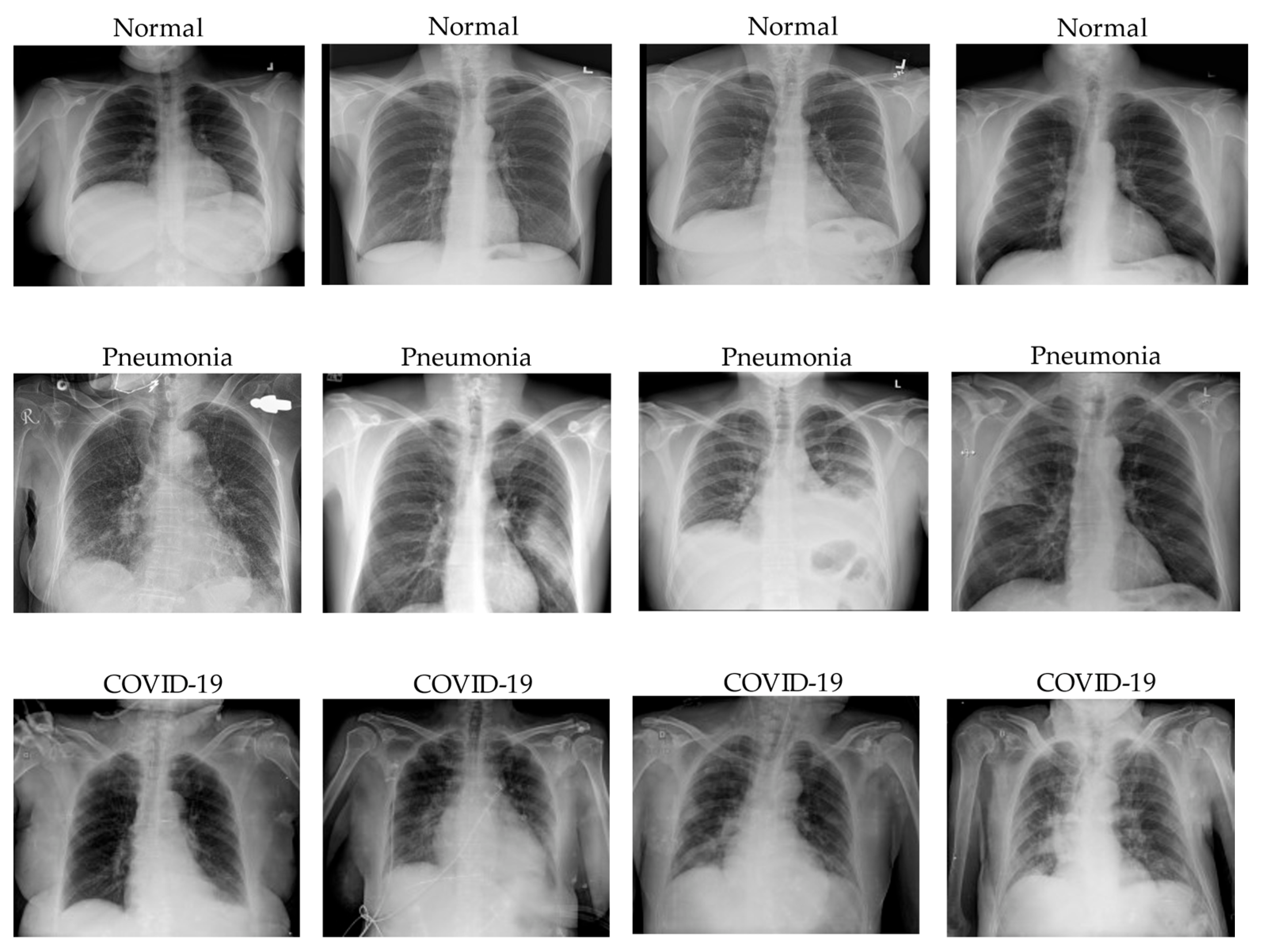
3.1. COVID-19 Classification Results
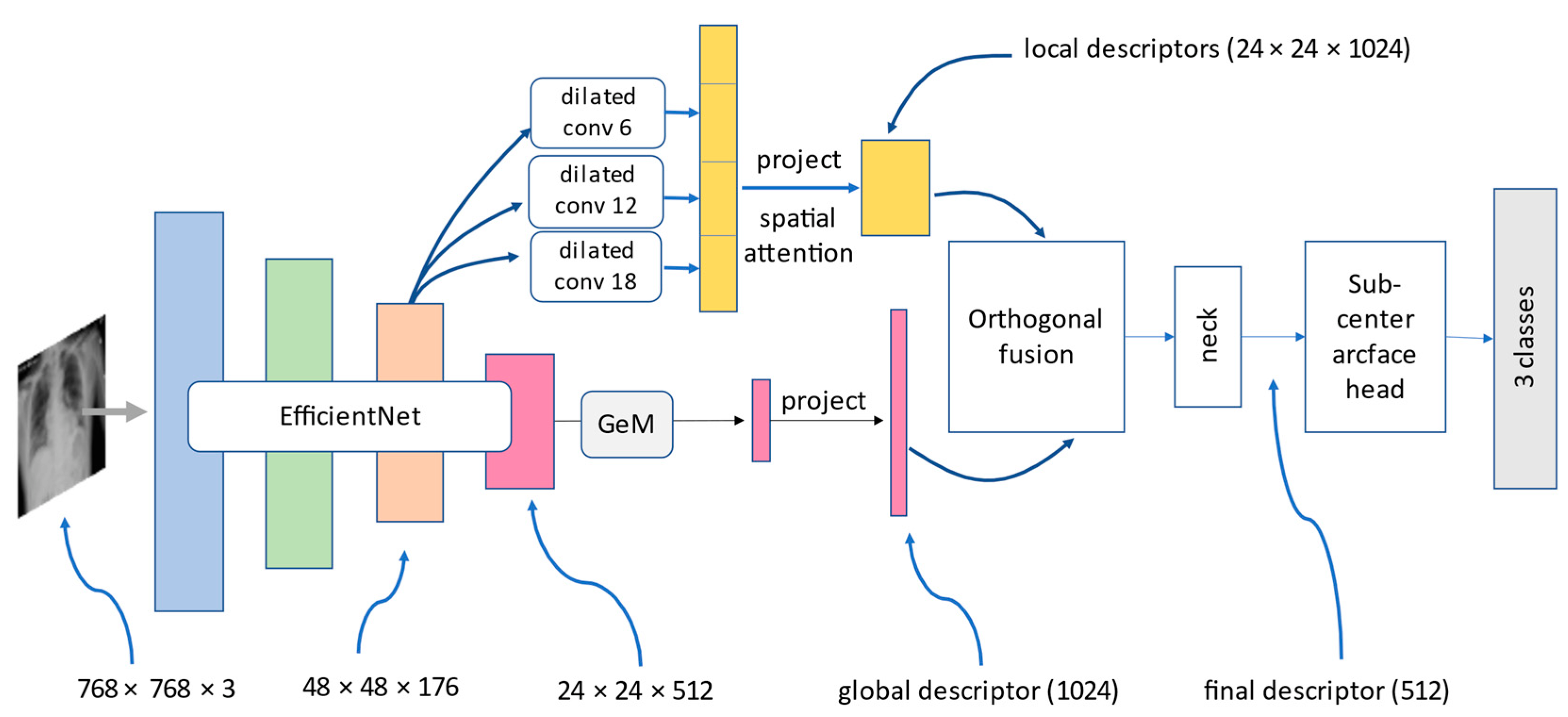
We trained three convolution-based and two transformer-based models on the customized COVID-19 classification dataset. The output results were evaluated based on three metrics, which were precision, recall, and F1-score with respect to COVID-19, normal, pneumonia, macro-average, and micro-average. Each tables below contained numerical results of three metrics and will be described in detail. The two models which showed the best results were DenseNet121 and Hybrid EfficientNet-DOLG.
