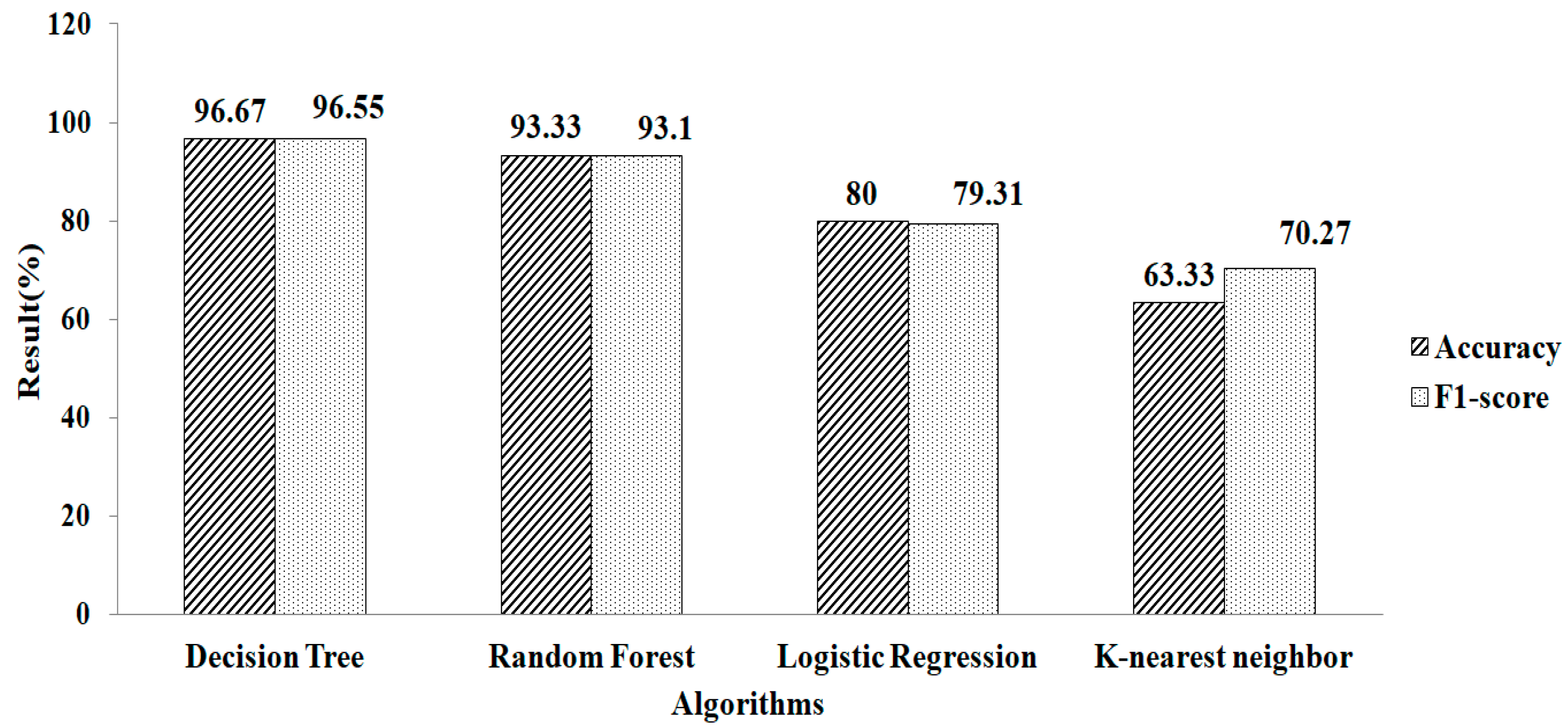
Features selection, based on using the GB classifier as an estimator, is shown in Table 2. Here, the total numbers of features selected are three. The experimental result showed that Random Forest gives a greater accuracy value and a higher F1-score value, of 95.12% and 93.75%, respectively, using the parameter values of n_estimators = 100 and criterion = ‘gini’, as shown in Figure 10. Features selected based on the GB classifier as an estimator with k-NN are shown in Table 2. Seven features are selected. The experimental result showed that the decision tree with parameter values of max_depth = 9 and random_ state = 100 give the greatest accuracy value and highest F1-score value, of 96.67% and 96.55%, respectivel, as shown in Figure 11.
