Featured Application
MR-derived cerebral metabolic rate of oxygen in contrast-enhancing and peritumoral non-enhancing regions, as calculated by an artificial neural network, allows for robust differentiation of glioblastoma and brain metastasis.
Abstract
Glioblastoma may appear similar to cerebral metastasis on conventional MRI in some cases, but their therapies differ significantly. This prospective feasibility study was aimed at differentiating them by applying the quantitative susceptibility mapping and quantitative blood-oxygen-level-dependent (QSM + qBOLD) model to these entities for the first time. We prospectively included 15 untreated patients with glioblastoma (n = 7, median age: 68 years, range: 54–84 years) or brain metastasis (n = 8, median age 66 years, range: 50–78 years) who underwent preoperative MRI including multi-gradient echo and arterial spin labeling sequences. Oxygen extraction fraction (OEF), cerebral blood flow (CBF) and cerebral metabolic rate of oxygen (CMRO2) were calculated in the contrast-enhancing tumor (CET) and peritumoral non-enhancing T2 hyperintense region (NET2), using an artificial neural network. We demonstrated that OEF in CET was significantly lower (p = 0.03) for glioblastomas than metastases, all features were significantly higher (p = 0.01) in CET than in NET2 for metastasis patients only, and the ratios of CET/NET2 for CBF (p = 0.04) and CMRO2 (p = 0.01) were significantly higher in metastasis patients than in glioblastoma patients. Discriminative power of a support-vector machine classifier was highest with a combination of two features, yielding an area under the receiver operating characteristic curve of 0.94 with 93% diagnostic accuracy. QSM + qBOLD allows for robust differentiation of glioblastoma and cerebral metastasis while yielding insights into tumor oxygenation.
1. Introduction
Glioblastoma (GBM) and cerebral metastasis (cMET) are the most common brain tumors in adult patients [1]. Reliably differentiating GBM and cMET based on their conventional magnetic resonance imaging (MRI) characteristics has proven difficult [2,3], as both tumor types can show necrotic centers, contrast-enhancing peripheral areas and peritumoral edema (Figure 1) [4]. However, studies employing advanced MR-imaging techniques focusing on the tumor microenvironment and hypoxia-induced changes in the microvasculature found that an elevated cerebral blood flow (CBF) and proxies for increased metabolic activity including a higher resulting cerebral metabolic rate of oxygen (CMRO2) were associated with high-grade gliomas [5,6]. Therefore, we performed a prospective feasibility study to differentiate between GBM and cMET by using an artificial neural network (ANN) approach for non-invasive estimation of CMRO2, combining quantitative susceptibility mapping (QSM) and the quantitative blood-oxygenation-level-dependent effect (qBOLD). To the best of the authors’ knowledge, the concept of using the combined QSM + qBOLD approach for estimation of CMRO2 is new for differentiating these two entities.
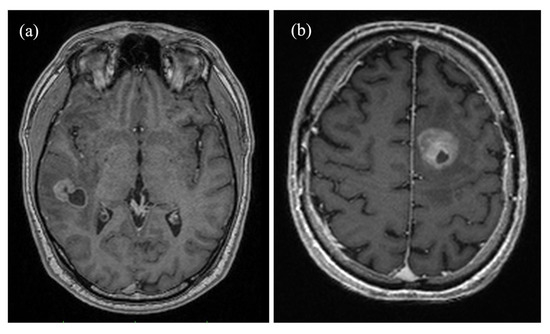
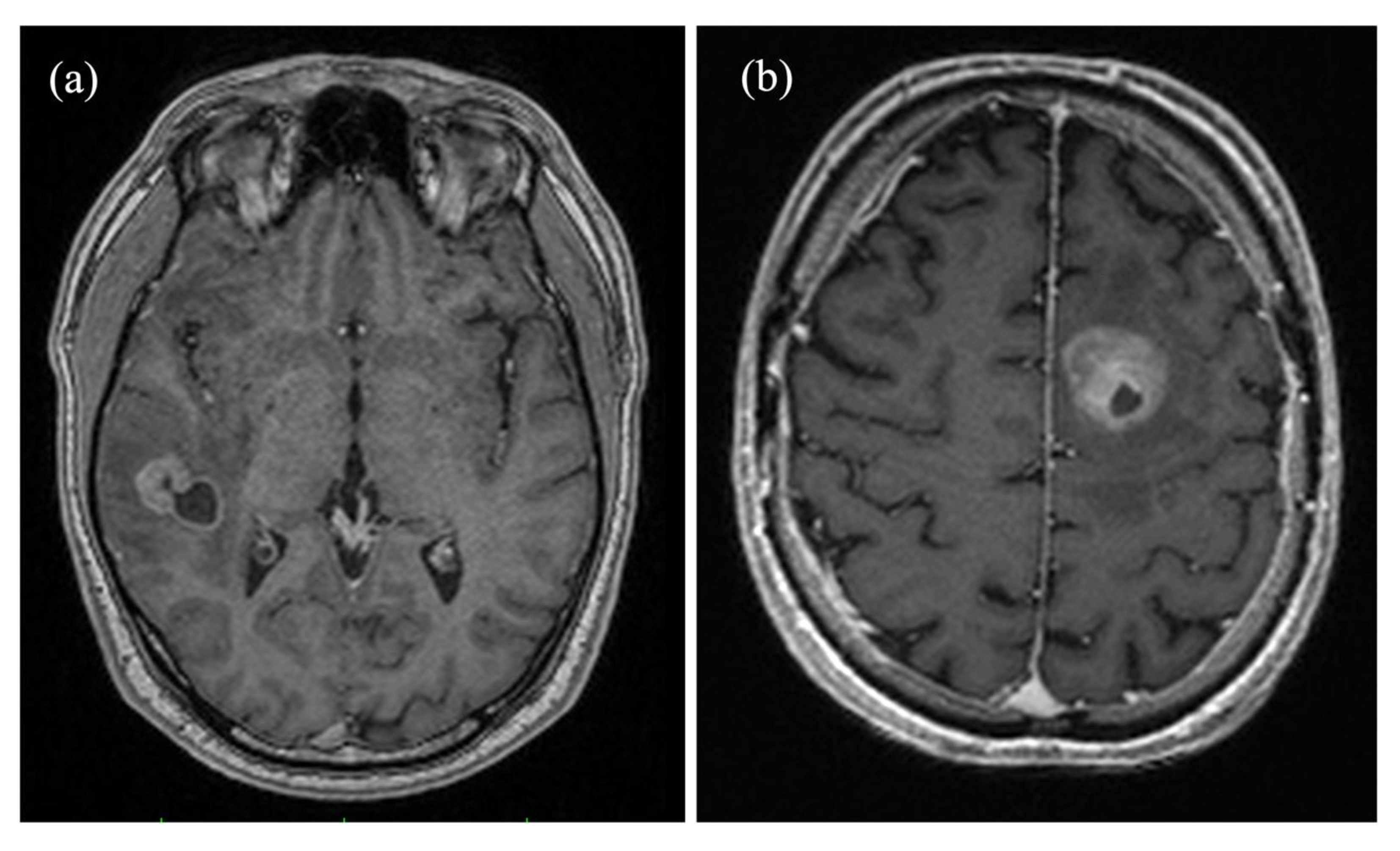
Figure 1.
Axial contrast-enhanced magnetization prepared rapid gradient echo (MP-RAGE) brain images comparing (a) a right temporal metastasis from known esophageal cancer in a 50-year-old male patient and (b) a glioblastoma in the frontal lobe of the left hemisphere of a 71-year-old male patient. Both tumors show cystic elements and peripheral contrast enhancement, complicating a differentiation based on solely morphological criteria.
GBMs constitute between 60% and 70% of all malignant gliomas [7]. Median survival for this highly malignant, infiltratively growing tumor is between 12 and 15 months with optimal treatment [7,8,9]. A higher median survival of 22 months has been reported for patients with recurrent glioblastoma undergoing a second surgical therapy [10]. The metabolism of GBM cells is adapted to the increased uptake of nutrients by utilizing aerobic glycolysis [11], thereby linking glucose metabolism to oxygen metabolism [12]. Although benefiting from a greater supply of components for cell upkeep and increased perfusion, GBMs frequently grow too fast for their vasculature, eventually resulting in the typical central necrosis with peripheral ring-enhancement [7,8].
