Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of Bacteria
2.3. Morphological and Biochemical Characterization of Isolates
2.4. Determination of Probiotic and Antibacterial Potential
2.4.1. Enzymatic Potential
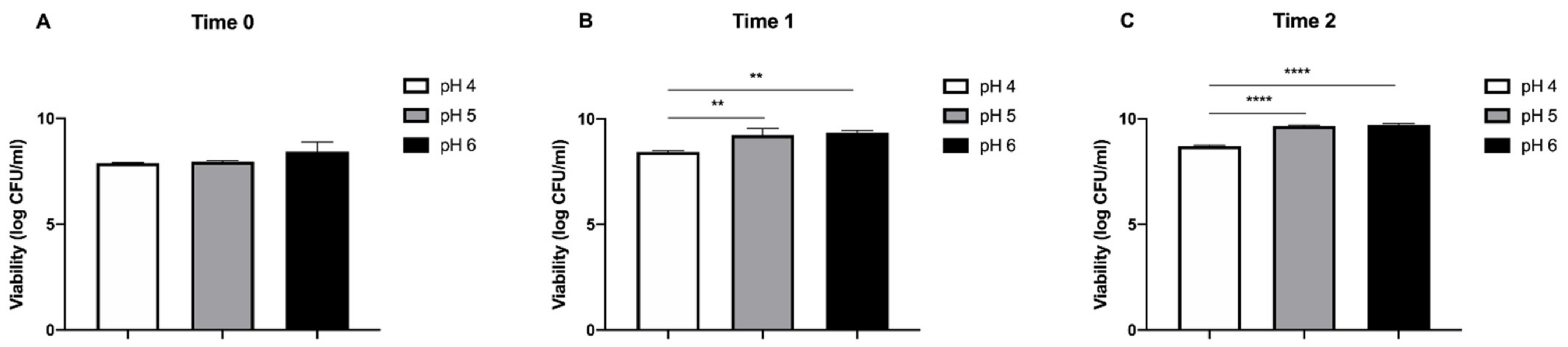
2.4.2. Acid Tolerance
2.4.3. Antibiotic Resistance and Antimicrobial Activity
2.4.4. DNA Extraction and Amplification
2.5. Sequencing Analysis
2.6. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rossi, M.; Amaretti, A.; Raimondi, S. Folate Production by Probiotic Bacteria. Nutrients 2011, 3, 118–134. [Google Scholar] [CrossRef] [Green Version]
- Blaiotta, G.; Murru, N.; Di Cerbo, A.; Romano, R.; Aponte, M. Production of probiotic bovine salami using Lactobacillus plantarum 299v as adjunct. J. Sci. Food Agric. 2018, 98, 2285–2294. [Google Scholar] [CrossRef] [PubMed]
- Di Cerbo, A.; Palmieri, B. Review: The market of probiotics. Pak. J. Pharm. Sci. 2015, 28, 2199–2206. [Google Scholar]
- Di Cerbo, A.; Palmieri, B.; Aponte, M.; Morales-Medina, J.C.; Iannitti, T. Mechanisms and therapeutic effectiveness of lactobacilli. J. Clin. Pathol. 2016, 69, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Fijan, S. Microorganisms with Claimed Probiotic Properties: An Overview of Recent Literature. Int. J. Environ. Res. Public Health 2014, 11, 4745–4767. [Google Scholar] [CrossRef] [PubMed]
- Amado, I.R.; Fuciños, C.; Fajardo, P.; Pastrana, L. Pediocin SA-1: A selective bacteriocin for controlling Listeria monocytogenes in maize silages. J. Dairy Sci. 2016, 99, 8070–8080. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.I.; Din, A.; Ali, G.M.; Khan, S.I.; Arif, I.; Riaz, M.N.; Ghazanfar, S. Screening of Lactic Acid Bacteria for Their Use as Buffalo Probiotic. J. Anim. Plant Sci. 2020, 30, 1357–1365. [Google Scholar]
- Woese, C.R.; Fox, G.E. Phylogenetic structure of the prokaryotic domain: The primary kingdoms. Proc. Natl. Acad. Sci. USA 1977, 74, 5088–5090. [Google Scholar] [CrossRef] [Green Version]
- Van Nieuwenhove, C.P.; Oliszewski, R.; Gonzalez, S.N.; Chaia, A.B.P. Conjugated linoleic acid conversion by dairy bacteria cultured in MRS broth and buffalo milk. Lett. Appl. Microbiol. 2007, 44, 467–474. [Google Scholar] [CrossRef]
