Differential Response of the Proteins Involved in Amino Acid Metabolism in Two Saccharomyces cerevisiae Strains during the Second Fermentation in a Sealed Bottle
Abstract
1. Introduction
2. Materials and Methods
2.1. Microorganisms and Fermentation Conditions
2.2. Proteomic Analysis
2.3. Statistical Analysis
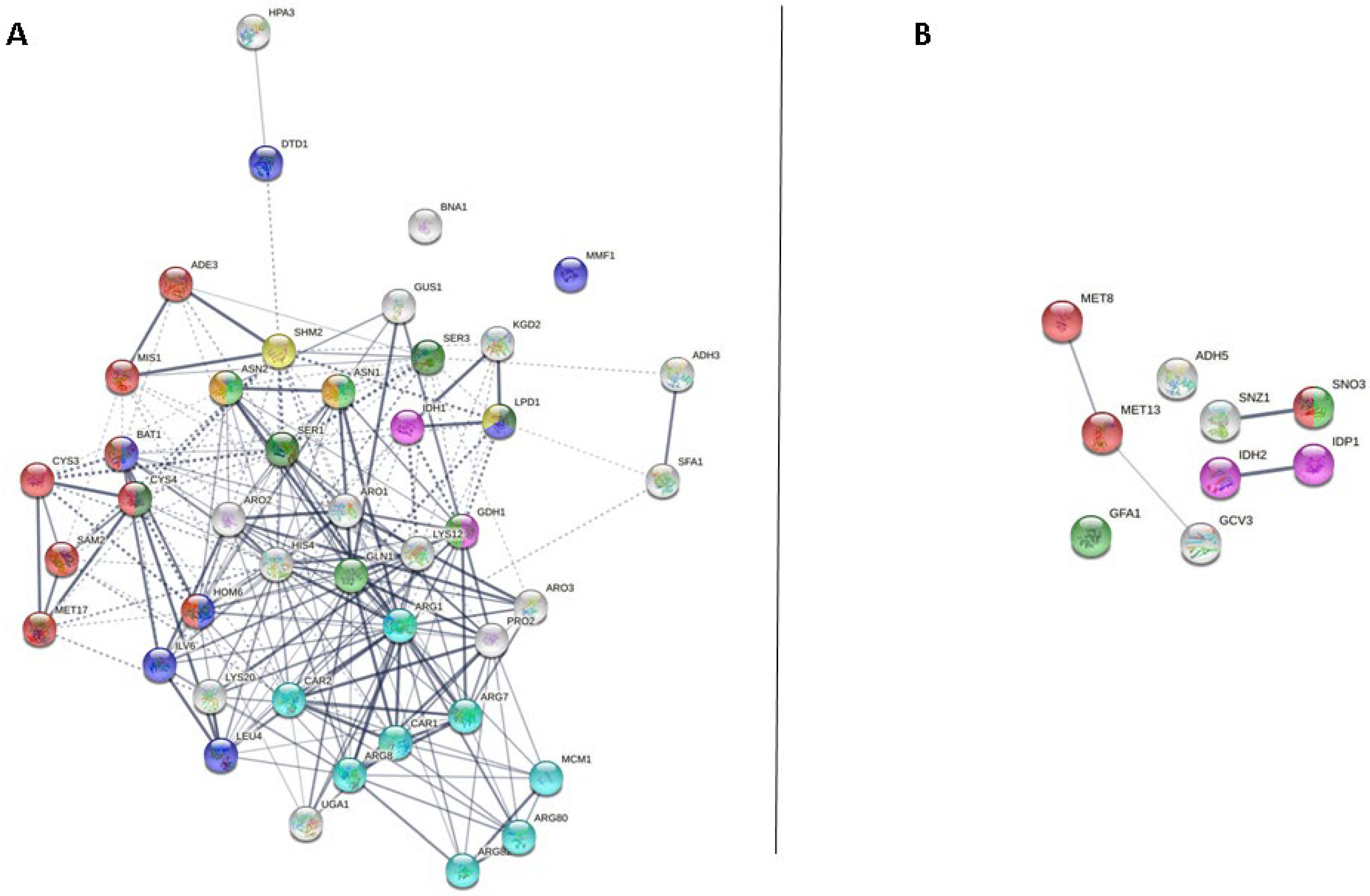
2.4. Protein Network Reconstruction
3. Results and Discussion
3.1. Amino Acid Transport
3.2. tRNA Aminoacylation for Protein Translation
3.3. Cellular Amino Acid Metabolic Process
3.3.1. Cellular Amino Acid Metabolic Process at the Middle of the Second Fermentation
3.3.2. Cellular Amino Acid Metabolic Process at the End of the Second Fermentation
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mauricio, J.C.; Ortega, J.M.; Salmon, J.M. Sugar uptake by three strains of Saccharomyces cerevisiae during alcoholic fermentation at different initial ammoniacal nitrogen concentrations. Int. Symp. Vitic. Enol. 1993, 388, 197–202. [Google Scholar] [CrossRef]
- Moreira, N.; De Pinho, P.G.; Santos, C.; Vasconcelos, I. Relationship between nitrogen content in grapes and volatiles, namely heavy sulphur compounds, in wines. Food Chem. 2011, 126, 1599–1607. [Google Scholar] [CrossRef] [PubMed]
- Giudici, P.; Kunkee, R.E. The effect of nitrogen deficiency and sulfur-containing amino acids on the reduction of sulfate to hydrogen sulfide by wine yeasts. Am. J. Enol. Viticult. 1994, 45, 107–112. [Google Scholar]
- Barbosa, C.; Mendes-Faia, A.; Mendes-Ferreira, A. The nitrogen source impacts major volatile compounds released by Saccharomyces cerevisiae during alcoholic fermentation. Int. J. Food Microbiol. 2012, 160, 87–93. [Google Scholar] [CrossRef]
- Su, Y.; Seguinot, P.; Sanchez, I.; Ortiz-Julien, A.; Heras, J.M.; Querol, A.; Camarasa, C.; Guillamón, J.M. Nitrogen sources preferences of non-Saccharomyces yeasts to sustain growth and fermentation under winemaking conditions. Food Microbial. 2020, 85, 103287. [Google Scholar] [CrossRef] [PubMed]
- Marsit, S.; Sanchez, I.; Galeote, V.; Dequin, S. Horizontally acquired oligopeptide transporters favour adaptation of Saccharomyces cerevisiae wine yeast to oenological environment. Environ. Microbiol. 2016, 18, 1148–1161. [Google Scholar] [CrossRef]