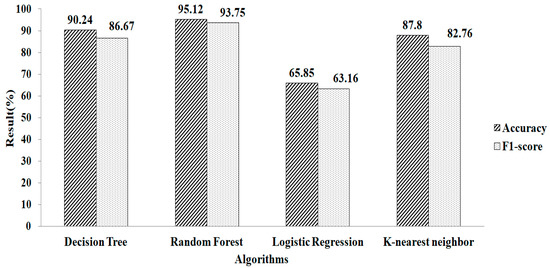
Figure 10.
Gradient boosting method for mean and mode.
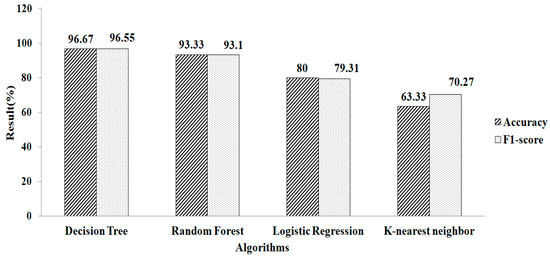
Figure 11.
Gradient Boosting method for k-NN.
Features selected based on the Random Forest classifier as an estimator, with mean and mode as the missing-data-handling method, are shown in Table 2. Seventeen features are selected. Features selected based on the Random Forest classifier as an estimator are shown in Table 2. Nine features are chosen using this method. It is noticed that the RF provides a greater accuracy value and a higher F1-score value, of 95.12% and 94.11%, respectively, when using the parameters of n_estimators = 100 and criterion = ‘gini’, as depicted in Figure 12. Similarly, the experimental result exhibits that the RF produced a 95.01% accuracy value and a 94.54% F1-score value, while using the default values of the parameters as n_estimators = 100 and criterion = ‘gini’, as shown in Figure 13.
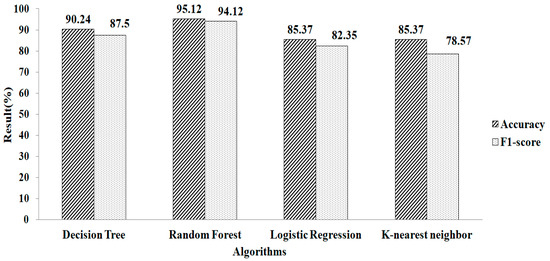
Figure 12.
Random Forest method for mean and mode.
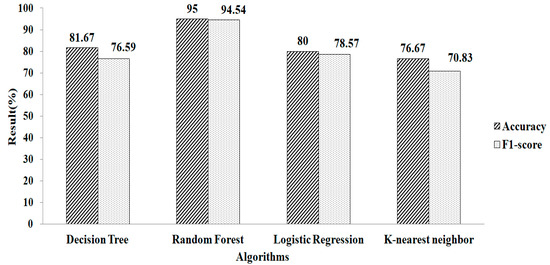
Figure 13.
Random Forest method for k-NN.
Comparing all feature-selection and classifier methods, the recursive feature-selection method using the GB classifier as an estimator is the best technique to increase the predictive model’s performance for a smaller number of selected features, with accuracy and F1-score values of 96.66% and 96.55%. RFE-GB gives fewer selected features with better accuracy with the essential elements, such as symptoms, age, alpha-fetoproteins, hemoglobin, aspartate-transaminase, iron, and ferritin, when using the k-NN method for missing-value replacement. Table 3 depicts the comparison of the feature-selection method’s performance.

Table 3.
Comparative analysis of different feature-selection techniques.
5. Comparison with Other Methods
Table 4 explores a comparative study between the present study with other proposed models.

Table 4.
Comparison of the present study with other state-of-the-art methods.