Similarly, cMETs can also become ring-enhancing after developing a necrotic center due to them outgrowing their blood and nutrient supply [13,14]. Over 20% of cancer patients develop disseminations to the central nervous system [15]. One study identified that 55% of cMET cases had no known primary at diagnosis [16], while between 30% to 50% of cMETs have been found to first appear as solitary lesions, further complicating their correct identification [2,17,18]. Accurately discriminating between GBM and cMET is of great clinical importance because therapy approach and surgical decisions are quite different and directly affect patient outcomes [9,17,19]. The current diagnostic standard is an invasive tissue biopsy with subsequent histopathological examination [4], a procedure that is not without inherent risks with a complication rate of about 6% [20].
Hence, the discrimination of GBM and cMET has been attempted many times using a variety of different radiological approaches [4,19,21,22,23,24]. Perfusion-based studies showed relative cerebral blood volume (rCBV) in the proximal peritumoral non-enhancing T2 FLAIR hyperintensity (NET2) to be significantly higher in GBM than in cMET [21,22,23,24]. In the distal parts of NET2, GBM and cMET showed similar rCBV values which may reflect a lack of GBM cell infiltration and angiogenesis, supporting the notion that angiogenesis follows a gradient around the tumor and is highest on the surface of the contrast-enhancing tumor region (CET) [24,25]. With regards to oxygenation, tissue hypoxia is widely accepted as a predictor of therapy resistance to radiation and chemotherapy in gliomas [5]. Hypoxia has been found to stimulate the growth of new blood vessels via the induction and release of vascular endothelial growth factor [10]. This neovascularization leads to a dilated and tortuous vessel configuration, abnormal branching and arteriovenous shunts [26]. The resulting inefficiencies in the tumor vasculature were found to be correlated with greatly increased CBF, lower oxygen extraction fraction (OEF) and, in sum, a higher CMRO2 [6].
In this work, tissue oxygenation was estimated using a combined QSM + qBOLD model that was introduced in 2018 and which utilizes both signal magnitude and phase of a 3D multi-gradient echo (mGRE) sequence [27,28,29]. We built upon an existing artificial neural network approach to perform the QSM + qBOLD analysis [29]. The artificial neural network was previously used only for mapping OEF in healthy individuals, making this study its first application in a clinical setting by employing it for the differential diagnosis of two brain tumor entities. Perfusion was measured with pseudocontinuous arterial spin labeling.
The purpose of this study was, therefore, to apply the QSM + qBOLD method for the first time to a prospectively recruited collective of GBM and cMET patients and to compare their cerebral oxygenation and perfusion. Based on the hypothesis that the infiltrative growth of GBM and the lack thereof in cMET would create differences in CET and NET2, a machine learning classifier was trained to differentiate the two entities.
2. Materials and Methods
2.1. Patients
Between December 2019 and October 2020, 15 patients with primary GBM (n = 7; median age: 68 years, range: 54–84 years) or cMET (n = 8; median age: 66 years, range: 50–78 years) before resection, radiation or chemotherapy were prospectively included in this study as a convenience sample. Ten patients were male, five were female. The cMETs, as determined by histopathology, originated from four lung carcinomas, one esophageal carcinoma and three cancers of unknown primary. Ethics committee approval was granted before recruitment of patients (reference: 2017-666N-MA). Written informed consent was obtained from every participant prior to MRI measurements and the acquired image data was anonymized before further processing. The study was conducted in compliance with the ethical standards of the Declaration of Helsinki of 2013.
2.2. Image Acquisition
All MRI scans were performed on a 3T MAGNETOM Trio system (Siemens Healthcare GmbH, Erlangen, Germany) at the Department of Neuroradiology of the Medical Faculty Mannheim. The perfusion data of the first two patients (one cMET, one GBM) were acquired with a 32-channel head coil. Due to practical reasons in clinical day-to-day MRI scans, a 12-channel head coil was used for all subsequent study participants. The MRI protocol included a 3D multi-gradient echo, an unbalanced axial two-dimensional pseudocontinuous arterial spin labeling (three-dimensional pseudocontinuous arterial spin labeling was employed for the first two patients that were scanned with a 32-channel head coil) and a magnetization prepared rapid gradient echo (MP-RAGE) sequence. The gadolinium-based contrast agent Dotarem® (Guerbet, Villepinte, France) was administered as bolus injection for the T1-weighted sequence at a dosage of 0.1 mL/kg body weight. The labeling plane for the arterial spin labeling sequence was placed circa 85 mm inferior to the anterior commissure-posterior commissure line, approximately perpendicular to the feeding arteries, in line with the consensus recommendation for arterial spin labeling imaging [30]. The sequences and specific parameters used in this study have all been described in detail in a previously published study [29].
2.3. Image Processing
The MRI images were registered using the statistical parametric mapping software SPM12 (Wellcome Centre for Human Neuroimaging, UCL, London, UK) using default values. Correct registration was verified with the open-source medical image viewer ITK-SNAP (http://www.itksnap.org/pmwiki/pmwiki.php, accessed on 13 October 2021). Post-processing was performed with MATLAB (Mathworks, Natick, MA, USA). Three regions of interest (ROI) were outlined manually (H.B.) for calculation of oxygenation and perfusion parameters: CET and central necrosis on all slices of the acquired images where the respective region was present, as well as NET2 (Figure 2). The propensity of the central necrotic region to hemorrhage and its very low to non-existent perfusion can lead to an unphysiological spike in OEF (cf. Figure 3) [5]. Therefore, the necrotic region was subtracted from CET to exclude non-vital parts of the tumor in metabolic assessment. For the peritumoral edematous area, a 15–20 mm wide ROI was defined in NET2 on three consecutive FLAIR images with the largest peritumoral hyperintensity, leaving an approximately 3 mm wide margin to avoid partial volume effects [31]. All ROIs were adapted to the tumor border, subtracting a cerebrospinal fluid mask of ventricles and sulci generated automatically in SPM12. The ROIs were audited by an experienced neuroradiologist (H.W.).