- Henschke, P.A.; Jiranek, V. Yeast-metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Langhorne, PA, USA, 1994; pp. 77–164. [Google Scholar]
- Moreno-Arribas, M.V.; Polo, M.C. Amino acids and biogenic amines. In Wine Chemistry and Biochemistry; Springer: New York, NY, USA, 2009; pp. 163–189. [Google Scholar]
- Kevvai, K.; Kütt, M.L.; Nisamedtinov, I.; Paalme, T. Simultaneous utilization of ammonia, free amino acids and peptides during fermentative growth of Saccharomyces cerevisiae: Simultaneous utilization of ammonia, free amino acids and peptides. J. Inst. Brew. 2016, 122, 110–115. [Google Scholar] [CrossRef]
- Large, P.J. Degradation of organic nitrogen compounds by yeasts. Yeast 1986, 2, 1–34. [Google Scholar] [CrossRef]
- Zhong, X.; Wang, A.; Zhang, Y.; Wu, Z.; Li, B.; Lou, H.; Huang, G.; Wen, H. Reducing higher alcohols by nitrogen compensation during fermentation of Chinese rice wine. Food Sci. Biotechnol. 2020, 29, 805–816. [Google Scholar] [CrossRef]
- González-Jiménez, M.C.; Moreno-García, J.; García-Martínez, T.; Moreno, J.J.; Puig-Pujol, A.; Capdevilla, F.; Mauricio, J.C. Differential analysis of proteins involved in ester metabolism in two Saccharomyces cerevisiae strains during the second fermentation in sparkling wine elaboration. Microorganisms 2020, 8, 403. [Google Scholar] [CrossRef]
- Porras-Agüera, J.A.; Moreno-García, J.; González-Jiménez, M.C.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. Autophagic proteome in two Saccharomyces cerevisiae strains during second fermentation for sparkling wine elaboration. Microorganisms 2020, 8, 523. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, R.; Roldán-Romero, Y.; Moreno, J.; Puig-Pujol, A.; Mauricio, J.C.; García-Martínez, T. Use of a flor yeast strain for the second fermentation of sparkling wines: Effect of endogenous CO2 over-pressure on the volatilome. Food Chem. 2020, 308, 125555. [Google Scholar] [CrossRef] [PubMed]
- Martínez-García, R.; García-Martínez, T.; Puig-Pujol, A.; Mauricio, J.C.; Moreno, J. Changes in sparkling wine aroma during the second fermentation under CO2 pressure in sealed bottle. Food Chem. 2017, 237, 1030–1040. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A Rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance Index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Bianchi, F.; van’t Klooster, J.S.; Ruiz, S.J.; Poolman, B. Regulation of amino acid transport in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2019, 83, e00024-19. [Google Scholar] [CrossRef] [PubMed]