This section shows how well our feature-selection model works compared to the previous works in this field. Książek, Gandor et al. [47] used a genetic algorithm with logistic regression for feature selection and used logistic regression for classification. They used data for training and testing, achieving accuracy and F1-scores of 94.55% and 93.56%, respectively, with 22 features. Table 4 shows the results obtained from different works. The result of a work [36] based on the usage of neural networks is 75.19% accurate. The use of genetic algorithms to optimize models [46,47] resulted in a substantial improvement in the results obtained; the best accuracy of such models is 94.55%. The authors of [38] introduced a novel hybrid model that combined neighborhood-components analysis, a genetic algorithm, and a support-vector machine classifier (NCA-GA-SVM). The results revealed a classification accuracy of 96.36% and an F1-score of 95.52%. Experimental results and other research regarding HCC demonstrated that RFE-GB-RF exhibits acceptable performance. Therefore, while analyzing various metrics, it is concluded that the proposed work shows better results than other works in the literature. We used the same data for training and testing and achieved accuracy and F1-score values of 96.66% and 95.66%, respectively, with minimal features.Many other related work can be found in [38,39,46,47].
6. Statistical Analysis
To validate the performance of algorithms, we have employed the Wilcoxon test. This measure helps to assess the performance of different missing-value-replacement methods statistically. Here, the null hypothesis (H0) represents that the performance of the methods is the same, whereas the alternate hypothesis (H1) confirms a significant difference in their performance. A significance level of 5% (α = 0.05) is used to perform the test, and a p-value is used to confirm the hypothesis. The null hypothesis is rejected when the p-value is low. Otherwise, it is accepted. Table 5 depicts the p-values of this test for RFE-GB against each method.

Table 5.
p-values of Wilcoxon test of the methods.
We observe that RFE-GB-RF with k-NN data imputation performed better than other methods.
7. Conclusions
The main objective of this work is to improve the survival classification of hepatocellular carcinoma, with minimal features. Initially, missing values are replaced using mean and mode, and k-NN is performed. Then, different feature-selection methods using the wrapper and embedded methods are applied. The wrapper method is based on the RFE with GB and random forests. The embedded method is based on logistic regression using LASSO and ridge regression. Experimental results show that the embedded method using the RFE-GB estimator gives the most accurate results, compared to other feature-selection methods, with accuracy and F1-score values of 96.66% and 95.66%, respectively. Future work would be to identify different cancer variants using deep learning and feature-selection techniques on large datasets.
Author Contributions
Conceptualization, F.R.P.P., R.R.R., S.M., S.S., D.S.A.E., I.A.E.G., L.A., R.O. and M.A.S.A.; data curation, K.S. and M.A.S.A.; formal analysis, F.R.P.P., R.R.R., S.M., S.S., K.S., D.S.A.E., I.A.E.G., L.A., R.O. and M.A.S.A.; funding acquisition, F.R.P.P. and M.A.S.A.; investigation, F.R.P.P., R.R.R. and D.S.A.E.; methodology, F.R.P.P., R.R.R., S.M., S.S., K.S., D.S.A.E., I.A.E.G., L.A., R.O. and M.A.S.A.; project administration, F.R.P.P., D.S.A.E. and M.A.S.A.; resources, F.R.P.P., S.M., S.S., K.S., D.S.A.E. and I.A.E.G.; software, S.M. and L.A.; supervision, F.R.P.P., R.R.R., S.S., K.S., D.S.A.E. and R.O.; validation, S.M., L.A. and M.A.S.A.; visualization, F.R.P.P., S.M., D.S.A.E. and I.A.E.G.; writing—original draft, F.R.P.P., R.R.R., S.M., S.S., K.S., D.S.A.E., I.A.E.G., L.A., R.O. and M.A.S.A.; writing—review and editing, F.R.P.P., R.R.R., S.M., S.S., K.S., D.S.A.E., I.A.E.G., L.A., R.O. and M.A.S.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Deanship of Scientific Research, King Faisal University, grant number GRANT712, and the APC was funded by the Deanship of Scientific Research, King Faisal University.
