Hyperspectral Chemical Imaging of Single Bacterial Cell Structure by Raman Spectroscopy and Machine Learning
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Cultivation
2.2. Sample Preparation for Raman Analysis
2.3. Bacterial Characterization by Raman Imaging
2.4. Data Analysis and Multivariate Modelling
3. Results and Discussion
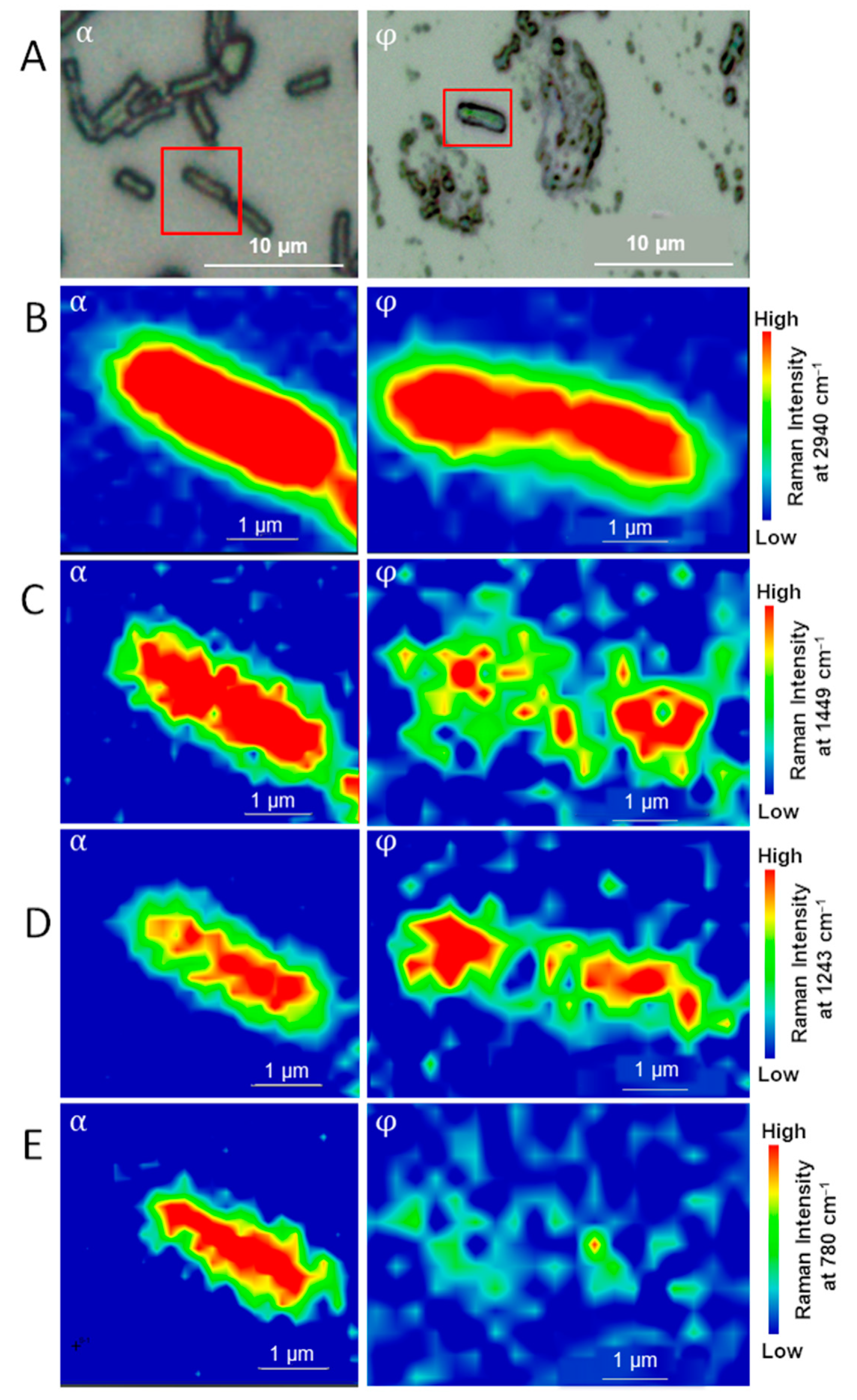
3.1. Sub-Diffraction Raman Imaging on Single Cells of E. coli
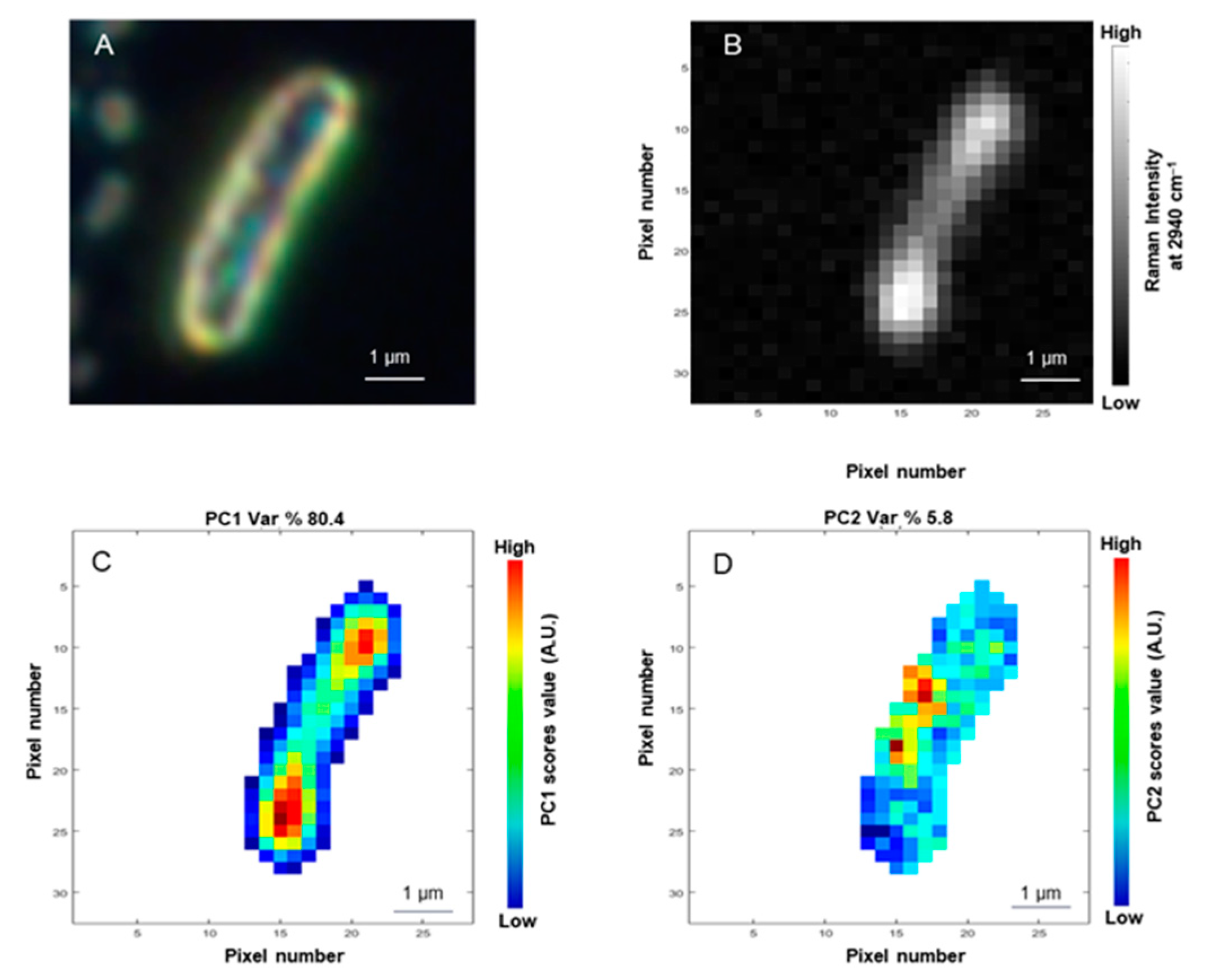
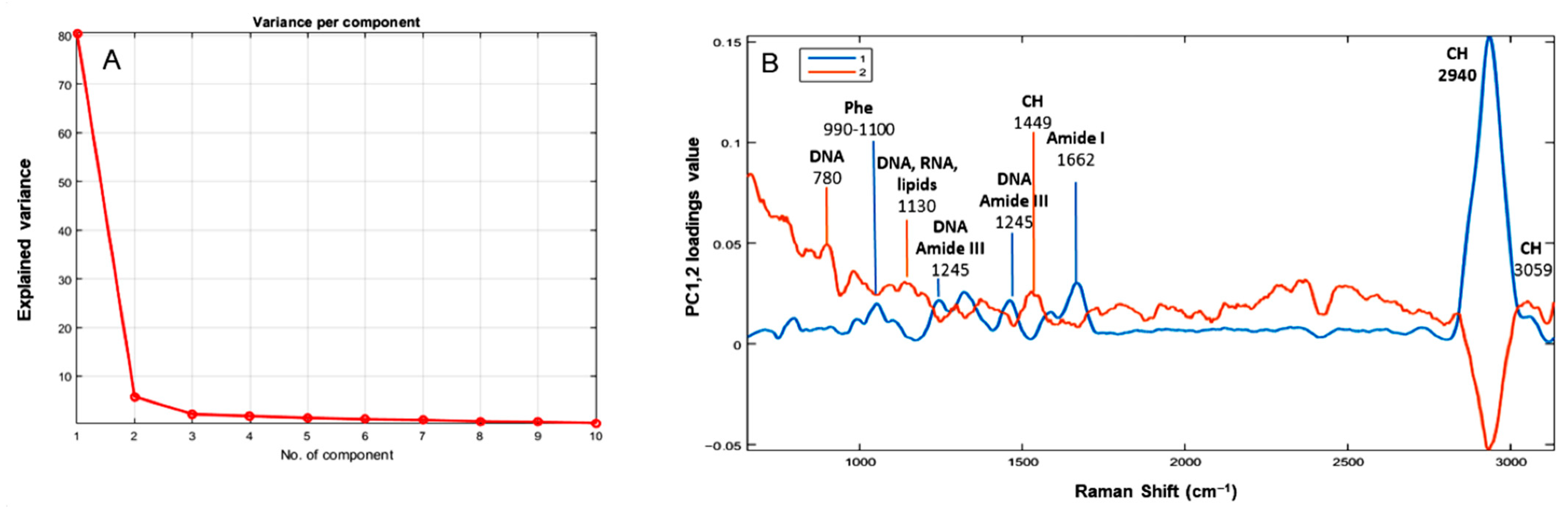
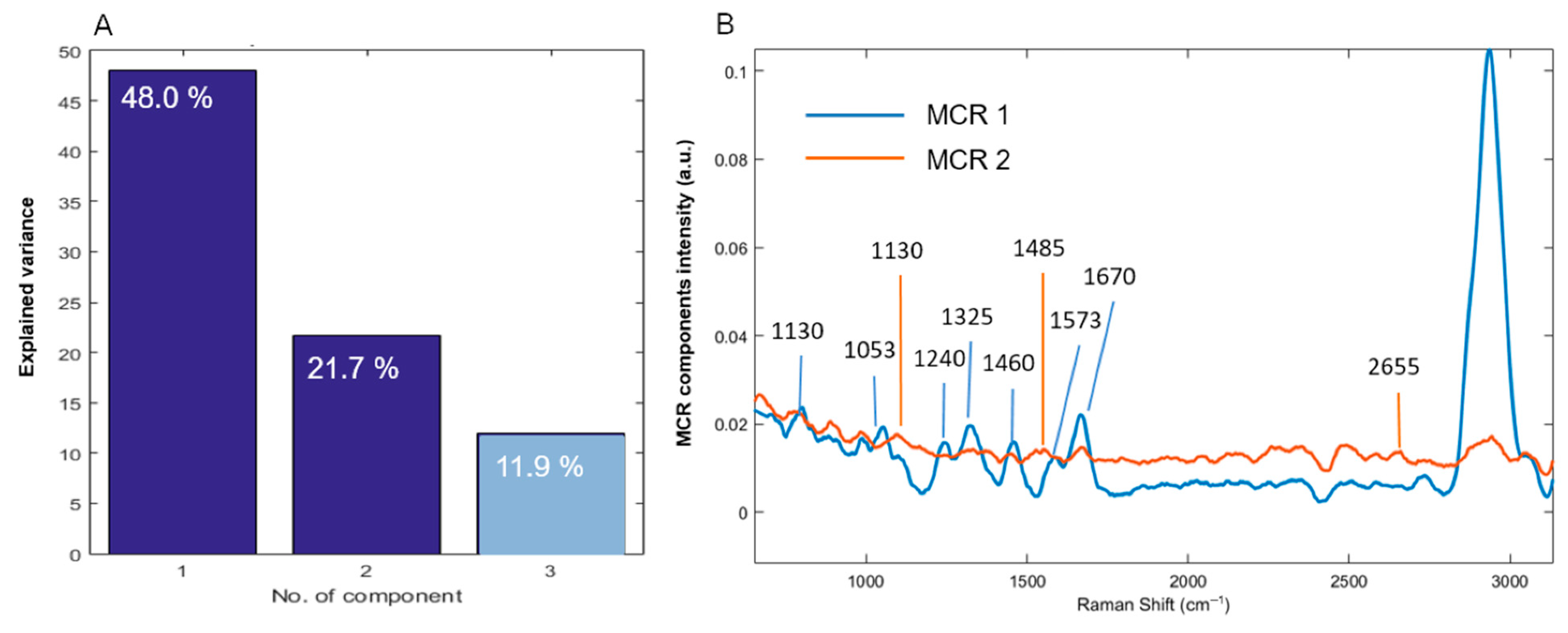
3.2. Principal Components Analysis
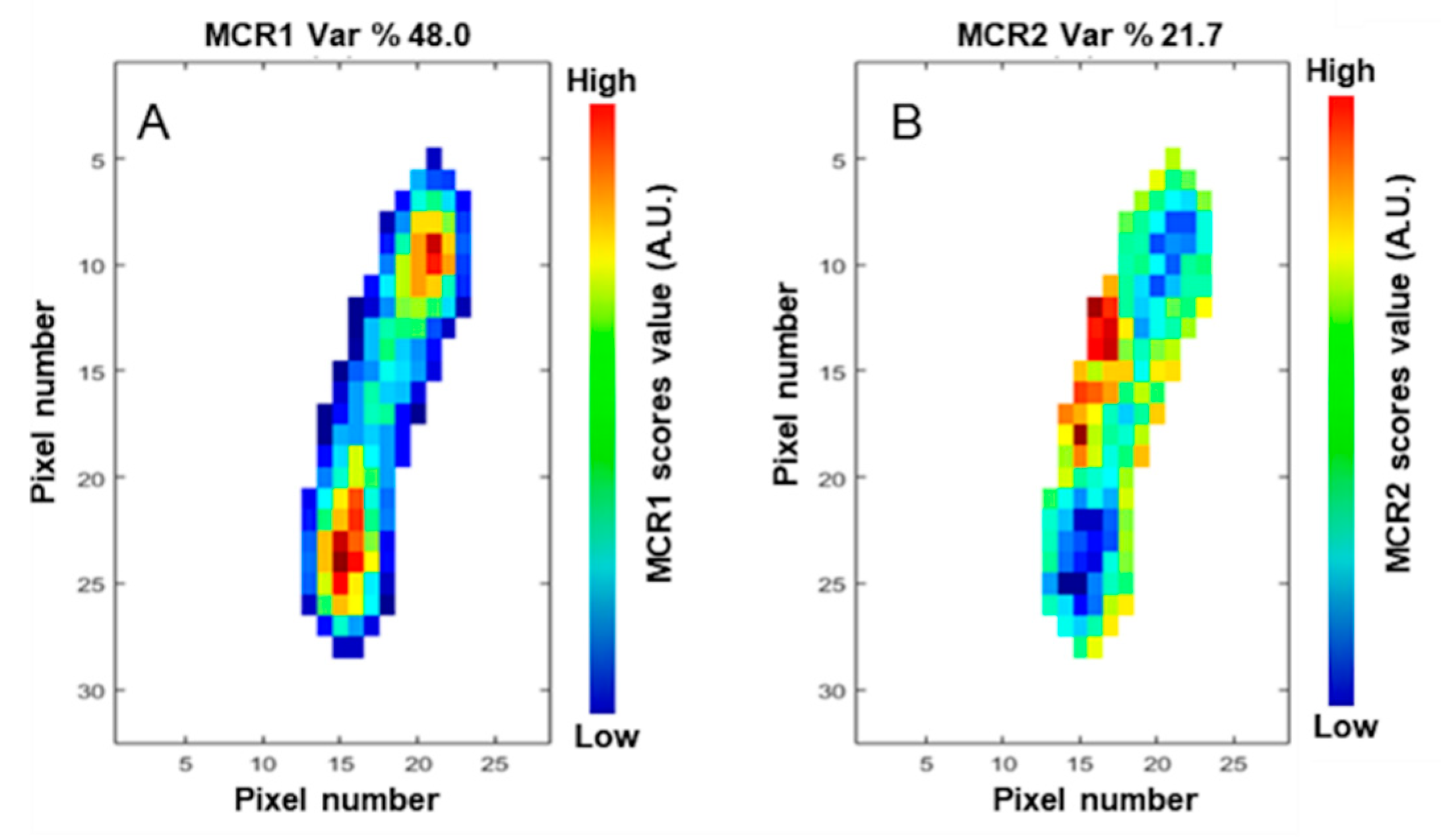
3.3. Multivariate Curve Resolution–Alternating Least Squares
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Uzunbajakava, N.; Lenferink, A.; Kraan, Y.; Volokhina, E.; Vrensen, G.; Greve, J.; Otto, C. Nonresonant confocal Raman imaging of DNA and protein distribution in apoptotic cells. Biophys. J. 2003, 84, 368–3981. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Xu, J.; Romero-Gonzalez, M.; Banwart, S.A.; Huang, W.E. Single cell Raman spectroscopy for cell sorting and imaging. Curr. Opin. Biotechnol. 2012, 23, 56–63. [Google Scholar] [CrossRef]