- Mulero, J.J.; Rosenthal, J.K.; Fox, T.D. PET112, a Saccharomyces cerevisiae nuclear gene required to maintain rho+ mitochondrial DNA. Curr. Genet. 1994, 25, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, M.; Ugliano, M.; Varela, C.; Siebert, T.; Pretorius, I.S.; Henschke, P.A. Assimilable nitrogen utilisation and production of volatile and non-volatile compounds in chemically defined medium by Saccharomyces cerevisiae wine yeasts. Appl. Microbiol. Biotechnol. 2007, 77, 145–157. [Google Scholar] [CrossRef]
- Fernández-Cruz, E.; Álvarez-Fernández, M.A.; Valero, E.; Troncoso, A.M.; García-Parrilla, M.C. Melatonin and derived L-tryptophan metabolites produced during alcoholic fermentation by different wine yeast strains. Food Chem. 2017, 217, 431–437. [Google Scholar] [CrossRef]
- Seaston, A.; Inkson, C.; Eddy, A.A. The absorption of protons with specific amino acids and carbohydrates by yeast. Biochem. J. 1973, 134, 1031–1043. [Google Scholar] [CrossRef]
- Chianelli, M.S. Sistemas De Transporte De Aminoácidos Neutros En saccharomyces Cerevisiae, Cepas Silvestres Y Mutantes Transporte-Defectivas. Ph.D. Thesis, Universidad de Buenos Aires (Facultad de Ciencias Exactas y Naturales), Buenos Aires, Argentina, 1998. [Google Scholar]
- Russnak, R.; Konczal, D.; McIntire, S.L. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J. Biol. Chem. 2001, 276, 23849–23857. [Google Scholar] [CrossRef]
- Tone, J.; Yamanaka, A.; Manabe, K.; Murao, N.; Kawano-Kawada, M.; Sekito, T.; Kakinuma, Y. A vacuolar membrane protein Avt7p is involved in transport of amino acid and spore formation in Saccharomyces cerevisiae. Biosci. Biotech. Bioch. 2015, 79, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Chattopadhyay, S.; Locke, S.; Pearce, D.A. Interaction among Btn1p, Btn2p, and Ist2p reveals potential interplay among the vacuole, amino acid levels, and ion homeostasis in the yeast Saccharomyces cerevisiae. Eukaryot. Cell. 2005, 4, 281–288. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Delarue, M. Aminoacyl-tRNA synthetases. Curr. Opin. Struct. Biol. 1995, 5, 48–55. [Google Scholar] [CrossRef]