Informed Consent Statement
Not applicable.
Acknowledgments
This work was supported by the Deanship of Scientific Research, King Faisal University, Saudi Arabia, grant number GRANT712.
Conflicts of Interest
The authors declare no potential conflict of interest.
Abbreviations
| DT | Decision Tree |
| FCNN | Fuzzy Convolutional Neural Network (FCNN) |
| GA | Genetic Algorithm |
| GAO | Genetic Algorithm Optimization |
| HCC | Hepatocellular Carcinoma |
| k-NN | k-Nearest Neighbor |
| LASSO | Least Absolute Shrinkage and Selection Operator |
| NCA | Neighborhood Components Analysis |
| RF | Random Forest |
| RFE | Recursive Feature Elimination |
| RFE-GB | Recursive Feature Elimination with Gradient Boosting |
| SMOTE | Synthetic Minority Over-sampling Technique |
References
- Abdar, M.; Zomorodi-Moghadam, M.; Das, R.; Ting, I.-H. Performance analysis of classification algorithms on early detection of liver disease. Expert Syst. Appl. 2017, 67, 239–251. [Google Scholar] [CrossRef]
- Akter, L.; Islam, M.M. Hepatocellular Carcinoma Patient’s Survival Prediction Using Oversampling and Machine Learning Techniques. In Proceedings of the 2021 2nd International Conference on Robotics, Electrical and Signal Processing Techniques (ICREST), Dhaka, Bangladesh, 5–7 January 2021; IEEE: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Ali, L.; Khelil, K.; Wajid, S.K.; Hussain, Z.U.; Shah, M.A.; Howard, A.; Adeel, A.; Shah, A.A.; Sudhakar, U.; Howard, N. Machine learning based computer-aided diagnosis of liver tumours. In Proceedings of the 2017 IEEE 16th International Conference on Cognitive Informatics & Cognitive Computing (ICCI* CC), Oxford, UK, 26–28 July 2017; IEEE: New York, NY, USA, 2017. [Google Scholar] [CrossRef]
- Amutha, M.J.; Soundar, K.R.; Piramu, M.; Murugesan, K. A Survey on Machine Learning Algorithms for Cardiovascular Diseases Predic-tion. IJIRMPS 2021, 9, 45–48. [Google Scholar] [CrossRef]
- Bralet, M.-P.; Regimbeau, J.-M.; Pineau, P.; Dubois, S.; Loas, G.; Degos, F.; Valla, D.; Belghiti, J.; Degott, C.; Terris, B. Hepatocellular carcinoma occurring in nonfibrotic liver: Epidemiologic and histopathologic analysis of 80 French cases. Hepatology 2000, 32, 200–204. [Google Scholar] [CrossRef] [PubMed]
- Cawley, G.C. Causal & non-causal feature selection for ridge regression. In Proceedings of the Workshop on the Causation and Prediction Challenge at WCCI, Hong Kong, China, 1–6 June 2008; PMLR: Bristol, UK, 2008. [Google Scholar]
- Chandrakar, P.; Shrivas, A.; Sahu, N. Design of a Novel Ensemble Model of Classification Technique for Gene-Expression Data of Lung Cancer with Modified Genetic Algorithm. EAI Endorsed Trans. Pervasive Health Technol. 2021, 7, e2. [Google Scholar] [CrossRef]
- Chaturvedi, A.; Gupta, A.; Rajpoot, V. Parameterized Comparison of Regularized Regression Models to Develop Models for Real Estate. In IOP Conference Series: Materials Science and Engineering; IOP Publishing: Bristol, UK, 2021. [Google Scholar] [CrossRef]