Table 4 compares the performance of five deep learning models based on the precision metric score. The numerical results illustrate each category and on macro- and micro-average. For the COVID-19 precision score, the best model was found to be Swin Transformer with a score of 0.99, and for normal images, we found Hybrid EfficientNet-DOLG a top score of 0.93, and for pneumonia, the DensNet121 model produced the highest result (0.94). For macro-average and micro-average, the Hybrid EfficientNet-DOLG was found to lead both metrics with scores of 0.95 and 0.96, respectively. The hierarchical architecture of Swin Transformer computed image representation in different scales which works best on COVID-19. On the other hand, the concatenate mechanism of DensNet boosted the pneumonia detection of DenseNet. The hybrid architecture helped Hybrid EfficientNet-DOLG perform best results on normal, macro average, and micro average.
As shown in
Table 5, we could see the outstanding performance of Hybrid EfficientNet-DOLG as it produced the highest score on three metrics: pneumonia, macro-average, and micro-average, with scores of 0.95, 0.96, and 0.96, respectively. For the COVID-19 category, we found the DenseNet121 to have a recall score of 0.98 and the second highest score was found to be Hybrid EfficientNet Transformer which was 0.97, only smaller than a 0.01 gap. For normal, we found Swin Transformer to have a score of 0.97 and the second score was DenseNet 121 and Hybrid EfficientNet-DOLG, which was 0.94. The other models, including ResNet50 and InceptionNet, also computed comparable