- Arnez, J.G.; Moras, D. Structural and functional considerations of the aminoacylation reaction. Trends Biochem. Sci. 1997, 22, 211–216. [Google Scholar] [CrossRef]
- González-Jiménez, M.D.C.; García-Martínez, T.; Puig-Pujol, A.; Capdevila, F.; Moreno-García, J.; Moreno, J.; Mauricio, J.C. Biological processes highlighted in Saccharomyces cerevisiae during the sparkling wines elaboration. Microorganisms 2020, 8, 1216. [Google Scholar] [CrossRef]
- Eriani, G.; Delarue, M.; Poch, O.; Gangloff, J.; Moras, D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 1990, 347, 203–206. [Google Scholar] [CrossRef] [PubMed]
- Porras-Agüera, J.A.; Moreno-García, J.; Mauricio, J.C.; Moreno, J.; García-Martínez, T. First proteomic approach to identify cell death biomarkers in oenological yeasts during sparkling wine production. Microorganisms 2019, 7, 542. [Google Scholar] [CrossRef] [PubMed]
- Olin-Sandoval, V.; Shu Lim Yu, J.; Miller-Fleming, L.; Tauqeer Alam, M.; Kamrad, S.; Correia-Melo, C.; Haas, R.; Segal, J.; Peña Navarro, D.A.; Herrera-Dominguez, L.; et al. Lysine harvesting is an antioxidant strategy and triggers underground polyamine metabolism. Nature 2019, 572, 249–253. [Google Scholar] [CrossRef]
- Huang, C.W.; Walker, M.E.; Fedrizzi, B.; Gardner, R.C.; Jiranek, V. Hydrogen sulfide and its role in Saccharomyces cerevisiae in a wine-making context. Yeast Res. FEMS 2017, 17, fox058. [Google Scholar] [CrossRef] [PubMed]
- Kessi-Pérez, E.I.; Molinet, J.; Martínez, C. Disentangling the genetic bases of Saccharomyces cerevisiae nitrogen consumption and adaptation to low nitrogen environments in wine fermentation. Biol. Res. 2020, 53, 1–10. [Google Scholar] [CrossRef]
- Eldarov, M.A. Metabolic engineering of wine strains of Saccharomyces cerevisiae. Genes 2020, 11, 964. [Google Scholar] [CrossRef] [PubMed]
- Björkeroth, J.; Campbell, K.; Malina, C.; Yu, R.; Di Bartolomeo, F.; Nielsen, J. Proteome reallocation from amino acid biosynthesis to ribosomes enables yeast to grow faster in rich media. PNAS 2020, 117, 21804–21812. [Google Scholar] [CrossRef] [PubMed]