- Ding, Y.; Wilkins, D. Improving the Performance of SVM-RFE to Select Genes in Microarray Data. In BMC Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2006. [Google Scholar] [CrossRef]
- Dong, R.; Yang, X.; Zhang, X.; Gao, P.; Ke, A.; Sun, H.-C.; Zhou, J.; Fan, J.; Cai, J.; Shi, G. Predicting overall survival of patients with hepatocellular carcinoma using a three-category method based on DNA methylation and machine learning. J. Cell. Mol. Med. 2019, 23, 3369–3374. [Google Scholar] [CrossRef]
- Duan, K.-B.; Rajapakse, J.C.; Nguyen, M.N. One-Versus-One and One-Versus-All Multiclass SVM-RFE for Gene Selection in Cancer Classification. In European Conference on Evolutionary Computation, Machine Learning and Data Mining in Bioinformatics; Springer: Berlin/Heidelberg, Germany, 2007. [Google Scholar] [CrossRef]
- Etzioni, R.; Urban, N.; Ramsey, S.D.; McIntosh, M.W.; Schwartz, S.M.; Reid, B.J.; Radich, J.P.; Anderson, G.; Hartwell, L. The case for early detection. Nat. Cancer 2003, 3, 243–252. [Google Scholar] [CrossRef]
- Ghazikhani, A.; Yazdi, H.S.; Monsefi, R. Class imbalance handling using wrapper-based random oversampling. In Proceedings of the 20th Iranian Conference on Electrical Engineering (ICEE2012), Tehran, Iran, 15–17 May 2012; IEEE: New York, NY, USA, 2012. [Google Scholar] [CrossRef]
- Grando-Lemaire, V.; Guettier, C.; Chevret, S.; Beaugrand, M.; Trinchet, J.-C. Hepatocellular carcinoma without cirrhosis in the West: Epidemiological factors and histopathology of the non-tumorous liver. J. Hepatol. 1999, 31, 508–513. [Google Scholar] [CrossRef]
- Granitto, P.; Furlanello, C.; Biasioli, F.; Gasperi, F. Recursive feature elimination with random forest for PTR-MS analysis of agroindustrial products. Chemom. Intell. Lab. Syst. 2006, 83, 83–90. [Google Scholar] [CrossRef]
- Guo, Y.; Chung, F.-L.; Li, G.; Zhang, L. Multi-Label Bioinformatics Data Classification with Ensemble Embedded Feature Selection. IEEE Access 2019, 7, 103863–103875. [Google Scholar] [CrossRef]
- Hashem, S.; ElHefnawi, M.; Habashy, S.; El-Adawy, M.; Esmat, G.; El-Akel, W.; Abdelazziz, A.O.; Nabeel, M.M.; Abdelmaksoud, A.H.; Elbaz, T.M.; et al. Machine Learning Prediction Models for Diagnosing Hepatocellular Carcinoma with HCV-related Chronic Liver Disease. Comput. Methods Programs Biomed. 2020, 196, 105551. [Google Scholar] [CrossRef]
- Hjerpe, A. Computing Random Forests Variable Importance Measures (Vim) on Mixed Numerical and Categorical Data; DiVA: Umeå, Sweden, 2016. [Google Scholar]
- Jeyalakshmi, K. Weighted Synthetic Minority Over-Sampling Technique (WSMOTE) Algorithm and Ensemble Classifier for Hepatocellular Carcinoma (HCC) In Liver Disease System. Turk. J. Comput. Math. Educ. 2021, 12, 7473–7487. [Google Scholar] [CrossRef]