- Wenning, M.; Scherer, S. Identification of microorganisms by FTIR spectroscopy: Perspectives and limitations of the method. Appl. Microbiol. Biotechnol. 2013, 97, 7111–7120. [Google Scholar] [CrossRef]
- Palonpon, A.F.; Ando, J.; Yamakoshi, H.; Dodo, K.; Sodeoka, M.; Kawata, S.; Fujita, K. Raman and SERS microscopy for molecular imaging of live cells. Nat. Protoc. 2013, 8, 677–692. [Google Scholar] [CrossRef] [PubMed]
- Mandrile, L.; Zeppa, G.; Giovannozzi, A.M.; Rossi, A.M. Controlling protected designation of origin of wine by Raman spectroscopy. Food Chem. 2016, 244, 16–24. [Google Scholar] [CrossRef]
- Lu, X.; Al-Qadiri, H.M.; Lin, M.; Rasco, B.A. Application of mid-infrared and Raman Spectroscopy to the study of bacteria. Food Bioproc. Technol. 2011, 4, 919–935. [Google Scholar] [CrossRef]
- de Juan, A.; Tauler, R. Multivariate curve resolution (MCR) from 2000: Progress in concepts and applications. Crit. Rev. Anal. Chem. 2006, 36, 163–176. [Google Scholar] [CrossRef]
- Piqueras, S.; Duponchel, L.; Tauler, R.; De Juan, A. Resolution and segmentation of hyperspectral biomedical images by Multivariate Curve Resolution-Alternating Least Squares. Anal. Chim. Acta 2011, 36, 163–176. [Google Scholar] [CrossRef]