- Manhar, A.K.; Bashir, Y.; Saikia, D.; Nath, D.; Gupta, K.; Konwar, B.K.; Kumar, R.; Namsa, N.D.; Mandal, M. Cellulolytic potential of probiotic Bacillus Subtilis AMS6 isolated from traditional fermented soybean (Churpi): An in-vitro study with regards to application as an animal feed additive. Microbiol. Res. 2016, 186–187, 62–70. [Google Scholar] [CrossRef]
- Marlida, R.; Suprayudi, M.; Widanarni, H.E.; Harris, E. Isolation, Selection and Application of Probiotic Bacteria for Improvement the Growth Performance of Humpback Groupers (Cromileptes altivelis). Int. J. Sci. Basic Appl. Res. 2014, 16, 364–379. [Google Scholar]
- Hassanzadazar, H.; Ehsani, A.; Mardani, K.; Hesari, J. Investigation of antibacterial, acid and bile tolerance properties of lactobacilli isolated from Koozeh cheese. Veter Res. Forum Int. Q. J. 2012, 3, 181–185. [Google Scholar]
- Olatunde, O.O.; Obadina, A.O.; Omemu, A.M.; Oyewole, O.B.; Olugbile, A.; Olukomaiya, O.O. Screening and molecular identification of potential probiotic lactic acid bacteria in effluents generated during ogi production. Ann. Microbiol. 2018, 68, 433–443. [Google Scholar] [CrossRef]
- Shakira, G.; Qubtia, M.; Ahmed, I.; Hasan, F.; Anjum, M.; Imran, M. Effect of indigenously isolated Saccharomyces cerevisiae probiotics on milk production, nutrient digestibility, blood chemistry and fecal microbiota in lactating dairy cows. J. Anim. Plant Sci. 2018, 28, 407–420. [Google Scholar]
- Ghazanfar, S.; Anjum, M.; Azim, A.; Ahmed, I.I. Effects of Dietary Supplementation of Yeast (Saccharomyces Cerevisiae) Culture on Growth Performance, Blood Parameters, Nutrient Digestibility and Fecal Flora of Dairy Heifers. J. Anim. Plant Sci. 2015, 25, 53–59. [Google Scholar]
- Coenye, T.; Vandamme, P. Intragenomic heterogeneity between multiple 16S ribosomal RNA operons in sequenced bacterial genomes. FEMS Microbiol. Lett. 2003, 228, 45–49. [Google Scholar] [CrossRef]
- Ghazanfar, S.; Ali, G.M. Pediococcus acidilactici strain NMCC-11 16S ribosomal RNA gene, partial sequence. 2019. Available online: www.ncbi.nlm.nih.gov/search/all/?term=MK606112 (accessed on 25 August 2021).