- Mauricio, J.C.; Millán, C.; Ortega, J.M. Influence of oxygen on the biosynthesis of cellular fatty acids, sterols and phospholipids during alcoholic fermentation by Saccharomyces cerevisiae and Torulaspora delbrueckii. World J. Microbiol. Biotechnol. 1998, 14, 405–410. [Google Scholar] [CrossRef]
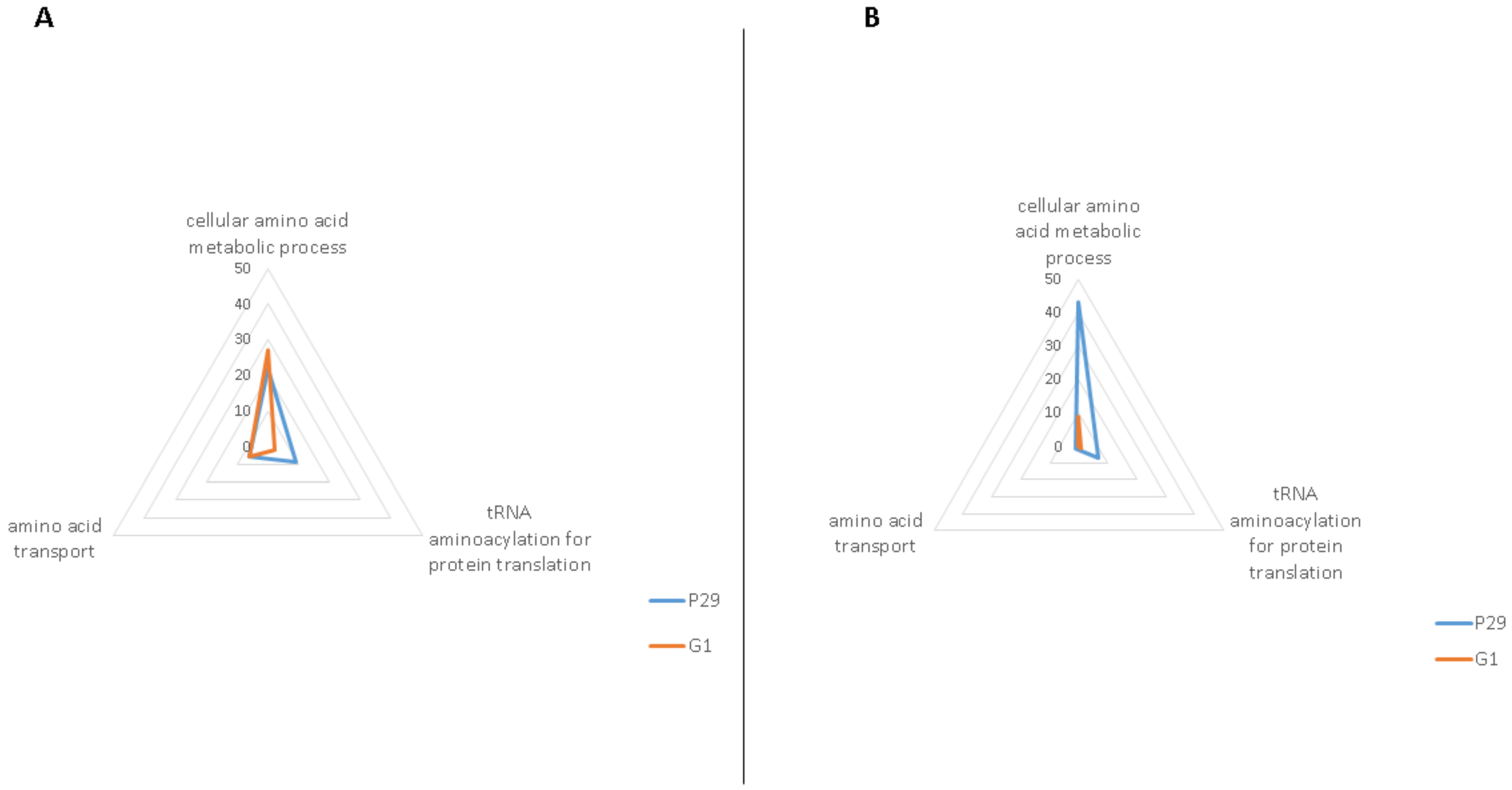

| Middle of the Second Fermentation, MF | ||||
| P29 Strain | G1 Strain | |||
| Biological Process | Specific Proteins | Specific/Total Proteins | Specific Proteins | Specific/Total Proteins |
| Cellular amino acid metabolic process (GO: 0006520) | Aat1p, Ald2p, Arg81p, Aro9p, Dal81p, Gcv2p, Gdh3p, Ilv6p, Leu9p, Lys1p, Lys4p, Met1p, Mis1p, Put1p, Snz3p, Ths1p, Uga2p, Uga3p, Ura7p, Utr4p, Xbp1p, Ydl168wp, Yml082wp, Ymr084wp | 24/103 | Aco2p, Adi1p, Aro10p, Bna4p, Cpa2p, Ehd3p, Gcv1p, Gfa1p, Gln4p, Gly1p, His5p, Idp1p, Lap3p, Lys21p, Mae1p, Met12p, Met16p, Mri1p, Nit3p, Pet112p, Ser1p, Sfa1p, Sno3p, Tum1p, Uga1p, Yhr112cp, Yhr208wp, Yll058wp | 28/107 |
| tRNA aminoacylation for protein translation (GO: 0006418) | Aim10p, Cdc60p, Ded81p, Dia4p, Dps1p, Msk1p, Slm5p, Ths1p, Wrs1p | 9/23 | Gln4p, Nam2p | 2/16 |
| Amino acid transport (GO: 0006865) | Avt7p, Bap2p, Lst4p, Put4p, Uga4p, Ydl119cp | 6/9 | Alp1p, Avt4p, Btn2p, Gnp1p, Ssy1p | 5/8 |
| End of the second fermentation, EF | ||||
| P29 Strain | G1 Strain | |||
| Biological Process | Specific Proteins | Specific/Total Proteins | Specific Proteins | Specific/Total Proteins |
| Cellular amino acid metabolic process (GO: 0006520) | Ade3p, Adh3p, Arg1p, Arg7p, Arg8p, Arg80p, Arg82p, Aro1p, Aro2p, Aro3p, Asn1p, Asn2p, Bna1p, Car1p, Car2p, Cys3p, Cys4p, Dtd1p, Gdh1p, Gln1p, Gus1p, His4p, Hom6p, Hpa3p, Idh1p, Ilv6p, Kgd2p, Leu4p, Lpd1p, Lys12p, Lys20p, Mcm1p, Met17p, Mis1p, Mmf1p, Pro2p, Sam2p, Ser1p, Ser3p, Sfa1p, Shm2p, Uga1p, Yhr208wp | 43/56 | Gcv3p, Gfa1p, Idh2p, Idp1p, Met13p, Met8p, Sno3p, Snz1p, Ybr145wp | 9/22 |
| tRNA aminoacylation for protein translation (GO: 0006418) | Arc1p, Grs1p, Gus1p, Ism1p, Msk1p, Ses1p, Vas1p | 7/8 | Msm1p | 1/2 |
| Amino acid transport (GO: 0006865) | Npr2p | 1/1 | ||
| MF | EF | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| P29 Strain | G1 Strain | P29 Strain | G1 Strain | ||||||||
| Proteins | mol% | SD | Proteins | mol% | SD | Proteins | mol% | SD | Proteins | mol% | SD |