results. With respect to recall, the dense architecture of DenseNet worked best on COVID-19 images, and different from precision metric, the hierarchy architecture of Swin Transformer achieved the best results on normal images. The combination of EfficientNet and DOLG took the best place on pneumonia, macro average, and micro average.
The third metric was the F1-score; in this metric, we could see the pattern that Hybrid EfficientNet produced the best results on all scores except the normal category, in which the top result belonged to DenseNet with a score of 0.95. For COVID-19, pneumonia, macro-average, and micro-average the Hybrid EfficientNet-DOLG produced the highest results with scores of 0.99, 0.94, 0.95, and 0.96, respectively. For COVID-19, pneumonia, macro-average, and micro-average, the second highest model was DensNet121, which produced scores of 0.98, 0.93, 0.94, and 0.95 sequentially. Different from precision and recall, the classification performance of DenseNet secured best on normal images, this might be because the dense connections performed well for normal cases regarding the F1-score, then the compound of EfficientNet and DOLG did efficiently on other categories.
From
Table 6, we can conclude that Hybrid EfficientNet-DOLG and DenseNet models are the best models for COVID-19 classification tasks on our customized dataset.
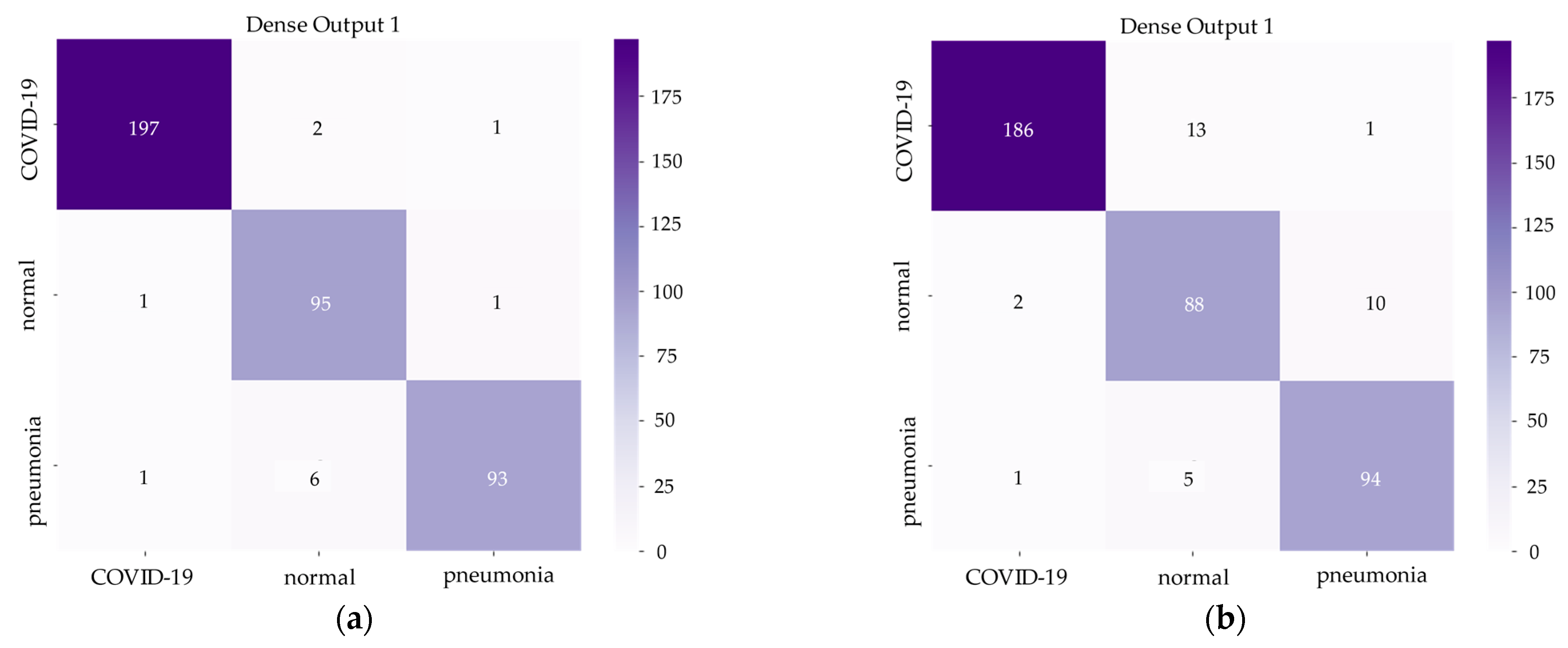
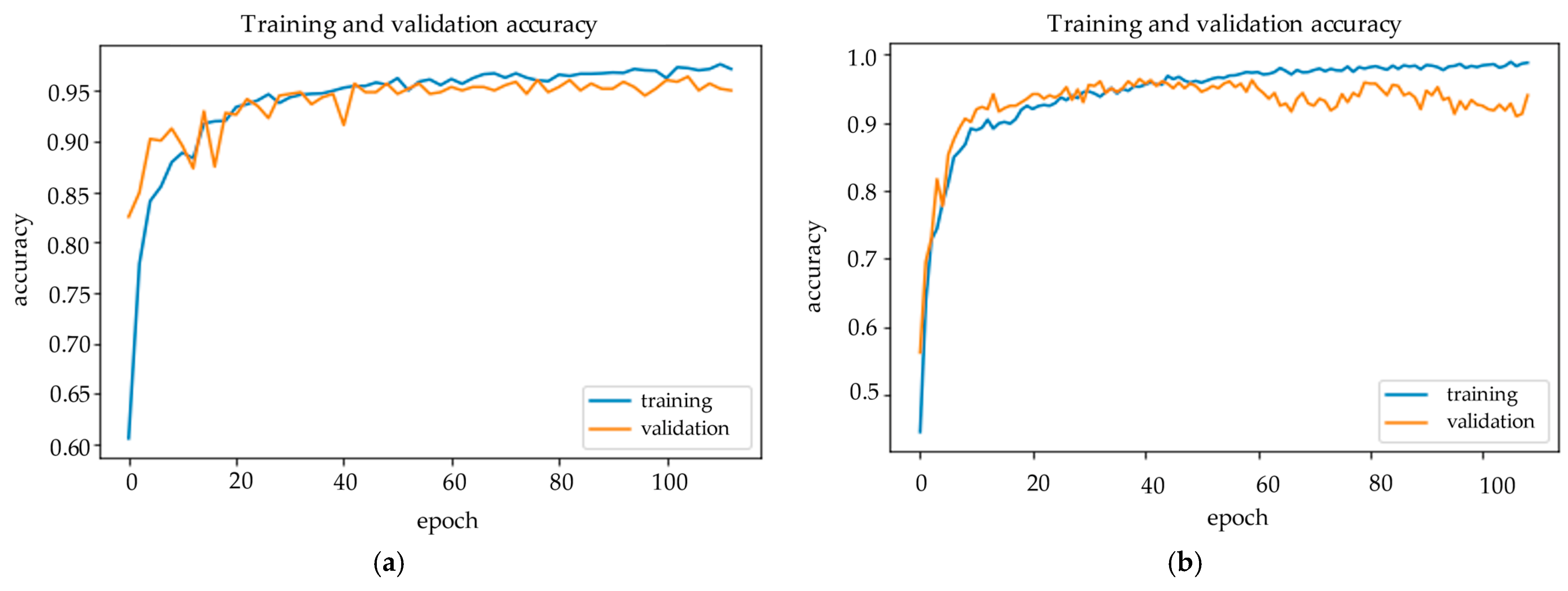
Figure 6 and
Figure 7 demonstrate the confusion matrix of inference results on the test set and training history of the Hybrid EfficientNet-DOLG and DenseNet models.