- Kumar, S.; Tamura, K.; Nei, M. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 1994, 10, 189–191. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Brooks, J.C.; Grunwald, N.J. Novel R tools for analysis of genome-wide population genetic data with emphasis on clonality. Front. Genet. 2015, 6, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shoukat, S.; Anwar, M.; Ali, S.; Begum, S.; Aqeel, M.; Zia, M.; Shah, S.; Ali, G. Estimation of Morphological and Molecular Diversity of Seventy-Two Advanced Pakistani Cotton Genotypes Using Simple Sequence Repeats. Appl. Ecol. Environ. Res. 2020, 18, 2043–2056. [Google Scholar] [CrossRef]
- Di Cerbo, A. Presencia de Antibióticos Inusual en el Gimnasio Entrenando Sujetos. Nutr. Hosp. 2014, 30, 395–398. [Google Scholar] [CrossRef]
- Iseppi, R.; Di Cerbo, A.; Messi, P.; Sabia, C. Antibiotic Resistance and Virulence Traits in Vancomycin-Resistant Enterococci (VRE) and Extended-Spectrum β-Lactamase/AmpC-producing (ESBL/AmpC) Enterobacteriaceae from Humans and Pets. Antibiot. 2020, 9, 152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghazanfar, S.; Khalid, N.; Ahmed, I.; Imran, I.A. Probiotic Yeast: Mode of Action and Its Effects on Ruminant Nutrition. In Yeast Industrial Applications; Morata, A., Loira, I., Eds.; IntechOpen: Rijeka, Croatia, 2017. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez-Palacios, A.; Staempfli, H.; Duffield, T.; Weese, J. Isolation of bovine intestinalLactobacillus plantarumandPediococcus acidilacticiwith inhibitory activity againstEscherichia coliO157 and F5. J. Appl. Microbiol. 2009, 106, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Ahmed, I.; Ahmed, S.; Ahmed, Z.; Riaz, M.N.; Ghazanfar, S. Screening of cattle gut associated Bacillus strains for their potential use as animal probiotic. Indian J. Anim. Res. 2018. [Google Scholar] [CrossRef]
- Morandi, S.; Brasca, M.; Alfieri, P.; Lodi, R.; Tamburini, A. Influence of pH and temperature on the growth of Enterococcus faecium and Enterococcus faecalis. Lait 2005, 85, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Abbasiliasi, S.; Tan, J.S.; Bashokouh, F.; Ibrahim, T.A.T.; Mustafa, S.; Vakhshiteh, F.; Sivasamboo, S.; Ariff, A.B. In vitro assessment of Pediococcus acidilactici Kp10 for its potential use in the food industry. BMC Microbiol. 2017, 17, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ho, J.C.K.; Sze, L.Y. Isolation, identification and characterization of enzyme-producing lactic acid bacteria from traditional fermented foods. Biosci. Horizons: Int. J. Stud. Res. 2018, 11, 11. [Google Scholar] [CrossRef]
- Osmanağaoğlu, Ö.; Kiran, F.; Ataoğlu, H. Evaluation of in vitro Probiotic Potential of Pediococcus pentosaceus OZF Isolated from Human Breast Milk. Probiotics Antimicrob. Proteins 2010, 2, 162–174. [Google Scholar] [CrossRef]
- Parvez, S.; Malik, K.; Kang, S.A.; Kim, H.-Y. Probiotics and their fermented food products are beneficial for health. J. Appl. Microbiol. 2006, 100, 1171–1185. [Google Scholar] [CrossRef]
- Fenchel, T.; Finlay, B.J. The diversity of microbes: Resurgence of the phenotype. Philos. Trans. R. Soc. B: Biol. Sci. 2006, 361, 1965–1973. [Google Scholar] [CrossRef] [Green Version]
- Kamvar, Z.N.; Tabima, J.F.; Grünwald, N.J. Poppr: An R package for genetic analysis of populations with clonal, partially clonal, and/or sexual reproduction. PeerJ 2014, 2, e281. [Google Scholar] [CrossRef] [Green Version]
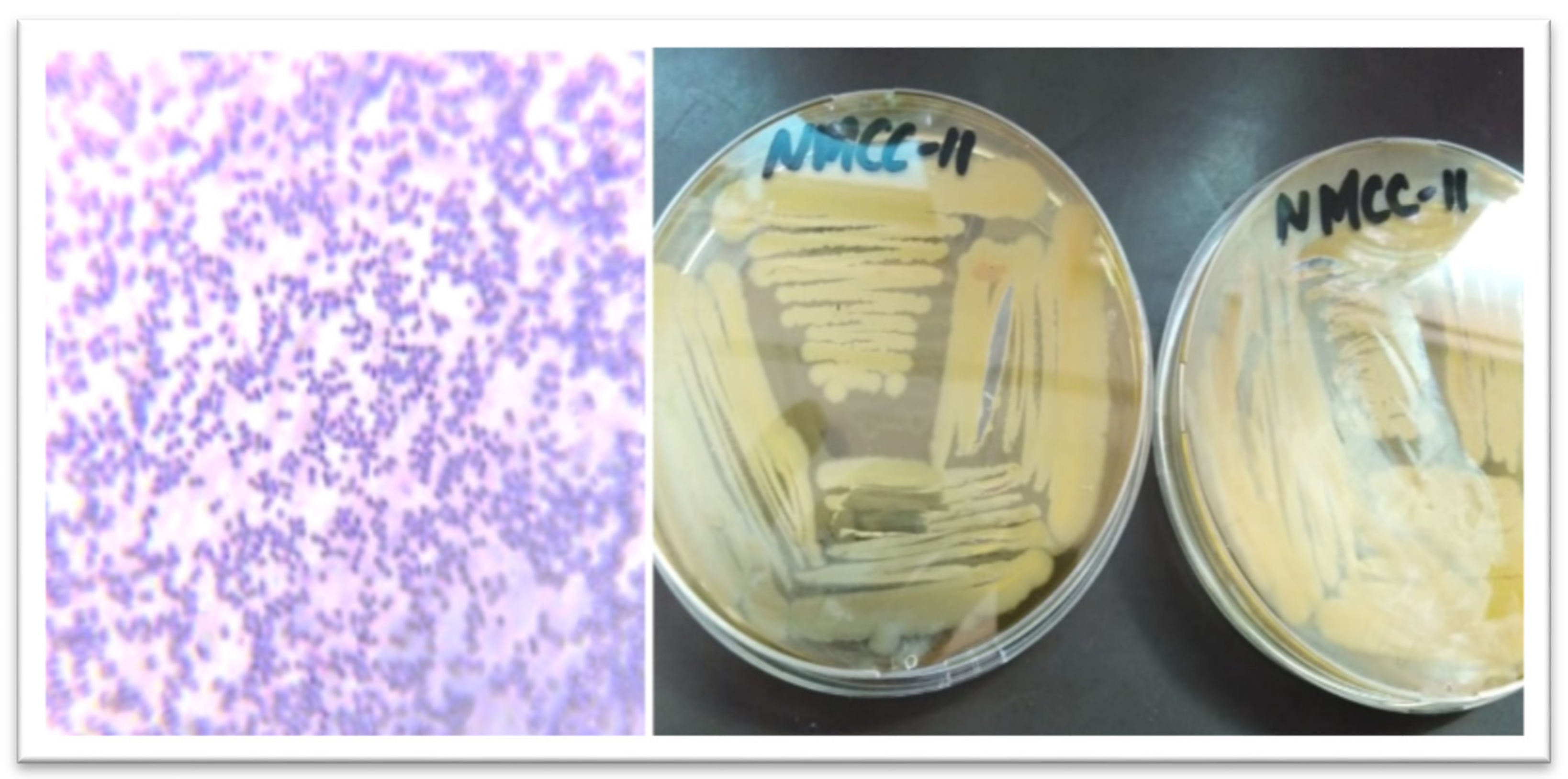
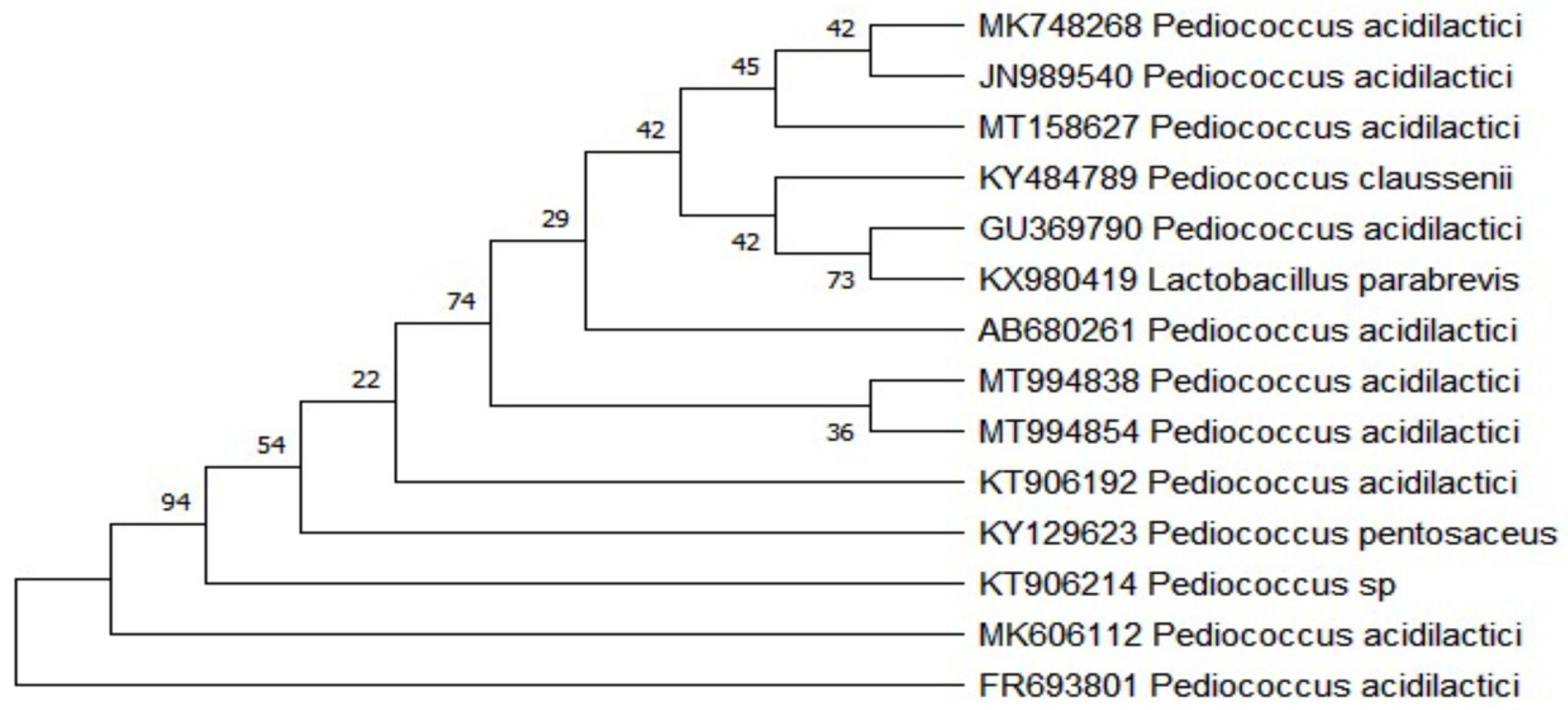
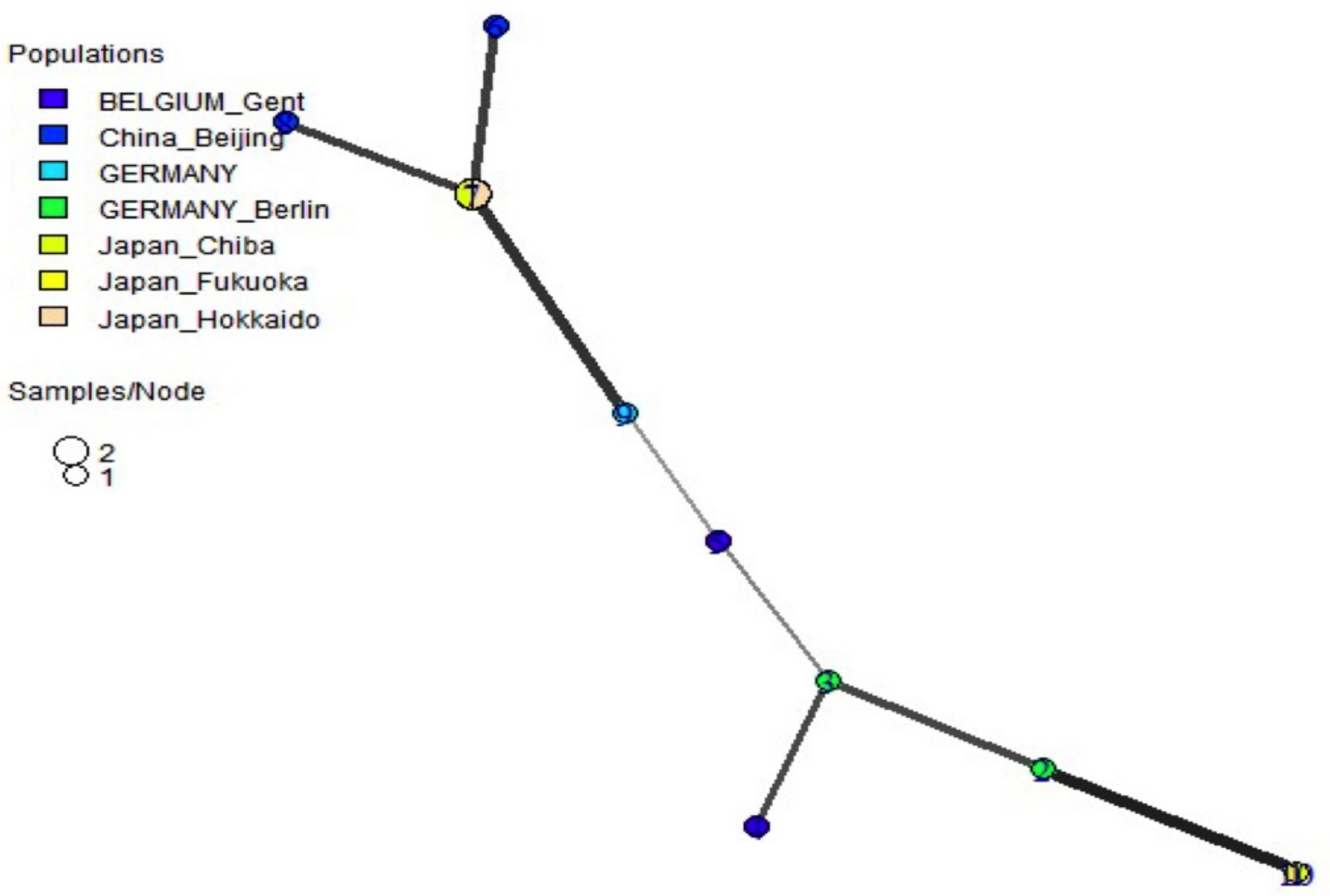
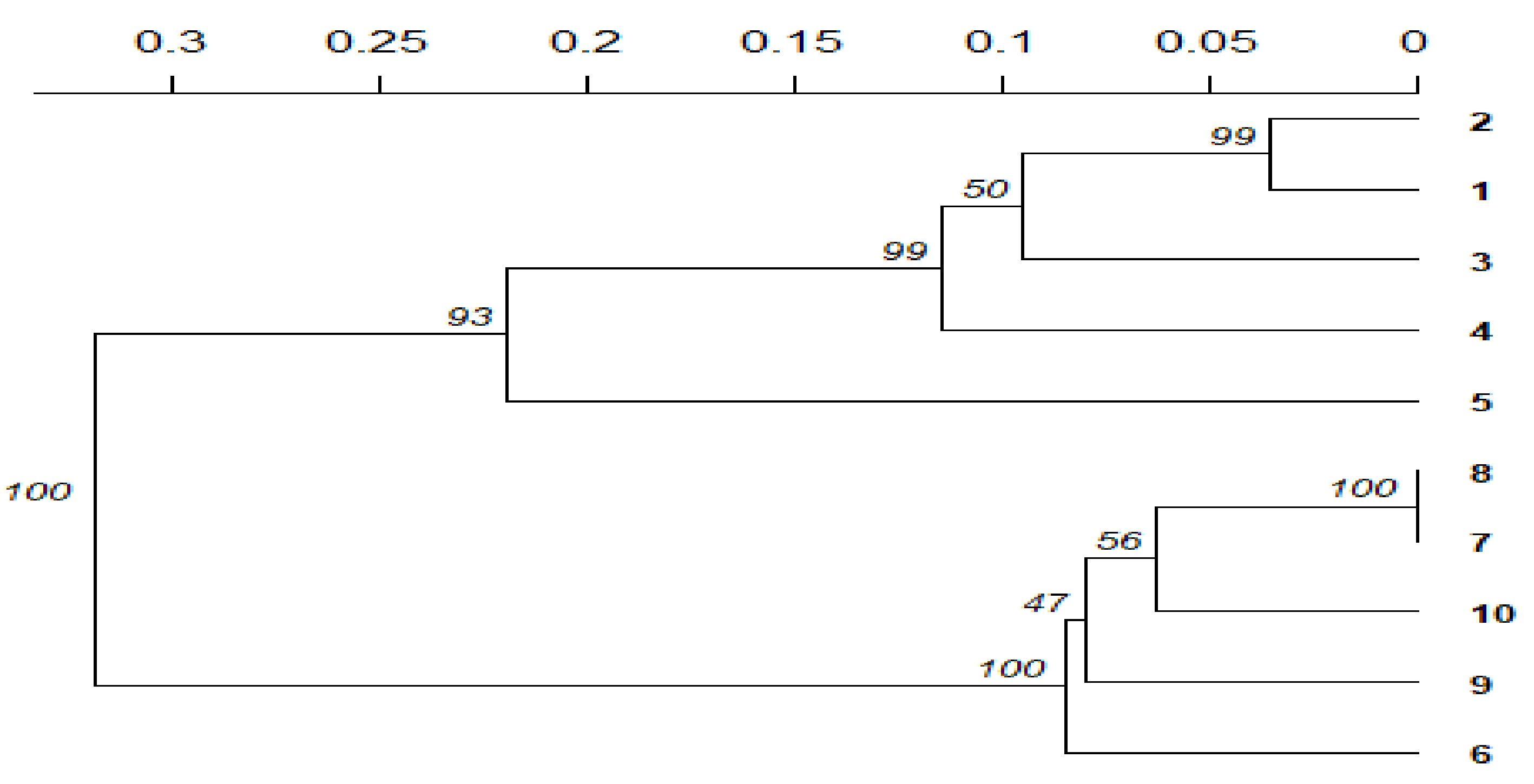
| Colony Morphological Characterization | Identification Based on 16S rRNA Sequence | ||
|---|---|---|---|
| Gram staining | Positive cocci | Strain ID | NMCC-11 |
| Shape | Circular | ||
| Surface | Smooth | Probiotic strain genus | Pediococcus acidilactici |
| Size | >0.098 mm | ||
| Color | White | Length of 16S rRNA gene | 634 bp |
| Margin | Entire | ||
| Elevation | Raised | Accession number | MK606112 |
| Biochemical characterization | |||
| Catalase | Negative | Closely related published taxa | Pediococcus acidilactici |
| Oxidase | Negative | ||