| Aat1p | 0.024 | 0.0002 | Aco2p | 0.028 | 0.0003 | Ade3p | 0.007 | 0.0001 | Gcv3p | 0.427 | 0.004 |
| Ald2p | 0.010 | 0.0001 | Adi1p | 0.048 | 0.0005 | Adh3p | 0.078 | 0.0008 | Gfa1p | 0.033 | 0.0008 |
| Arg81p | 0.006 | 0.0001 | Aro10p | 0.010 | 0.0001 | Arg1p | 0.114 | 0.0011 | Idh2p | 0.081 | 0.001 |
| Aro9p | 0.028 | 0.0003 | Bna4p | 0.010 | 0.0001 | Arg7p | 0.055 | 0.0006 | Idp1p | 0.0465 | 0.0005 |
| Dal81p | 0.007 | 0.0001 | Cpa2p | 0.028 | 0.0003 | Arg8p | 0.013 | 0.0001 | Met13p | 0.051 | 0.001 |
| Gcv2p | 0.005 | 0.0001 | Ehd3p | 0.011 | 0.0001 | Arg80p | 0.039 | 0.0004 | Met8p | 0.069 | 0.0007 |
| Gdh3p | 0.057 | 0.0006 | Gcv1p | 0.013 | 0.0001 | Arg82p | 0.0187 | 0.0002 | Sno3p | 0.106 | 0.0011 |
| Ilv6p | 0.016 | 0.0002 | Gfa1p | 0.015 | 0.0001 | Aro1p | 0.004 | 0.0000 | Snz1p | 0.076 | 0.0006 |
| Leu9p | 0.048 | 0.0005 | Gln4p | 0.005 | 0.0003 | Aro2p | 0.150 | 0.0015 | |||
| Lys1p | 0.052 | 0.0005 | Gly1p | 0.108 | 0.0011 | Aro3p | 0.276 | 0.0028 | |||
| Lys4p | 0.015 | 0.003 | His5p | 0.060 | 0.0006 | Asn1p | 0.082 | 0.0008 | |||
| Met1p | 0.015 | 0.0001 | Idp1p | 0.031 | 0.0003 | Asn2p | 0.036 | 0.0004 | |||
| Mis1p | 0.005 | 0.0000 | Lap3p | 0.046 | 0.0005 | Bna1p | 0.039 | 0.0004 | |||
| Put1p | 0.011 | 0.0001 | Lys21p | 0.138 | 0.0014 | Car1p | 0.134 | 0.0013 | |||
| Snz3p | 0.016 | 0.0002 | Mae1p | 0.016 | 0.0002 | Car2p | 0.055 | 0.0006 | |||
| Ths1p | 0.011 | 0.0001 | Met12p | 0.017 | 0.0002 | Cys3p | 0.113 | 0.0011 | |||
| Uga2p | 0.011 | 0.0001 | Met16p | 0.022 | 0.0002 | Cys4p | 0.131 | 0.0013 | |||
| Uga3p | 0.010 | 0.0001 | Mri1p | 0.014 | 0.0001 | Dtd1p | 0.114 | 0.0011 | |||
| Ura7p | 0.017 | 0.0002 | Nit3p | 0.016 | 0.0002 | Gdh1p | 0.139 | 0.0014 | |||
| Utr4p | 0.029 | 0.0003 | Pet112p | 0.009 | 0.0005 | Gln1p | 0.305 | 0.0031 | |||
| Yml082wp | 0.008 | 0.0001 | Ser1p | 0.043 | 0.0004 | Gus1p | 0.041 | 0.0004 | |||
| Ymr084wp | 0.019 | 0.0002 | Sno3p | 0.023 | 0.0002 | His4p | 0.093 | 0.0009 | |||
| Put4p | 0.013 | 0.0001 | Tum1p | 0.015 | 0.0002 | Hom6p | 0.158 | 0.0016 | |||
| Uga4p | 0.019 | 0.0004 | Uga1p | 0.081 | 0.0008 | Hpa3p | 0.116 | 0.0011 | |||
| Lst4p | 0.012 | 0.0001 | Yhr112cp | 0.014 | 0.0001 | Idh1p | 0.079 | 0.0008 | |||
| Bap2p | 0.012 | 0.0001 | Yll058wp | 0.008 | 0.0001 | Ilv6p | 0.068 | 0.0007 | |||