- Jeyalakshmi, R.R. Intelligence Ensemble-Based Feature Selection (Iefs) Algorithm and Fuzzy Convolutional Neural Network (Fcnn) for Hepatocellular Carcinoma (Hcc) in Liver Disease System. Ann. Rom. Soc. Cell Biol. 2021, 25, 4759–4782. [Google Scholar]
- Joshi, J.; Doshi, R.; Patel, J. Diagnosis and prognosis breast cancer using classification rules. Int. J. Eng. Res. Gen. Sci. 2014, 2, 315–323. [Google Scholar]
- Karegowda, A.G.; Manjunath, A.S.; Jayaram, M.A. Feature Subset Selection Problem using Wrapper Approach in Supervised Learning. Int. J. Comput. Appl. 2010, 1, 13–17. [Google Scholar] [CrossRef]
- Khan, I.U.; Aslam, N.; Alshehri, R.; Alzahrani, S.; Alghamdi, M.; Almalki, A.; Balabeed, M. Cervical Cancer Diagnosis Model Using Extreme Gradient Boosting and Bioinspired Firefly Optimization. Sci. Program. 2021, 2021, 5540024. [Google Scholar] [CrossRef]
- Kim, W.R. Epidemiology of hepatitis B in the United States. Hepatology 2009, 49, S28–S34. [Google Scholar] [CrossRef] [PubMed]
- Koh, K.; Kim, S.-J.; Boyd, S. An interior-point method for large-scale l1-regularized logistic regression. J. Mach. Learn. Res. 2007, 8, 1519–1555. [Google Scholar]
- Ali, M.A.; Balasubramanian, K.; Krishnamoorthy, G.D.; Muthusamy, S.; Pandiyan, S.; Panchal, H.; Mann, S.; Thangaraj, K.; El-Attar, N.E.; Abualigah, L.; et al. Classification of Glaucoma Based on Elephant-Herding Optimization Algorithm and Deep Belief Network. Electronics 2022, 11, 1763. [Google Scholar] [CrossRef]
- Houssein, E.H.; Abdelminaam, D.S.; Ibrahim, I.E.; Hassaballah, M.; Wazery, Y.M. A hybrid heartbeats classification approach based on marine predators algorithm and convolution neural networks. IEEE Access 2021, 9, 86194–86206. [Google Scholar] [CrossRef]
- Liu, Y.-X.; Liu, X.; Cen, C.; Li, X.; Liu, J.-M.; Ming, Z.-Y.; Yu, S.-F.; Tang, X.-F.; Zhou, L.; Yu, J.; et al. Comparison and development of advanced machine learning tools to predict nonalcoholic fatty liver disease: An extended study. Hepatobiliary Pancreat. Dis. Int. 2021, 20, 409–415. [Google Scholar] [CrossRef]
- Marinho, R.T.; Giria, J.; Moura, M.C. Rising costs and hospital admissions for hepatocellular carcinoma in Portugal (1993-2005). World J. Gastroenterol. WJG 2007, 13, 1522–1527. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Muthukrishnan, R.; Rohini, R. LASSO: A feature selection technique in predictive modeling for machine learning. In Proceedings of the 2016 IEEE International Conference on Advances in Computer Applications (ICACA), Coimbatore, India, 24 October 2016; IEEE: New York, NY, USA, 2016. [Google Scholar] [CrossRef]
- Venkatachalam, K.; Prabhu, P.; Balaji, B.S.; Abouhawwash, M.; Rajadev, R. Recursive Feature Elimination with Ridge Regression (L2) Machine Learning Hybrid Feature Selection Algorithm for Diabetic Prediction Using Random Forest Classifer. 2021. Available online: https://www.researchsquare.com/article/rs-742641/v1 (accessed on 17 May 2022).