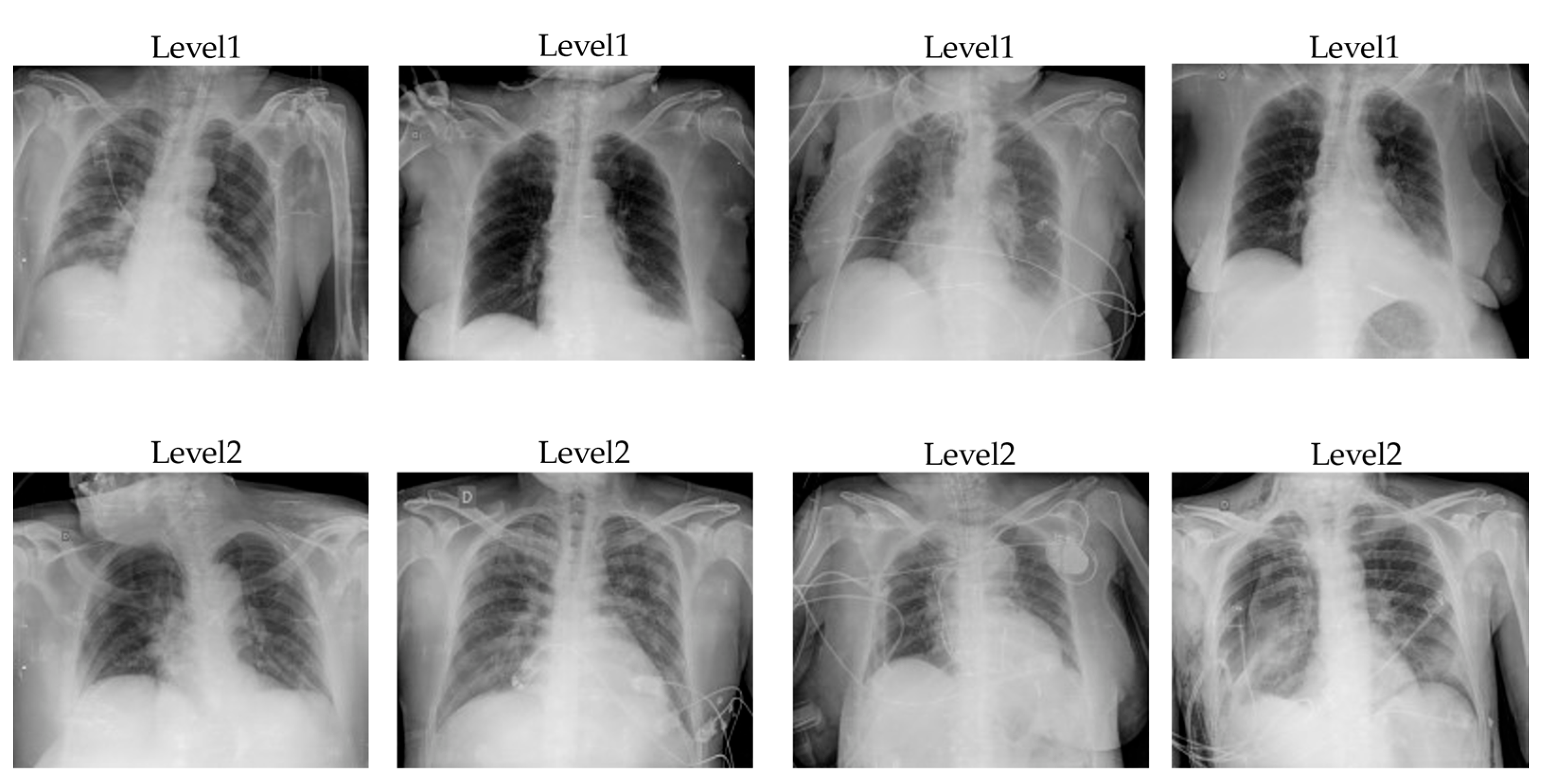
3.2. Severity Assessment Results
With the COVID-19 severity assessment task, we also trained the customized COVID-19 severity dataset with five models: DenseNet121, ResNet50, InceptionNet, Swin Transformer, and Hybrid EffificientNet-DOLG. The dataset contains images of two categories, level1 and level2, in which level1 indicates the patient severity is normal and the patient can self-quarantine at home without requiring a further treatment response. Level2 indicates that the patients need to have further support and need to go to the hospital for a treatment response because the pneumonia extent of COVID-19 damage is large and severe.