- Barzan, G.; Sacco, A.; Mandrile, L.; Giovannozzi, A.M.; Brown, J.; Portesi, C.; Alexander, M.R.; Williams, P.; Hardie, K.R.; Rossi, A.M. New frontiers against antibiotic resistance: A Raman-based approach for rapid detection of bacterial susceptibility and biocide-induced antibiotic cross-tolerance. Sens. Actuat. B Chem. 2020, 309, 127774. [Google Scholar] [CrossRef]
- Münchberg, U.; Rösch, P.; Bauer, M.; Popp, J. Raman spectroscopic identification of single bacterial cells under antibiotic influence. Anal. Bioanal. Chem. 2014. [Google Scholar] [CrossRef]
- Polisetti, S.; Bible, A.N.; Morrell-Falvey, J.L.; Bohn, P.W. Raman chemical imaging of the rhizosphere bacterium Pantoea spp. YR343 and its co-culture with Arabidopsis thaliana. Analyst 2016, 141, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Mandrile, L.; Amato, G.; Marchis, D.; Martra, G.; Rossi, A.M. Species-specific detection of processed animal proteins in feed by Raman spectroscopy. Food Chem. 2017, 229, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Mandrile, L.; Rotunno, S.; Miozzi, L.; Vaira, A.M.; Giovannozzi, A.M.; Rossi, A.M.; Noris, E. Nondestructive Raman spectroscopy as a tool for early detection and discrimination of the infection of tomato plants by two economically important viruses. Anal. Chem. 2019, 91, 9025–9031. [Google Scholar] [CrossRef] [PubMed]