- Pan, R.; Yang, T.; Cao, J.; Lu, K.; Zhang, Z. Missing data imputation by K nearest neighbours based on grey relational structure and mutual information. Appl. Intell. 2015, 43, 614–632. [Google Scholar] [CrossRef]
- Ren, S.; Qi, Q.; Liu, S.; Duan, S.; Mao, B.; Chang, Z.; Zhang, Y.; Wang, S.; Zhang, L. Preoperative prediction of pathological grading of hepatocellular carcinoma using machine learning-based ultrasomics: A multicenter study. Eur. J. Radiol. 2021, 143, 109891. [Google Scholar] [CrossRef] [PubMed]
- Santos, M.S.; Abreu, P.H.; García-Laencina, P.J.; Simão, A.; Carvalho, A. A new cluster-based oversampling method for improving survival prediction of hepatocellular carcinoma patients. J. Biomed. Inform. 2015, 58, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Sawhney, R.; Mathur, P.; Shankar, R. A Firefly Algorithm Based Wrapper-Penalty Feature Selection Method for Cancer Diagnosis. In International Conference on Computational Science and Its Applications; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Książek, W.; Abdar, M.; Acharya, U.R.; Pławiak, P. A novel machine learning approach for early detection of hepatocellular carcinoma patients. Cogn. Syst. Res. 2019, 54, 116–127. [Google Scholar] [CrossRef]
- Ali, L.; Wajahat, I.; Golilarz, N.A.; Keshtkar, F.; Bukhari, S.A.C. LDA–GA–SVM: Improved hepatocellular carcinoma prediction through dimensionality reduction and genetically optimized support vector machine. Neural Comput. Appl. 2021, 33, 2783–2792. [Google Scholar] [CrossRef]
- Dessie, E.Y.; Tu, S.-J.; Chiang, H.-S.; Tsai, J.J.; Chang, Y.-S.; Chang, J.-G.; Ng, K.-L. Construction and Validation of a Prognostic Gene-Based Model for Overall Survival Prediction in Hepatocellular Carcinoma Using an Integrated Statistical and Bioinformatic Approach. Int. J. Mol. Sci. 2021, 22, 1632. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, B.; Youn, S.Y.; Kim, H.; Choi, J.-I. Diagnostic Performance of KLCA-NCC 2018 Criteria for Hepatocellular Carcinoma Using Magnetic Resonance Imaging: A Systematic Review and Meta-Analysis. Diagnostics 2021, 11, 1763. [Google Scholar] [CrossRef]
- Książek, W.; Hammad, M.; Pławiak, P.; Acharya, U.R.; Tadeusiewicz, R. Development of novel ensemble model using stacking learning and evolutionary computation techniques for automated hepatocellular carcinoma detection. Biocybern. Biomed. Eng. 2020, 40, 1512–1524. [Google Scholar] [CrossRef]
- Hattab, M.; Maalel, A.; Ben Ghezala, H.H. Towards an Oversampling Method to Improve Hepatocellular Carcinoma Early Prediction. In Digital Health in Focus of Predictive, Preventive and Personalised Medicine; Springer: Berlin/Heidelberg, Germany, 2020; pp. 139–148. [Google Scholar] [CrossRef]
- Tuncer, T.; Ertam, F. Neighborhood component analysis and reliefF based survival recognition methods for Hepatocellular carcinoma. Phys. A Stat. Mech. Its Appl. 2020, 540, 123143. [Google Scholar] [CrossRef]
- Książek, W.; Gandor, M.; Pławiak, P. Comparison of various approaches to combine logistic regression with genetic algorithms in survival prediction of hepatocellular carcinoma. Comput. Biol. Med. 2021, 134, 104431. [Google Scholar] [CrossRef] [PubMed]
- Książek, W.; Turza, F.; Pławiak, P. NCA-GA-SVM: A new two-level feature selection method based on neighborhood component analysis and genetic algorithm in hepatocellular carcinoma (HCC) fatality prognosis. Int. J. Numer. Methods Biomed. Eng. 2022, 38, e3599. [Google Scholar] [CrossRef] [PubMed]
- Akter, L.; Islam, M.; Al-Rakhami, M.S.; Haque, M. Prediction of cervical cancer from behavior risk using machine learning techniques. SN Comput. Sci. 2021, 2, 1–10. [Google Scholar]
- Mroweh, M.; Decaens, T.; Marche, P.N.; Jilkova, Z.M.; Clément, F. Modulating the Crosstalk between the Tumor and Its Microenvironment Using RNA Interference: A Treatment Strategy for Hepatocellular Carcinoma. Int. J. Mol. Sci. 2020, 21, 5250. [Google Scholar] [CrossRef]
- Liu, Z.; Thapa, N.; Shaver, A.; Roy, K.; Siddula, M.; Yuan, X.; Yu, A. Using Embedded Feature Selection and CNN for Classification on CCD-INID-V1—A New IoT Dataset. Sensors 2021, 21, 4834. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).