| Avt7p | 0.028 | 0.0006 | Yhr208wp | 0.161 | 0.0016 | Kgd2p | 0.056 | 0.0006 | |||
| Ydl119cp | 0.014 | 0.001 | Alp1p | 0.016 | 0.0002 | Leu4p | 0.041 | 0.0004 | |||
| Gnp1p | 0.009 | 0.0001 | Lpd1p | 0.011 | 0.0001 | ||||||
| Avt4p | 0.009 | 0.0001 | Lys12p | 0.326 | 0.007 | ||||||
| Btn2p | 0.021 | 0.0002 | Lys20p | 0.028 | 0.0003 | ||||||
| Ssy1p | 0.008 | 0.0001 | Mcm1p | 0.214 | 0.0021 | ||||||
| Odc2p | 0.015 | 0.0002 | Met17p | 0.054 | 0.0005 | ||||||
| Mis1p | 0.006 | 0.0001 | |||||||||
| Mmf1p | 0.546 | 0.009 | |||||||||
| Pro2p | 0.040 | 0.0004 | |||||||||
| Sam2p | 0.205 | 0.003 | |||||||||
| Ser1p | 0.017 | 0.0002 | |||||||||
| Ser3p | 0.020 | 0.0002 | |||||||||
| Sfa1p | 0.080 | 0.0008 | |||||||||
| Shm2p | 0.039 | 0.0004 | |||||||||
| Uga1p | 0.013 | 0.0001 | |||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Jiménez, M.d.C.; Mauricio, J.C.; Moreno-García, J.; Puig-Pujol, A.; Moreno, J.; García-Martínez, T. Differential Response of the Proteins Involved in Amino Acid Metabolism in Two Saccharomyces cerevisiae Strains during the Second Fermentation in a Sealed Bottle. Appl. Sci. 2021, 11, 12165. https://doi.org/10.3390/app112412165
González-Jiménez MdC, Mauricio JC, Moreno-García J, Puig-Pujol A, Moreno J, García-Martínez T. Differential Response of the Proteins Involved in Amino Acid Metabolism in Two Saccharomyces cerevisiae Strains during the Second Fermentation in a Sealed Bottle. Applied Sciences. 2021; 11(24):12165. https://doi.org/10.3390/app112412165
Chicago/Turabian StyleGonzález-Jiménez, María del Carmen, Juan Carlos Mauricio, Jaime Moreno-García, Anna Puig-Pujol, Juan Moreno, and Teresa García-Martínez. 2021. "Differential Response of the Proteins Involved in Amino Acid Metabolism in Two Saccharomyces cerevisiae Strains during the Second Fermentation in a Sealed Bottle" Applied Sciences 11, no. 24: 12165. https://doi.org/10.3390/app112412165
APA StyleGonzález-Jiménez, M. d. C., Mauricio, J. C., Moreno-García, J., Puig-Pujol, A., Moreno, J., & García-Martínez, T. (2021). Differential Response of the Proteins Involved in Amino Acid Metabolism in Two Saccharomyces cerevisiae Strains during the Second Fermentation in a Sealed Bottle. Applied Sciences, 11(24), 12165. https://doi.org/10.3390/app112412165