- Mandrile, L.; Fusaro, I.; Amato, G.; Marchis, D.; Martra, G.; Rossi, A.M. Detection of insect′s meal in compound feed by Near Infrared spectral imaging. Food Chem. 2018, 267, 240–245. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amigo, J.M.; Martí, I.; Gowen, A. Hyperspectral Imaging and Chemometrics. A Perfect Combination for the Analysis of Food Structure, Composition and Quality. In Data Handling in Science and Technology; Elviser: Amsterdam, The Netherlands, 2013. [Google Scholar]
- Eriksson, L.; Antti, H.; Gottfries, J.; Holmes, E.; Johansson, E.; Lindgren, F.; Long, I.; Lundstedt, T.; Trygg, J.; Wold, S. Using chemometrics for navigating in the large data sets of genomics, proteomics, and metabonomics (gpm). Anal. Bioanal. Chem. 2004, 380, 419–429. [Google Scholar] [CrossRef]
- Sacco, A.; Imbraguglio, D.; Giovannozzi, A.M.; Portesi, C.; Rossi, A.M. Development of a candidate reference sample for the characterization of tip-enhanced Raman spectroscopy spatial resolution. RSC Adv. 2018, 8, 27863–27869. [Google Scholar] [CrossRef] [Green Version]
- Cara, E.; Mandrile, L.; Sacco, A.; Giovannozzi, A.M.; Rossi, A.M.; Celegato, F.; De Leo, N.; Hönicke, P.; Kayser, Y.; Beckhoff, B.; et al. Towards a traceable enhancement factor in surface-enhanced Raman spectroscopy. J. Mater. Chem. C 2020, 8, 16513–16519. [Google Scholar] [CrossRef]
- De Juan, A.; Jaumot, J.; Tauler, R. Multivariate Curve Resolution (MCR). Solving the mixture analysis problem. Anal. Methods 2014, 6, 4964–4976. [Google Scholar] [CrossRef]
- Harz, M.; Rösch, P.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Popp, J. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 2005, 130, 1543–1550. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; Rehman, I.U. Raman spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2007, 42, 493–541. [Google Scholar] [CrossRef]
- Harz, M.; Kiehntopf, M.; Stöckel, S.; Rösch, P.; Straube, E.; Deufer, T.; Popp, J. Direct analysis of clinical relevant single bacterial cells from cerebrospinal fluid during bacterial meningitis by means of micro-Raman spectroscopy. J. Biophotonics 2009, 2, 70–80. [Google Scholar] [CrossRef]
- Socrates, G. Infrared and Raman Characteristic Group Frequencies. Tables and Charts; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2001. [Google Scholar]