After training models for hours, we obtained the output results as shown in
Table 7,
Table 8 and
Table 9. We also evaluated severity assessment results on three metrics: precision, recall, and F1-score with respect to level1, level2, macro-average, and micro-average, the same as the COVID-19 classification task. A detailed analysis of deep learning models’ performance on COVID-19 severity assessment is presented for each metric under every table.
From
Table 7, we can see that with level1, DenseNet121 output had the top precision score result of 0.76, and level2 Hybrid EfficientNet-DOLG produced a precision score of 0.87. The Swin Transformer led the macro-average with a 0.81 precision score, and with respect to the micro-average, the Hybrid Efficient-DOLG also produced the highest score of 0.82. The dense connection of DenseNet helped to reduce gradients vanishing which improve the precision metric of DenseNet on level1 chest X-ray images. The Swin Transformer produced the best on macro average because the accuracy of level1 and level2 is very high, and Hybrid EfficientNet-DOLG surpassed other neural nets on micro average because its precision on level2 is the highest and on level1 almost equal DenseNet.
When comparing the precision score, we could see that the Hybrid EfficientNet-DOLG outperformed on all three categories: level1, macro-average, and micro-average with the score of 0.75, 0.80, and 0.82. The DenseNet121 produced the highest score of level2 (0.89) on the recall metric. The dense connection mechanism of DenseNet is sensitive to level2 chest X-ray images that made up the highest score for DenseNet on the recall metric. The EfficientNet encoder of the hybrid network produces overall superb performance on both level1, macro-average, and micro-average. Swin Transformer and other two convolution-based neural nets, ResNet50 and InceptionNet, also achieved comparable results with DenseNet and Hybrid EfficientNet-DOLG.
The last metric we analyzed was F1-score. In this metric, Hybrid EfficientNet-DOLG outperformed all four categories: level1, level2, macro-average, and micro-average, with scores of 0.74, 0.86, 0.80, and 0.82. The top results on the F1-score of Hybrid EfficientNet-DOLG are based on the robustness of EfficientNet as an encoder of the structure and combine with the global and local descriptor of DOLG. DenseNet also produced high output results with scores of 0.71 on level1, 0.86 on level2, 0.79 on macro-average, and 0.81 on micro-average. Output of DenseNet was only smaller than Hybrid EfficientNet-DOLG with 0.01 score gaps on level1, macro-average, and micro-average and produce the equal result on level2. Other neural nets also achieved comparable results on F1-score with DenseNet and Hybrid EfficientNet-DOLG.
Overall, we can conclude that Hybrid EfficientNet-DOLG and DenseNet are two models which produce the highest inference results on the COVID-19 severity assessment task; it is the same pattern as the performance on the COVID-19 classification task.