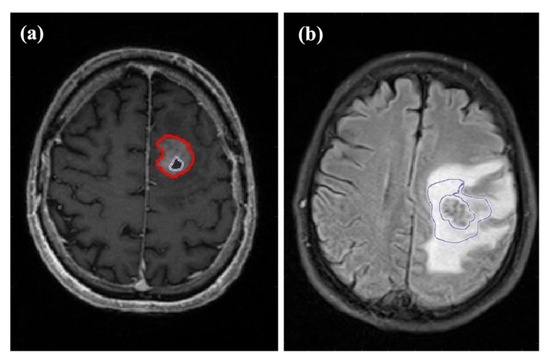
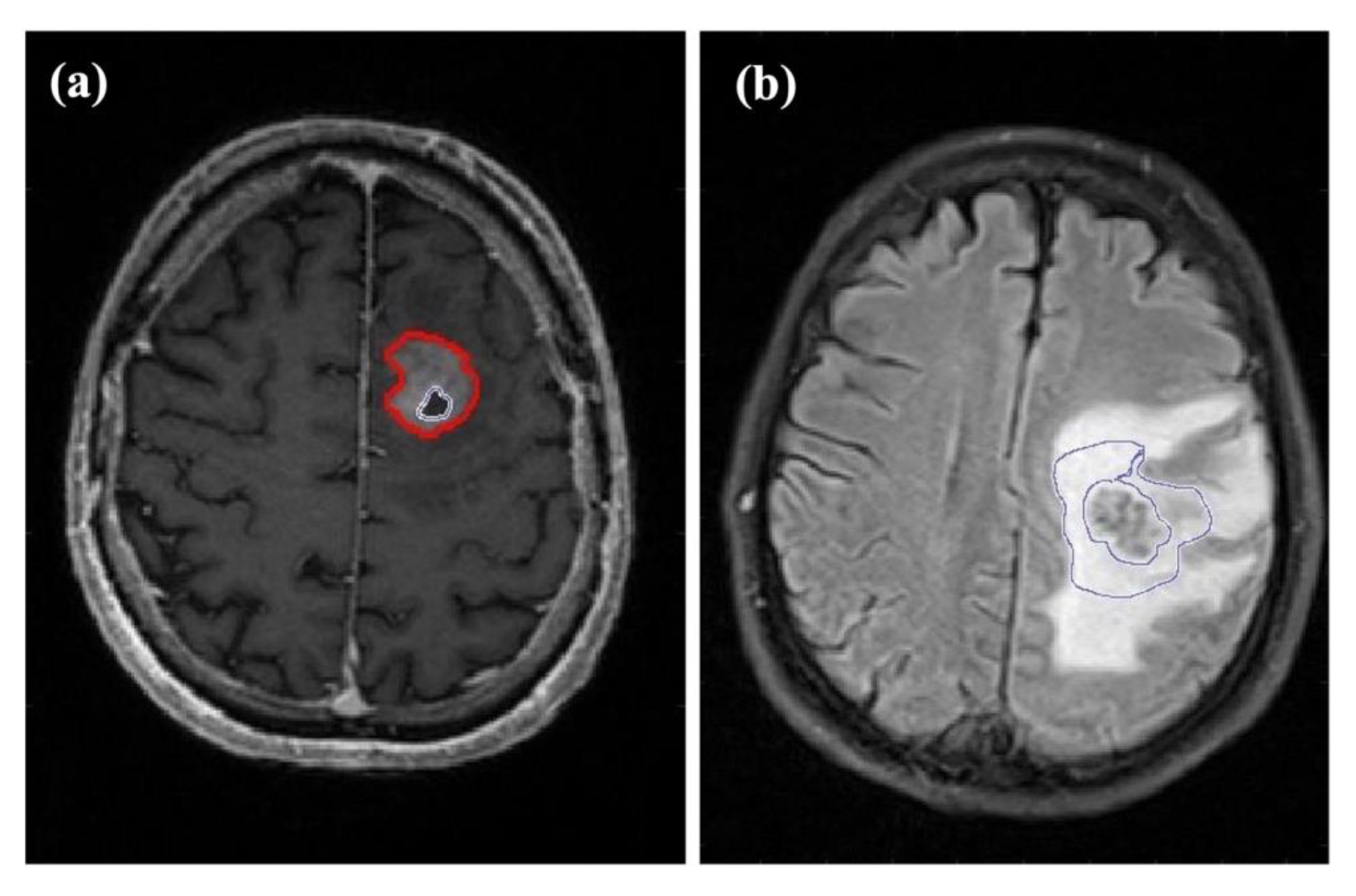
Figure 2.
Examples of the different regions of interest (ROIs) assessed in this study. (a) Axial T1-weighted contrast-enhanced image of a 71-year-old male patient with glioblastoma in the frontal lobe of the left hemisphere. The outer ROI marks contrast-enhancing tumor while the ROI inside the tumor indicates the central necrosis. (b) Axial FLAIR image of a 66-year-old male patient with a left hemispheric metastasis in the perirolandic region from unknown primary. The ROI marks the proximal edema bordering the solid-appearing tumor.
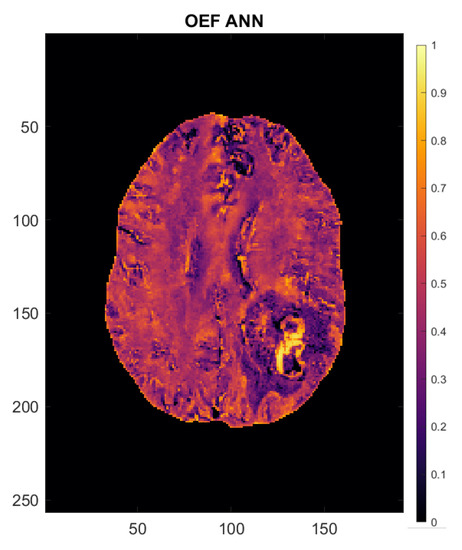
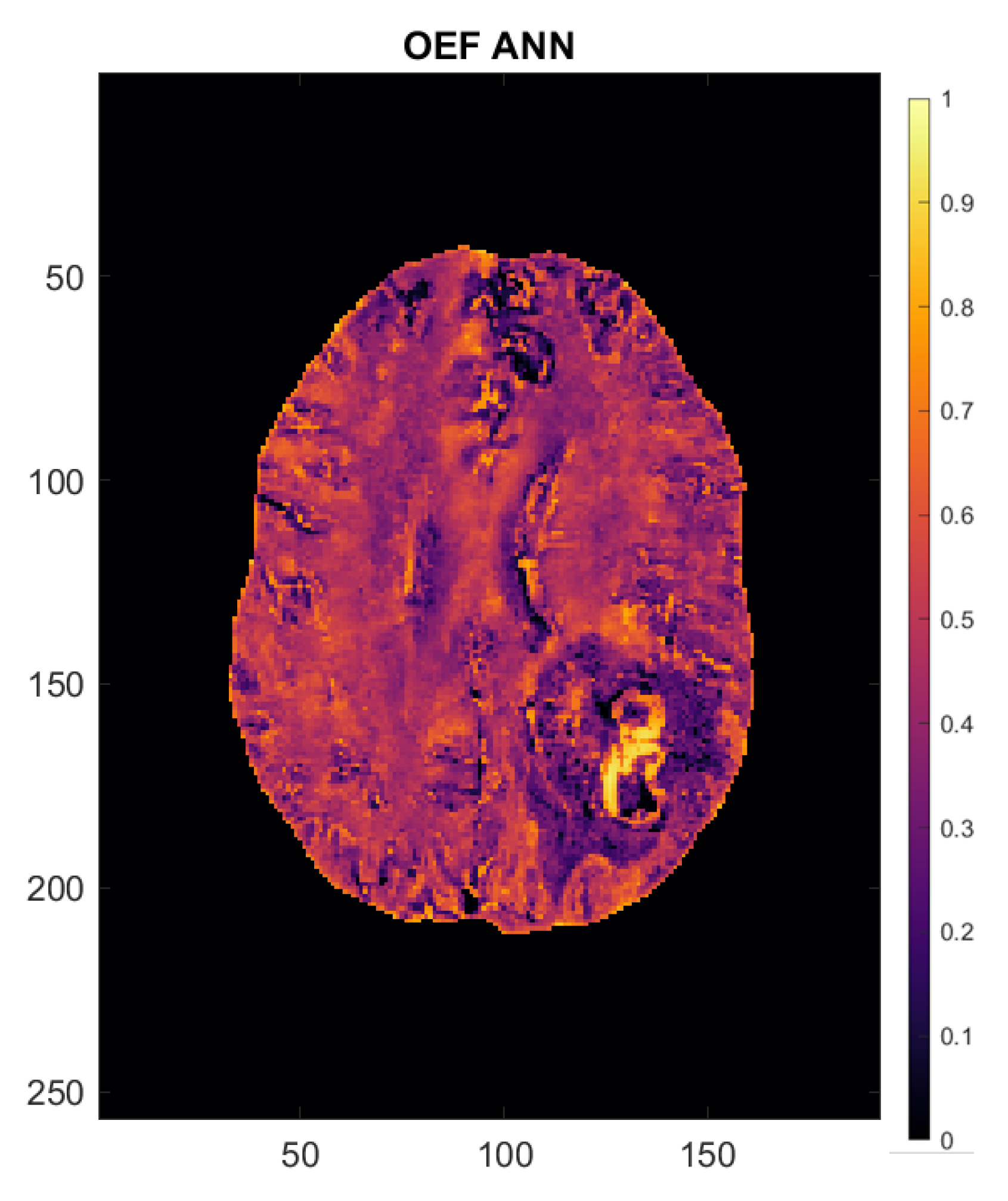
Figure 3.