- Kloß, S.; Kampe, B.; Sachse, S.; Rösch, P.; Straube, E.; Pfister, W.; Kiehntopf, M.; Popp, J. Culture independent Raman spectroscopic identification of urinary tract infection pathogens: A proof of principle study. Anal. Chem. 2013, 85, 9610–9616. [Google Scholar] [CrossRef]
- Tamotsu, K.; Shinji, F.; Akemi, F.; Wataru, S.; Masahira, H.; Jun, K.; Hiroshi, O.; Ohara, O. Multiple omics uncovers host-gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res. 2014, 21, 469–480. [Google Scholar] [CrossRef] [Green Version]
- Motegi, H.; Tsuboi, Y.; Saga, A.; Kagami, T.; Inoue, M.; Toki, H.; Minowa, O.; Noda, T.; Kikuchi, J. Identification of Reliable Components in Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS): A Data-Driven Approach across Metabolic Processes. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niki, H.; Yamaichi, Y.; Hiraga, S. Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev. 2000, 14, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.S.; Wright, A. DNA Segregation in Bacteria. Annu. Rev. Microbiol. 2000, 54, 681–708. [Google Scholar] [CrossRef] [PubMed]
- Woldringh, C.L.; Mulder, E.; Huls, P.G.; Vischer, N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res. Microbiol. 1991, 142, 309–320. [Google Scholar] [CrossRef]