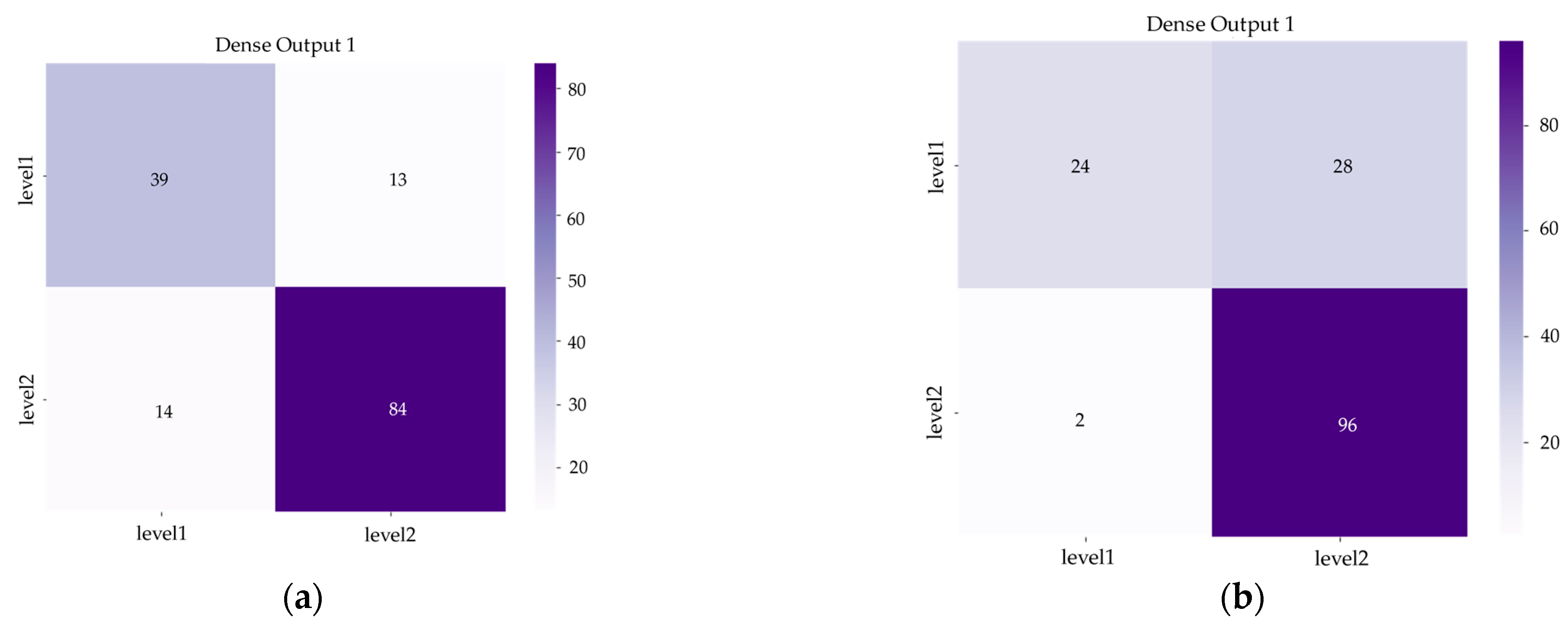
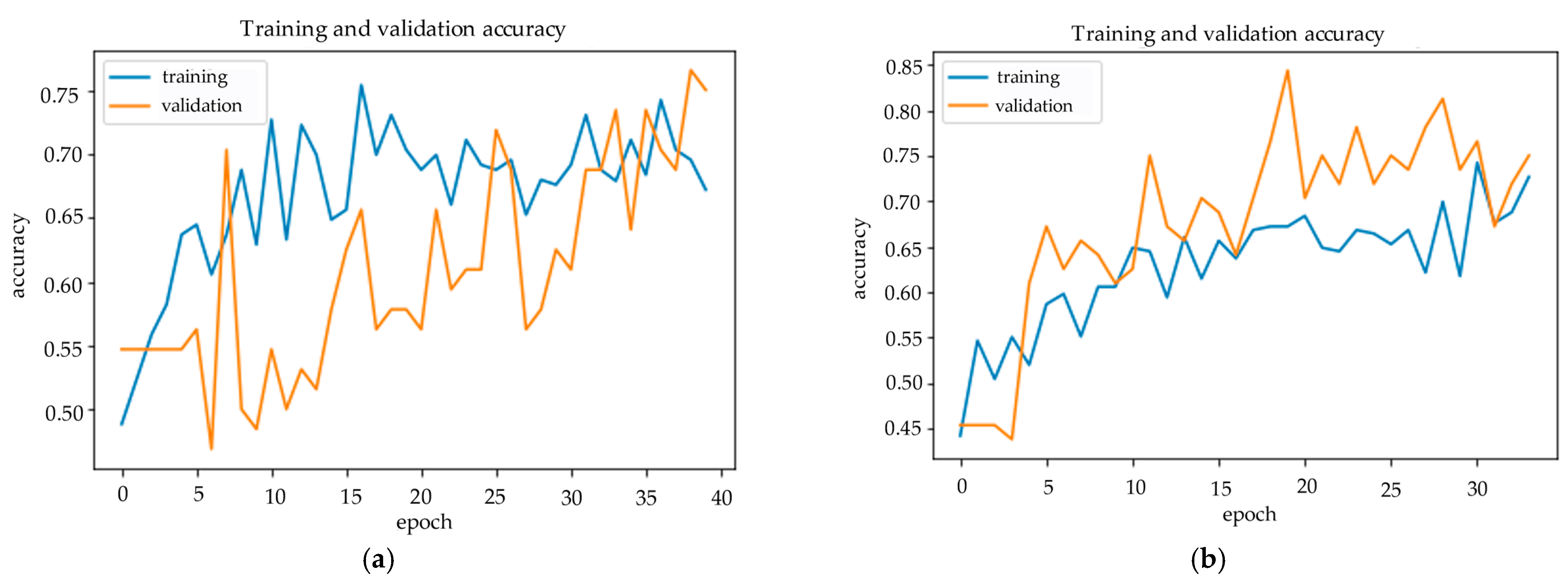
Figure 8 and
Figure 9 show us the confusion matrix and training history of DenseNet and Hybrid EfficientNet-DOLG after inferencing and training on the COVID-19 severity assessment dataset.
There are many deep learning architecture and deep transfer learning techniques that could apply to X-ray imagery as a potential method for classifying COVID-19 detection and severity assessment. Some of the architecture techniques include Wide Residual Networks [
61] (WRNs) and Visual Geometry Group [
62] (VGG). Wide Residual Networks are variants of ResNet, which both increase the width and decrease the depth of residual networks, and also create lightweight models with high performance. With only 16 layers, the network can outperform another convolutional neural networks with over 1000 layers on CIFAR and ImageNet datasets. The second network is VGG, which won the ImageNet Challenge 2014 and was first and second in terms of localization and classification, respectively. We have VGG16 and VGG19, which represent 16 layers and 19 layers of neural networks. In general, the architecture of VGG includes input layers, convolutional layers, hidden layers, and fully connected layers, and depending on the architecture we use, that the number of layers might be different. There is research that studies the application of deep transfer learning, including the work of Naushad et al. [
63] in which the author efficiently implemented deep transfer learning techniques for land use and land cover classification based on WSNs and VGG pre-trained models. Another study by Das et al. [