Oxygen extraction fraction (OEF) map of a 54-year-old female patient with left occipito-temporal glioblastoma. Axes are in mm. The scale to the right of the image indicates OEF (e.g., 0.5 = 50%).
2.4. Calculation of Perfusion and Oxygenation Parameters
In order to estimate cerebral perfusion, a quantitative perfusion map was created from the arterial spin labeling data using SPM12. Both the control and tag images from the pCASL sequence were averaged and used together with the proton density weighted image to calculate CBF in mL/100 g/min, using the consensus recommendation for implementation of arterial spin labeling perfusion MRI in clinical applications from Alsop et al. [30]. Time correction for multi-slice imaging was implemented to account for the different transit time of the labeled bolus depending on the time of slice acquisition [30].
CMRO2 in μmol/100 g/min was calculated as follows:
where Υa is the arterial oxygen saturation, assumed to be 98%, and [H] = 7.53 μmol/mL is the heme molar concentration in tissue blood assuming a hematocrit of Hct = 0.357 in arterioles [27,32].
An artificial neural network was employed for combined QSM + qBOLD analysis of the mGRE data to estimate Υ and calculate OEF [33]:
where Υ and Υa are venous and arterial blood oxygenation, the latter again assumed to be 98% [27].
2.5. Artificial Neural Network
The feed-forward artificial neural network used for estimation of Υ was designed in the Neural Network Toolbox in MATLAB (Mathworks, Natick, MA, USA), consisting of one input layer, one hidden layer with 10 nodes and one output layer. The normalized mGRE magnitude signal and the magnetic susceptibility from QSM were used as inputs. The artificial neural network has been described in detail in a previous study [26], where it was trained and used to emulate the solution of the qBOLD model for free induction decay [27], yielding an estimate of venous oxygen saturation Υ, transverse relaxation rate R2, deoxygenated blood volume ν and non-blood susceptibility χnb [29]. After doing this for every voxel, OEF maps were created with Equation (2). In order to make the qBOLD model more robust, QSM was added by calculating magnetic susceptibility [27], using the MEDI toolbox (Cornell MRI Research Lab, Cornell University, New York, NY, USA). The resulting parameters were used for a last fitting step by giving starting values for a quasi-Newton optimization that was stopped once the relative change was smaller than 0.001 or a maximum of 50 iterations was reached (this was mostly the case after approx. 10 iterations) [34].
2.6. Statistical Analysis
The statistical analyses were descriptive and performed in MATLAB R2020b (Mathworks, Natick, MA, USA). Variables were summarized using their median, minimum–maximum and interquartile range. Outliers were included in the statistical tests. For all patients, means and standard deviations of OEF, CBF and CMRO2 were calculated in the ROIs set out in Figure 2. In patients with multiple cMETs, perfusion and oxygenation parameters were not assessed on a metastasis-by-metastasis basis but averaged across all metastases present in the respective patient’s brain. GBM patients were not stratified according to IDH or MGMT promoter methylation status.
Non-parametric Mann–Whitney–Wilcoxon tests for two populations were applied to properly compare the distributions of explanatory variables between the two groups. Two-tailed Wilcoxon signed-rank tests were used for intra-individual comparisons between CET and NET2. p-values < 0.05 were considered significant. Due to the explorative nature of this study, p-values were not adjusted for multiple comparisons and power analysis for determination of required sample size was not performed [35].
The primary learning objective was the binary classification of GBM vs. cMET. For this, a well-established maximum margin classifier, a linear kernel support-vector machine [36], was fitted to the following features: OEF, CBF and CMRO2 in CET and NET2. Additional secondary measures, i.e., the ratios of said features in CET divided by NET2, were also calculated and used for fitting [17]. We performed five-fold cross-validation with 10 repeats to assess the robustness of these explanatory variables [37]. Receiver operating characteristic analysis was performed to calculate the area under the receiver operating characteristic curve (AUC) accuracy metrics and derivatives of the confusion matrix, averaged over the five folds [37]. In an effort to compare the classification performance of the linear kernel support-vector machine with other common binary classifiers, ROC analysis was performed for naïve Bayes, weighted k-nearest neighbor, decision trees and for quadratic as well as Gaussian kernel support-vector machines.
3. Results
Analysis of oxygenation and perfusion maps in both groups revealed OEF to be lower and CBF to be generally higher than in normal-appearing brain, leading to a net positive effect on CMRO2, meaning a higher oxygen metabolism in tumor tissue than in an unaffected contralateral brain, both in GBM and in cMET patients. The OEF map of a 54-year-old female patient with left occipito-temporal GBM supplied in Figure 3 gives an illustrative example of the oxygenation values emulated by the artificial neural network. The patient’s brain shows largely uniform OEF values across “healthy” brain matter. The solid-appearing region of the tumor displays a strong OEF signal compared to the surrounding tissue and the rest of the brain while OEF in the peritumoral edematous area appears to be lower than in contralateral white matter.
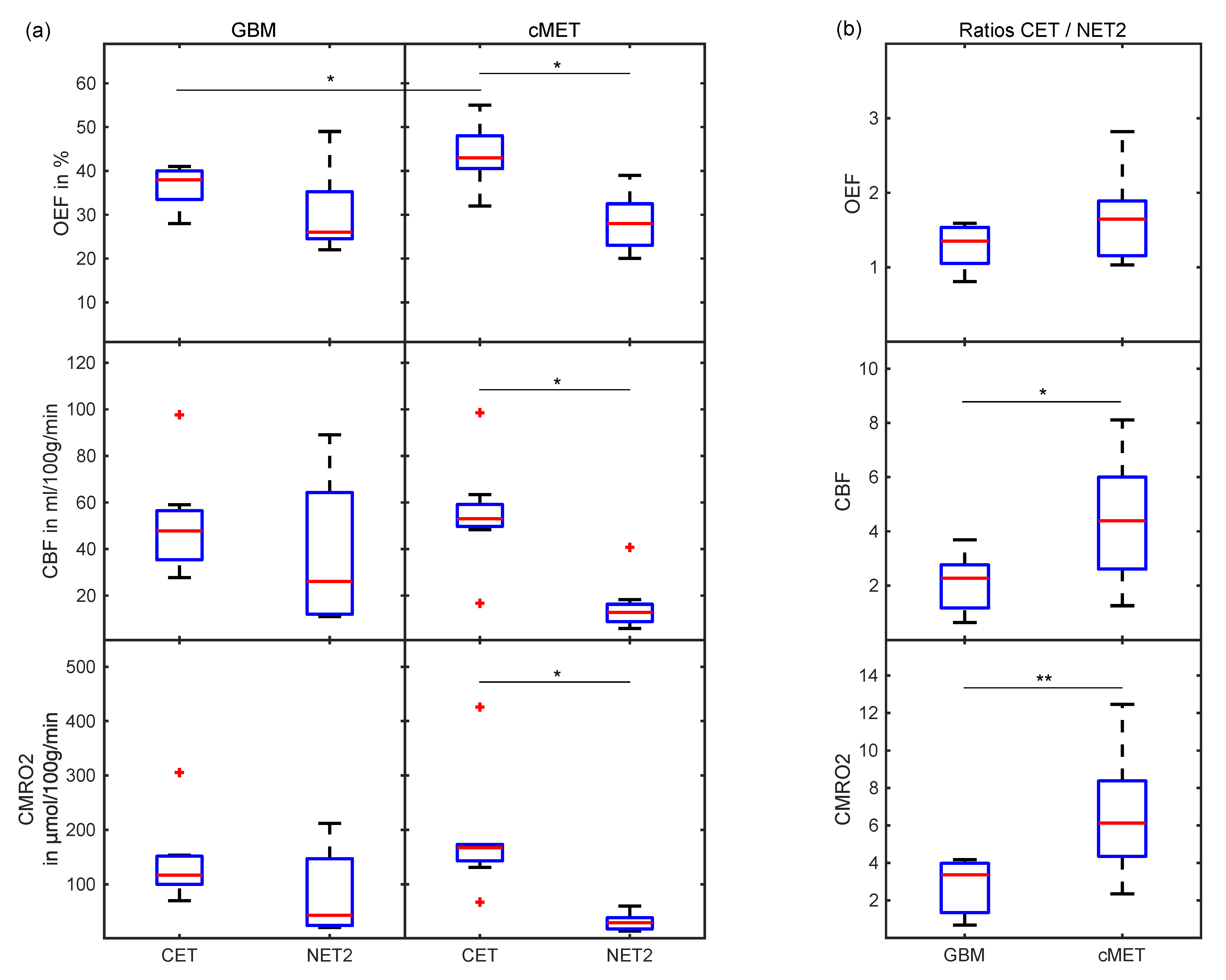
When comparing oxygenation and perfusion parameters between GBM and cMET, OEF in CET was found to be significantly (p = 0.03) lower in GBM than in cMET. No significant differences were found between CET of GBM and cMET in terms of CBF (p = 0.33) and CMRO2 (p = 0.15). For cMET patients, all parameters, i.e., OEF, CBF and CMRO2, were significantly (p = 0.01) higher in the CET region than in the NET2 region. Meanwhile, for the GBM group, neither the difference between CET and NET2 in OEF (p = 0.11), nor CBF (p = 0.15), nor CMRO2 (p = 0.08) was significant. A visual representation of this can be found in the boxplots supplied in Figure 4a. For an overview of oxygenation and perfusion parameters on a patient-by-patient basis, please consult Table S1 (supplement).