- Shapiro, L.; Losick, R. Dynamic spatial reglation in the bacterial cell. Cell 2000, 100, 89–98. [Google Scholar] [CrossRef] [Green Version]
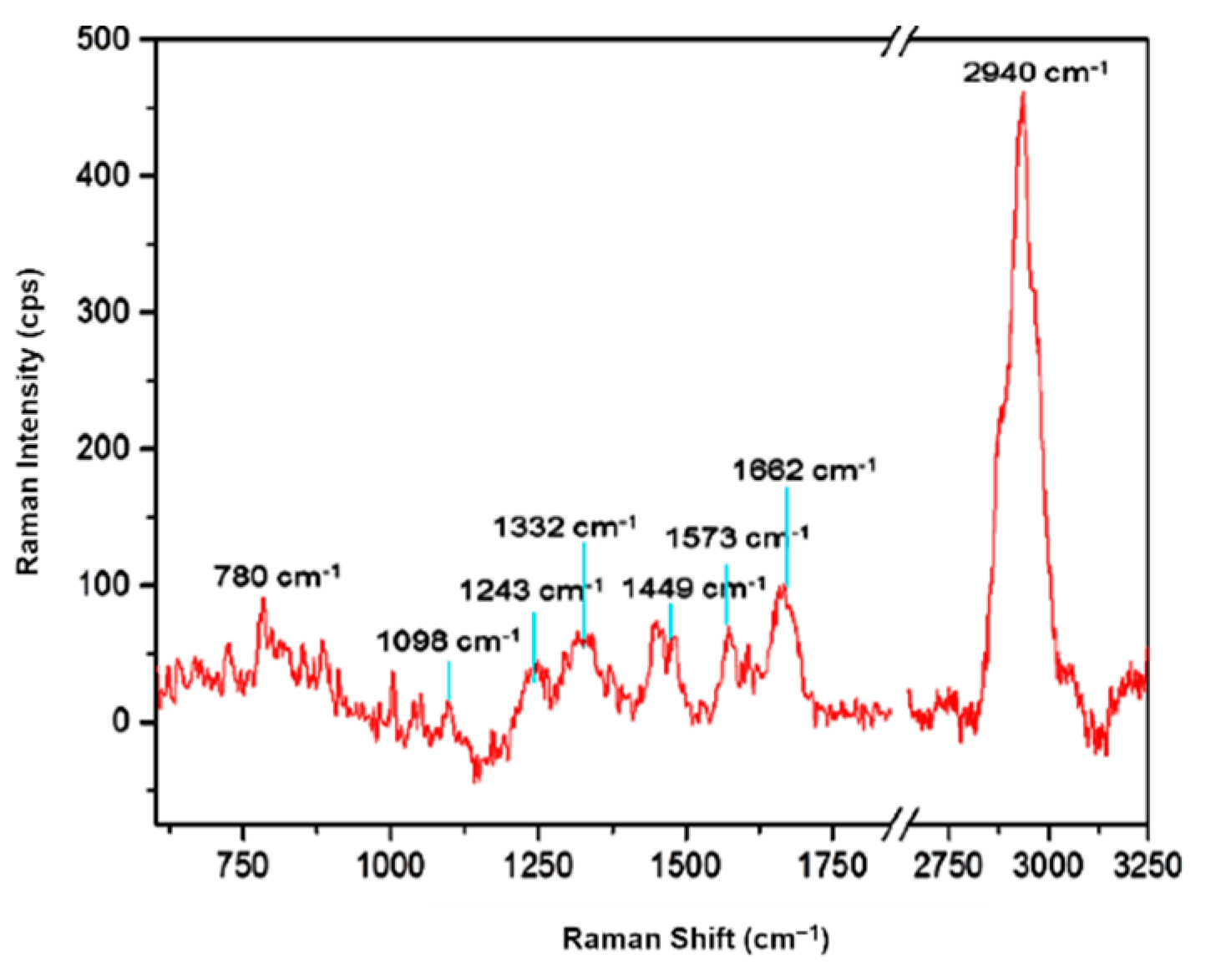
| Raman Shift (cm−1) | Assignment 1 | Chemical Compound 1 | Reference |
|---|---|---|---|
| 2940 | νCH2 asymmetric νCH3 symmetric | carbohydrates, amino acids, proteins, lipids | 2935 [20] 2935 [21] |
| 1662 | amide I band νC=C | nucleic acids | 1666 [22] 1663 [21] |
| 1573 | amide II band δNH; νCN | nucleic acids (G, A) | 1565 [23] 1575 [20] |
| 1440–1480 | δ(CH2/CH3) | lipids, amino acid, side chains of proteins and carbohydrates | 1440–1460 [20]; 1450 [22] |
| 1332 | ring vibration δCH2 | nucleic acids (G, A) and tryptophan proteins | 1337 [20] 1337 [20] |
| 1243 | νC–C, νC–O amide III band ν(PO2−) asymmetric νC–O–S | nucleic acids (A, T) DNA bases sulpholipids | 1250–1000 [23] 1245 [20] 1243 [20] 1240 [23] |
| 1098 | ring stretching νC–O–C symmetric glycosidic link; νC–N; νC–C | saccharide components (lipid, DNA and RNA backbone); phenylalanine | 1115–1005 [23] 1104 [21] 1125 [20]; 1099 [21] |