64] tried to apply deep transfer learning automatically to detect COVID-19 based on chest X-ray images.
This study has some limitations. First, we collected data from many open-source datasets; to some extent this might affect the model accuracy. Because X-ray images obtained from different machines have various image qualities, image color channels as well as resolutions, these factors have significantly impact on the model training pipeline. Another shortcoming of this study was the severity assessment levels; to have a more precise treatment response for patients, having many classes of severity is better than having few classes. In this study, we only focused on two classes, level1 and level2; this is not as detailed as it could be with more levels for severity diagnosis, which means a more appropriate treatment response could be designed. The last disadvantage of this paper is that we did not propose a deep learning model to customize for our chest X-ray dataset. We only used a built-in model from available libraries, which was then not as efficient as we trained on X-ray imagery. In future work, we aim to build a model that is lightweight, robust, and has a lower computational complexity for X-ray image classification tasks. We used five deep neural networks with computational complexity as follows: Swin Transformer 1038 (Giga FLOPs), InceptionNet (24.57 Giga FLOPs), Hybrid EfficientNet-DOLG (9.9 Giga FLOPs), DenseNet121 (5.69 Giga FLOPs), and ResNet50 (3.8 Giga FLOPs). We will design a network with computational complexity approximate to 10 Giga FLOPs for an efficient and robust model.