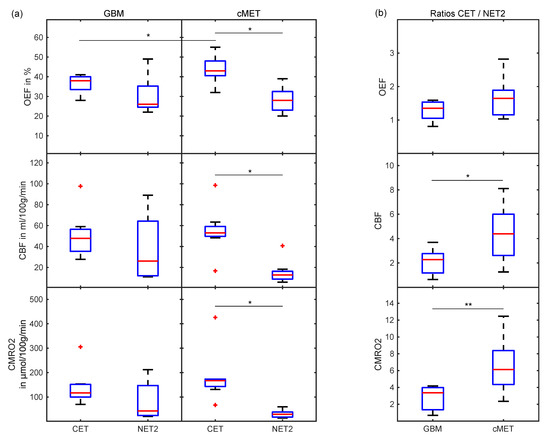
Figure 4.
Boxplots (a) comparing OEF, CBF and CMRO2 between CET and NET2 for GBM and cMET and (b) displaying the ratios of OEF, CBF and CMRO2 in CET divided by NET2 for GBM and cMET patients. Box: first to third quartile; whiskers: 1.5 times the interquartile distance or the maximum/minimum value, if contained therein; red line: median. Outliers are displayed as red crosses. Significant differences (p < 0.05) are marked with an asterisk, the highly significant difference (p = 0.01) is marked with two asterisks. OEF: oxygen extraction fraction; CBF: cerebral blood flow; CMRO2: cerebral metabolic rate of oxygen; CET: contrast-enhancing tumor; NET2: peritumoral non-enhancing T2 FLAIR hyperintensity.
The ratio of CET divided by NET2 was demonstrated to be another useful metric for differentiation of GBM and cMET. While OEF for CET/NET2 was not significantly different between GBM and cMET patients (p = 0.12), the ratio for CBF was significantly higher for cMET (p = 0.04), as was the ratio for CMRO2 (p = 0.01). Boxplots depicting these quotients are displayed in Figure 4b.
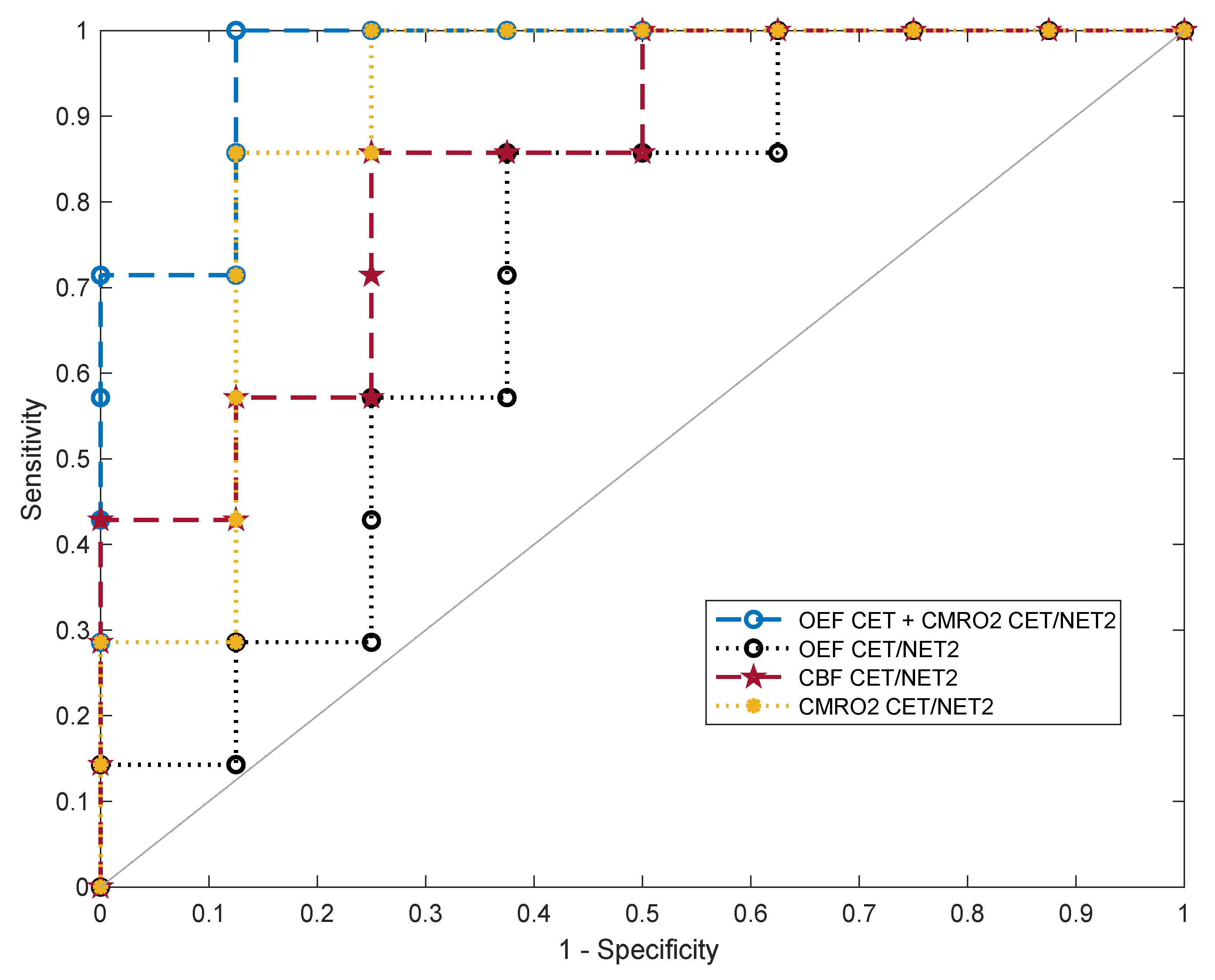
After fitting a classifier support-vector machine to the oxygenation and perfusion features, different metrics for binary classification of GBM and cMET were assessed. The receiver operating characteristic curves of the ratios of OEF, CBF and CMRO2 in CET divided by NET2 as well as the best overall feature combination in terms of AUC (OEF in CET and CMRO2 in CET/NET2) are exhibited in Figure 5.
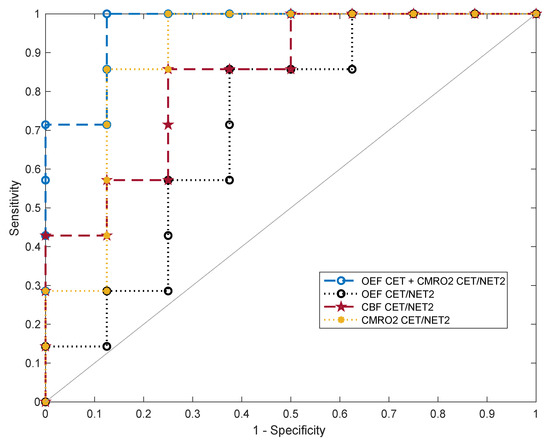
Figure 5.