| 780 | ring breathing modes | nucleic acids (C, T, U), phenylalanine, pyranose compounds | 786–780 [21] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barzan, G.; Sacco, A.; Mandrile, L.; Giovannozzi, A.M.; Portesi, C.; Rossi, A.M. Hyperspectral Chemical Imaging of Single Bacterial Cell Structure by Raman Spectroscopy and Machine Learning. Appl. Sci. 2021, 11, 3409. https://doi.org/10.3390/app11083409
Barzan G, Sacco A, Mandrile L, Giovannozzi AM, Portesi C, Rossi AM. Hyperspectral Chemical Imaging of Single Bacterial Cell Structure by Raman Spectroscopy and Machine Learning. Applied Sciences. 2021; 11(8):3409. https://doi.org/10.3390/app11083409
Chicago/Turabian StyleBarzan, Giulia, Alessio Sacco, Luisa Mandrile, Andrea Mario Giovannozzi, Chiara Portesi, and Andrea Mario Rossi. 2021. "Hyperspectral Chemical Imaging of Single Bacterial Cell Structure by Raman Spectroscopy and Machine Learning" Applied Sciences 11, no. 8: 3409. https://doi.org/10.3390/app11083409
APA StyleBarzan, G., Sacco, A., Mandrile, L., Giovannozzi, A. M., Portesi, C., & Rossi, A. M. (2021). Hyperspectral Chemical Imaging of Single Bacterial Cell Structure by Raman Spectroscopy and Machine Learning. Applied Sciences, 11(8), 3409. https://doi.org/10.3390/app11083409