Five-fold cross-validated ROC curves for the support-vector machine classifier predicting binary outcome (GBM or cMET). Curves are shown for the ratios of oxygen extraction fraction (OEFCET/NET2), cerebral blood flow (CBFCET/NET2) and cerebral metabolic rate of oxygen (CMRO2 CET/NET2) in contrast-enhancing tumor (CET) divided by peritumoral non-enhancing T2 FLAIR hyperintensity (NET2) as well as for the multivariable fit to OEFCET and CMRO2 CET/NET2. OEFCET: OEF in contrast-enhancing tumor.
For each parameter, values for accuracy, optimal sensitivity and specificity and AUC are listed in Table 1. CMRO2 CET/NET2 emerged as the best single feature for differentiation of GBM from cMET. The resulting model had an AUC of 0.85 with an accuracy of 83% at an optimal sensitivity and specificity of 85% and 82%, respectively. The next best single classification features in terms of AUC were the ratio CBFCET/NET2 (0.80) and OEF in CET (0.79). The highest discriminative power with the best diagnostic accuracy was achieved by combining OEFCET and CMRO2 CET/NET2 for fitting a support-vector machine (AUC = 0.94). This allowed for an accurate classification of the tumors in 93% of cases, with a sensitivity of 99% and a specificity of 88%. Averaged over all iterations, out of 15 tumor patients included in this study, 14 were correctly diagnosed and only one case was misclassified.

Table 1.
Receiver operating characteristic analysis results of a linear kernel support-vector machine trained on oxygenation and perfusion parameters.
We also assessed other machine learning classifiers that showed a lower discrimination performance including weighted k-nearest neighbor (AUC: 0.93, accuracy: 87%), naïve Bayes (AUC: 0.88, accuracy: 93%), and decision trees (AUC: 0.66, accuracy: 73%). Additionally, we investigated different variants of support-vector machines with quadratic (AUC: 0.89, accuracy: 87%) and Gaussian kernels (AUC: 0.86, accuracy: 87%). All of these achieved a smaller AUC with k-nearest neighbor coming closest to the linear kernel support-vector machine.
4. Discussion
In this feasibility study, we applied for the first time the MRI-based QSM + qBOLD approach for OEF and CMRO2 estimation to a prospectively recruited collective of GBM and cMET patients in order to distinguish the two entities based on their respective cerebral oxygenation and perfusion. Since the MRI protocol employed in this study for estimating cerebral tissue oxygenation and perfusion does not require the administration of an intravenous contrast agent per se, it offers the potential to facilitate metabolic imaging of cerebral tumors as well as non-invasive differential diagnosis of GBM and cMET. We demonstrated for the two patient groups that (i) OEF in the enhancing tumor was significantly lower in GBM than in cMET, (ii) the differences in perfusion and oxygenation between CET and NET2 were only significant for the cMET group, and (iii) the ratios of CBF and CMRO2 in CET divided by NET2 were significantly higher for cMET patients than for GBM patients.
In order to differentiate the two entities, a support-vector machine classifier was trained on oxygenation and perfusion parameters in CET and NET2. A support-vector machine was chosen since it emerged as the best classifier with the highest accuracy, in line with previous investigations that compared it to different approaches such as naïve Bayes, weighted k-nearest neighbor and decision trees for binary classification of glioblastoma and cerebral metastasis [4,23,38]. The described procedure could identify OEF in CET and the ratio of CMRO2 in CET versus NET2 as the most promising features for distinguishing GBM from cMET, achieving the highest discriminative power. With an accuracy of 93% and an AUC of 0.94, the two entities could be successfully differentiated.
Our results suggest that differentiation of GBM and cMET based solely on OEF in CET is good but not sufficient for reliably distinguishing the two groups. AUC and accuracy were higher than for any other individual parameter in CET or NET2 but lower than those obtained from using the ratios of CBF or CMRO2 in CET versus NET2. This may be explained by the tumor microenvironment, in particular angiogenesis, that is similar in the contrast-enhancing part of GBM and many hematogenous cMETs [17]. In both entities, a disruption of the blood-brain barrier is caused by tumor growth and angiogenesis: the hypoxic state of the tumor and its surroundings lead to hypoxia-inducible factors being activated and their gene product VEGF being expressed [39,40]. The improved classification performance with the ratios of CET versus NET2 is likely attributable to the dissimilarities in NET2 tissue structure between GBM and cMET. Higher vascularization and neoplastic cell growth are hallmarks of the region surrounding contrast-enhancing GBM [5,6,41], while the peritumoral edema around cMET is caused purely by vasogenic edema and does not show extensive tumor cell infiltration [42]. This may explain why OEF, CBF and CMRO2 were significantly higher in CET than in NET2 for cMET patients only.
We observed OEF in CET of GBM patients to be in line with the results of previous studies [26,27,43], reproducing the findings of low OEF, which is possibly a marker of less efficient oxygen extraction due to a physiologically inferior capillary configuration [27], coupled with high CBF. This gives plausibility to the parameter maps calculated with the QSM + qBOLD model that have in the past shown higher, more uniform OEF across the whole brain and more robust CMRO2 estimates than QSM or qBOLD alone [27]. It was surprising, however, to find discrepancies between the OEF values computed in NET2 of GBM patients: While we found lower OEF in NET2 than in CET, reproducing the results of a previous study [43], two other studies found higher OEF in the edema surrounding the tumor [5,6]. This might either be a cause of incongruent definitions of the ROIs “edema” and “NET2” or another underlying phenomenon. A PET/MR study with a sufficiently large number of patients might be useful to shed light on true OEF in the peritumoral region of GBM.
In addition to the good classification accuracy, the CMRO2 yielded by QSM + qBOLD serves as an important physiological parameter that has been shown to predict tumor response to antiangiogenic therapy as well as progression-free survival and overall survival in GBM patients [44] through association with intratumoral angiogenesis and oxygenation status [45]. Other possible applications of CMRO2 that warrant further investigation consist of monitoring tumor response after initial radiation or chemotherapy and early detection of recurrence. With regard to preoperative imaging, CMRO2 may also serve as a parameter to improve assessment of the surgical margins necessary for a more complete resection of glioblastomas, possibly adding complementary information to conventional MRI sequences for a “supratotal resection” [46].
This work is subject to a number of limitations. The QSM + qBOLD model requires a set of assumptions about physiologic parameters that were not measured for each patient individually. Among these assumptions are a constant tissue hematocrit and arterial oxygen saturation. Furthermore, the OEF values from QSM + qBOLD are prone to susceptibility artifacts from disturbances close to air-tissue bounds, e.g., the sinus frontalis, iron accumulations in deep grey matter or from blood degradation metabolites from hemorrhage, a phenomenon common in the necrotic centers of GBMs and cMETs, hence their exclusion from the assessment. Additional variance was introduced by changing the head coil after the first two patients. However, the perfusion values of these patients remained within a plausible range. Segmentation of ROIs was performed manually, adding a degree of intra- and inter-observer variability. Moreover, cMETs originating from various primary tumors may cause different oxygen metabolism characteristics. Subgroup analysis was not performed because of the relatively small sample size due to the explorative nature of this feasibility study. Thus, further research with larger patient populations is recommended before implementation of QSM + qBOLD into the clinical routine.
5. Conclusions
This study demonstrated that QSM + qBOLD allows for non-invasive differential diagnosis of GBM and cMET. In future studies, this differentiation could also be made without the use of an intravenous contrast agent. Going forward, our MRI approach for assessment of tissue oxygen metabolism might be helpful as a diagnostic tool that complements or replaces invasive stereotactic biopsies while yielding metabolic information about the tumor microenvironment and its surroundings, e.g., for predicting tumor response to therapy.
Supplementary Materials
The following is available online at https://www.mdpi.com/article/10.3390/app11219928/s1, Table S1: Oxygenation and perfusion parameters in CET and NET2 by individual patient.
Author Contributions
Conceptualization, H.B., A.F., L.R.S. and H.W.; methodology, H.B., S.H. and H.W.; software, H.B. and S.H.; validation, M.E.M. and S.A.M.; formal analysis, H.B., M.E.M. and S.A.M.; investigation, H.W.; resources, L.R.S. and H.W.; data curation, H.B. and S.A.M.; writing—Original draft preparation, H.B.; writing—Review and editing, S.H., S.A.M., M.E.M. and H.W.; visualization, H.B.; supervision, L.R.S. and H.W.; project administration, H.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Ethics Committee of the University Hospital Mannheim (reference number: 2017-666N-MA, date of approval: 6 August 2019).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. For a detailed breakdown of perfusion and oxygenation values by patient, please consult the supplementary material.
Acknowledgments
The authors are thankful for the technical help provided by the staff of the Institute for Computer Assisted Clinical Medicine at the Medical Faculty Mannheim.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Lee, E.J.; TerBrugge, K.; Mikulis, D.; Choi, D.S.; Bae, J.M.; Lee, S.K.; Moon, S.Y. Diagnostic value of peritumoral minimum apparent diffusion coefficient for differentiation of glioblastoma multiforme from solitary metastatic lesions. AJR Am. J. Roentgenol. 2011, 196, 71–76. [Google Scholar] [CrossRef]
- Bauer, A.H.; Erly, W.; Moser, F.G.; Maya, M.; Nael, K. Differentiation of solitary brain metastasis from glioblastoma multiforme: A predictive multiparametric approach using combined MR diffusion and perfusion. Neuroradiology 2015, 57, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, D.; Liao, S.; Guo, L.; Xiao, X.; Liu, X.; Xu, Y.; Hua, J.; Pillai, J.J.; Wu, Y. Discrimination between glioblastoma and solitary brain metastasis: Comparison of inflow-based vascular-space-occupancy and dynamic susceptibility contrast MR imaging. AJNR Am. J. Neuroradiol. 2020, 41, 583–590. [Google Scholar] [CrossRef]
- Artzi, M.; Bressler, I.; Ben Bashat, D. Differentiation between glioblastoma, brain metastasis and subtypes using radiomics analysis. J. Magn. Reason. Imaging 2019, 50, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Preibisch, C.; Shi, K.; Kluge, A.; Lukas, M.; Wiestler, B.; Gottler, J.; Gempt, J.; Ringel, F.; Al Jaberi, M.; Schlegel, J.; et al. Characterizing hypoxia in human glioma: A simultaneous multimodal MRI and PET study. NMR Biomed. 2017, 30, e3775. [Google Scholar] [CrossRef] [PubMed]
- Stadlbauer, A.; Zimmermann, M.; Kitzwogerer, M.; Oberndorfer, S.; Rossler, K.; Dorfler, A.; Buchfelder, M.; Heinz, G. MR imaging-derived oxygen metabolism and neovascularization characterization for grading and IDH gene mutation detection of gliomas. Radiology 2017, 283, 799–809. [Google Scholar] [CrossRef] [Green Version]
- Wen, P.Y.; Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noroxe, D.S.; Poulsen, H.S.; Lassen, U. Hallmarks of glioblastoma: A systematic review. ESMO Open 2016, 1, e000144. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Montemurro, N.; Fanelli, G.N.; Scatena, C.; Ortenzi, V.; Pasqualetti, F.; Mazzanti, C.M.; Morganti, R.; Paiar, F.; Naccarato, A.G.; Perrini, P. Surgical outcome and molecular pattern characterization of recurrent glioblastoma multiforme: A single-center retrospective series. Clin. Neurol. Neurosurg. 2021, 207, 106735. [Google Scholar] [CrossRef]
- Stadlbauer, A.; Oberndorfer, S.; Zimmermann, M.; Renner, B.; Buchfelder, M.; Heinz, G.; Doerfler, A.; Kleindienst, A.; Roessler, K. Physiologic MR imaging of the tumor microenvironment revealed switching of metabolic phenotype upon recurrence of glioblastoma in humans. J Cereb. Blood Flow Metab. 2020, 40, 528–538. [Google Scholar] [CrossRef]
- Montemurro, N.; Perrini, P.; Rapone, B. Clinical risk and overall survival in patients with diabetes mellitus, hyperglycemia and glioblastoma multiforme. A review of the current literature. Int. J. Environ. Res. Public Health 2020, 17, 8501. [Google Scholar] [CrossRef]
- Pope, W.B. Brain metastases: Neuroimaging. Handb. Clin. Neurol. 2018, 149, 89–112. [Google Scholar] [PubMed]
- Smirniotopoulos, J.G.; Murphy, F.M.; Rushing, E.J.; Rees, J.H.; Schroeder, J.W. Patterns of contrast enhancement in the brain and meninges. Radiographics 2007, 27, 525–551. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Wright, C.H.; Barnholtz-Sloan, J.S. Brain metastases: Epidemiology. Handb. Clin. Neurol. 2018, 149, 27–42. [Google Scholar] [PubMed]
- Giordana, M.T.; Cordera, S.; Boghi, A. Cerebral metastases as first symptom of cancer: A clinico-pathologic study. J. Neurooncol. 2000, 50, 265–273. [Google Scholar] [CrossRef]
- Server, A.; Orheim, T.E.; Graff, B.A.; Josefsen, R.; Kumar, T.; Nakstad, P.H. Diagnostic examination performance by using microvascular leakage, cerebral blood volume, and blood flow derived from 3-T dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging in the differentiation of glioblastoma multiforme and brain metastasis. Neuroradiology 2011, 53, 319–330. [Google Scholar]
- Blasel, S.; Jurcoane, A.; Franz, K.; Morawe, G.; Pellikan, S.; Hattingen, E. Elevated peritumoural rCBV values as a mean to differentiate metastases from high-grade gliomas. Acta Neurochir. 2010, 152, 1893–1899. [Google Scholar] [CrossRef]
- Lee, E.J.; Ahn, K.J.; Lee, E.K.; Lee, Y.S.; Kim, D.B. Potential role of advanced MRI techniques for the peritumoural region in differentiating glioblastoma multiforme and solitary metastatic lesions. Clin. Radiol. 2013, 68, e689–e697. [Google Scholar] [CrossRef]
- Malone, H.; Yang, J.; Hershman, D.L.; Wright, J.D.; Bruce, J.N.; Neugut, A.I. Complications following stereotactic needle biopsy of intracranial tumors. World Neurosurg. 2015, 84, 1084–1089. [Google Scholar] [CrossRef]
- Swinburne, N.C.; Schefflein, J.; Sakai, Y.; Oermann, E.K.; Titano, J.J.; Chen, I.; Tadayon, S.; Aggarwal, A.; Doshi, A.; Nael, K. Machine learning for semi-automated classification of glioblastoma, brain metastasis and central nervous system lymphoma using magnetic resonance advanced imaging. Ann. Transl. Med. 2019, 7, 232. [Google Scholar] [CrossRef]
- Askaner, K.; Rydelius, A.; Engelholm, S.; Knutsson, L.; Latt, J.; Abul-Kasim, K.; Sundgren, P.C. Differentiation between glioblastomas and brain metastases and regarding their primary site of malignancy using dynamic susceptibility contrast MRI at 3T. J. Neuroradiol. 2019, 46, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Tsolaki, E.; Svolos, P.; Kousi, E.; Kapsalaki, E.; Fountas, K.; Theodorou, K.; Tsougos, I. Automated differentiation of glioblastomas from intracranial metastases using 3T MR spectroscopic and perfusion data. Int. J. Comput. Assist. Radiol. Surg. 2013, 8, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, P.; Saliou, G.; de Marco, G.; Monet, P.; Souraya, S.E.; Bruniau, A.; Vallee, J.N.; Ducreux, D. Cerebral peritumoral oedema study: Does a single dynamic MR sequence assessing perfusion and permeability can help to differentiate glioblastoma from metastasis? Eur. J. Radiol. 2012, 81, 522–527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asgari, S.; Rohrborn, H.J.; Engelhorn, T.; Stolke, D. Intra-operative characterization of gliomas by near-infrared spectroscopy: Possible association with prognosis. Acta Neurochir. 2003, 145, 453–459. [Google Scholar] [CrossRef] [PubMed]
- Hardee, M.E.; Zagzag, D. Mechanisms of glioma-associated neovascularization. Am. J. Pathol. 2012, 181, 1126–1141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, J.; Kee, Y.; Spincemaille, P.; Nguyen, T.D.; Zhang, J.; Gupta, A.; Zhang, S.; Wang, Y. Cerebral metabolic rate of oxygen (CMRO2) mapping by combining quantitative susceptibility mapping (QSM) and quantitative blood oxygenation level-dependent imaging (qBOLD). Magn. Reason. Med. 2018, 80, 1595–1604. [Google Scholar] [CrossRef] [PubMed]
- Kurz, F.T.; Buschle, L.R.; Rotkopf, L.T.; Herzog, F.S.; Sterzik, A.; Schlemmer, H.P.; Kampf, T.; Bendszus, M.; Heiland, S.; Ziener, C.H. Dependence of the frequency distribution around a sphere on the voxel orientation. Z. Med. Phys. 2021. [Google Scholar] [CrossRef]
- Hubertus, S.; Thomas, S.; Cho, J.; Zhang, S.; Wang, Y.; Schad, L.R. Using an artificial neural network for fast mapping of the oxygen extraction fraction with combined QSM and quantitative BOLD. Magn. Reson. Med. 2019, 82, 2199–2211. [Google Scholar] [CrossRef]
- Alsop, D.C.; Detre, J.A.; Golay, X.; Gunther, M.; Hendrikse, J.; Hernandez-Garcia, L.; Lu, H.; MacIntosh, B.J.; Parkes, L.M.; Smits, M.; et al. Recommended implementation of arterial spin-labeled perfusion MRI for clinical applications: A consensus of the ISMRM perfusion study group and the European consortium for ASL in dementia. Magn. Reson. Med. 2015, 73, 102–116. [Google Scholar] [CrossRef] [Green Version]
- Dong, F.; Li, Q.; Jiang, B.; Zhu, X.; Zeng, Q.; Huang, P.; Chen, S.; Zhang, M. Differentiation of supratentorial single brain metastasis and glioblastoma by using peri-enhancing oedema region-derived radiomic features and multiple classifiers. Eur. Radiol. 2020, 30, 3015–3022. [Google Scholar] [CrossRef]
- Ma, Y.; Mazerolle, E.L.; Cho, J.; Sun, H.; Wang, Y.; Pike, G.B. Quantification of brain oxygen extraction fraction using QSM and a hyperoxic challenge. Magn. Reason. Med. 2020, 84, 3271–3285. [Google Scholar] [CrossRef]
- Hubertus, S.; Thomas, S.; Cho, J.; Zhang, S.; Wang, Y.; Schad, L.R. Comparison of gradient echo and gradient echo sampling of spin echo sequence for the quantification of the oxygen extraction fraction from a combined quantitative susceptibility mapping and quantitative BOLD (QSM+qBOLD) approach. Magn. Reason. Med. 2019, 82, 1491–1503. [Google Scholar] [CrossRef]
- Cho, J.; Zhang, S.; Kee, Y.; Spincemaille, P.; Nguyen, T.D.; Hubertus, S.; Gupta, A.; Wang, Y. Cluster analysis of time evolution (CAT) for quantitative susceptibility mapping (QSM) and quantitative blood oxygen level-dependent magnitude (qBOLD)-based oxygen extraction fraction (OEF) and cerebral metabolic rate of oxygen (CMRO2) mapping. Magn. Reason. Med. 2020, 83, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Wenz, H.; Maros, M.E.; Meyer, M.; Forster, A.; Haubenreisser, H.; Kurth, S.; Schoenberg, S.O.; Flohr, T.; Leidecker, C.; Groden, C.; et al. Image quality of 3rd generation spiral cranial dual-source CT in combination with an advanced model iterative reconstruction technique: A prospective intra-individual comparison study to standard sequential cranial CT using identical radiation dose. PLoS ONE 2015, 10, e0136054. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Maros, M.E.; Capper, D.; Jones, D.T.W.; Hovestadt, V.; von Deimling, A.; Pfister, S.M.; Benner, A.; Zucknick, M.; Sill, M. Machine learning workflows to estimate class probabilities for precision cancer diagnostics on DNA methylation microarray data. Nat. Protoc. 2020, 15, 479–512. [Google Scholar] [CrossRef]
- Shrot, S.; Salhov, M.; Dvorski, N.; Konen, E.; Averbuch, A.; Hoffmann, C. Application of MR morphologic, diffusion tensor, and perfusion imaging in the classification of brain tumors using machine learning scheme. Neuroradiology 2019, 61, 757–765. [Google Scholar] [CrossRef]
- Fanelli, G.N.; Grassini, D.; Ortenzi, V.; Pasqualetti, F.; Montemurro, N.; Perrini, P.; Naccarato, A.G.; Scatena, C. Decipher the glioblastoma microenvironment: The first milestone for new groundbreaking therapeutic strategies. Genes 2021, 12, 445. [Google Scholar] [CrossRef]
- Guan, Z.; Lan, H.; Cai, X.; Zhang, Y.; Liang, A.; Li, J. Blood-brain barrier, cell junctions, and tumor microenvironment in brain metastases, the biological prospects and dilemma in therapies. Front. Cell Dev. Biol. 2021, 9, 722917. [Google Scholar] [CrossRef] [PubMed]
- Sunwoo, L.; Yun, T.J.; You, S.H.; Yoo, R.E.; Kang, K.M.; Choi, S.H.; Kim, J.H.; Sohn, C.H.; Park, S.W.; Jung, C.; et al. Differentiation of glioblastoma from brain metastasis: Qualitative and quantitative analysis using arterial spin labeling MR imaging. PLoS ONE 2016, 11, e0166662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fink, K.R.; Fink, J.R. Imaging of brain metastases. Surg. Neurol. Int. 2013, 4, S209–S219. [Google Scholar] [CrossRef] [PubMed]
- Hubertus, S.; Thomas, S.; Cho, J.; Zhang, S.; Kovanlikaya, I.; Wang, Y.; Schad, L.R. In MRI-based oxygen extraction fraction and cerebral metabolic rate of oxygen mapping in high-grade glioma using a combined quantitative susceptibility mapping and quantitative blood oxygenation level-dependent approach. In Proceedings of the International Society for Magnetic Resonance in Medicine, Montréal, QC, Canada, 11–16 May 2019; p. 0391. [Google Scholar]
- Bonekamp, D.; Mouridsen, K.; Radbruch, A.; Kurz, F.T.; Eidel, O.; Wick, A.; Schlemmer, H.P.; Wick, W.; Bendszus, M.; Ostergaard, L.; et al. Assessment of tumor oxygenation and its impact on treatment response in bevacizumab-treated recurrent glioblastoma. J. Cereb. Blood Flow Metab. 2017, 37, 485–494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kickingereder, P.; Brugnara, G.; Hansen, M.B.; Nowosielski, M.; Pfluger, I.; Schell, M.; Isensee, F.; Foltyn, M.; Neuberger, U.; Kessler, T.; et al. Noninvasive characterization of tumor angiogenesis and oxygenation in bevacizumab-treated recurrent glioblastoma by using dynamic susceptibility MRI: Secondary analysis of the European Organization for Research and Treatment of Cancer 26101 trial. Radiology 2020, 297, 164–175. [Google Scholar] [CrossRef] [PubMed]
- Yordanova, Y.N.; Duffau, H. Supratotal resection of diffuse gliomas—An overview of its multifaceted implications. Neurochirurgie 2017, 63, 243–249. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).