| Glucose fermentation | Positive | Sequence similarity | 99.21% |
| Temperature | NaCl Concentration in MRS Broth | Enzymatic Potential | |||
|---|---|---|---|---|---|
| 5–10 °C | − | 0.5% | ++ | Amylolytic activity | + |
| 30–35 °C | + | 2% | − | Proteolytic activity | ++ |
| 37 °C | ++ | 3% | − | Cellulolytic activity | + |
| 45 °C | + | 6.5% | − | Bacteriocin production | + |
| 60 °C | − | 10% | − | ||
| Strain | Test Pathogen | ||||
|---|---|---|---|---|---|
| Pediococcus acidilactici (NMCC-11) | Escherichia coli | Pseudomonas aeruginosa | Staphylococcus aureus | Listeria monocytogenes | Bacillus cereus |
| − | ++ | ++ | ++ | ++ | |
| Antibiotic | Zone of Inhibition | Mean Zone Diameter |
|---|---|---|
| Ampicillin (5 ug) | (S) | 30 mm |
| Penicillin G (10 ug) | (S) | 22 mm |
| Lincomycin (10 ug) | (R) | 00 mm |
| Cloxacillin (10 ug) | (I) | 17 mm |
| Streptomycin (10 ug) | (I) | 16 mm |
| Erythromycin (15 ug) | (S) | 30 mm |
| Tetracycline (30 ug) | (S) | 31.5 mm |
| Oxytetracycline (10 ug) | (S) | 20 mm |
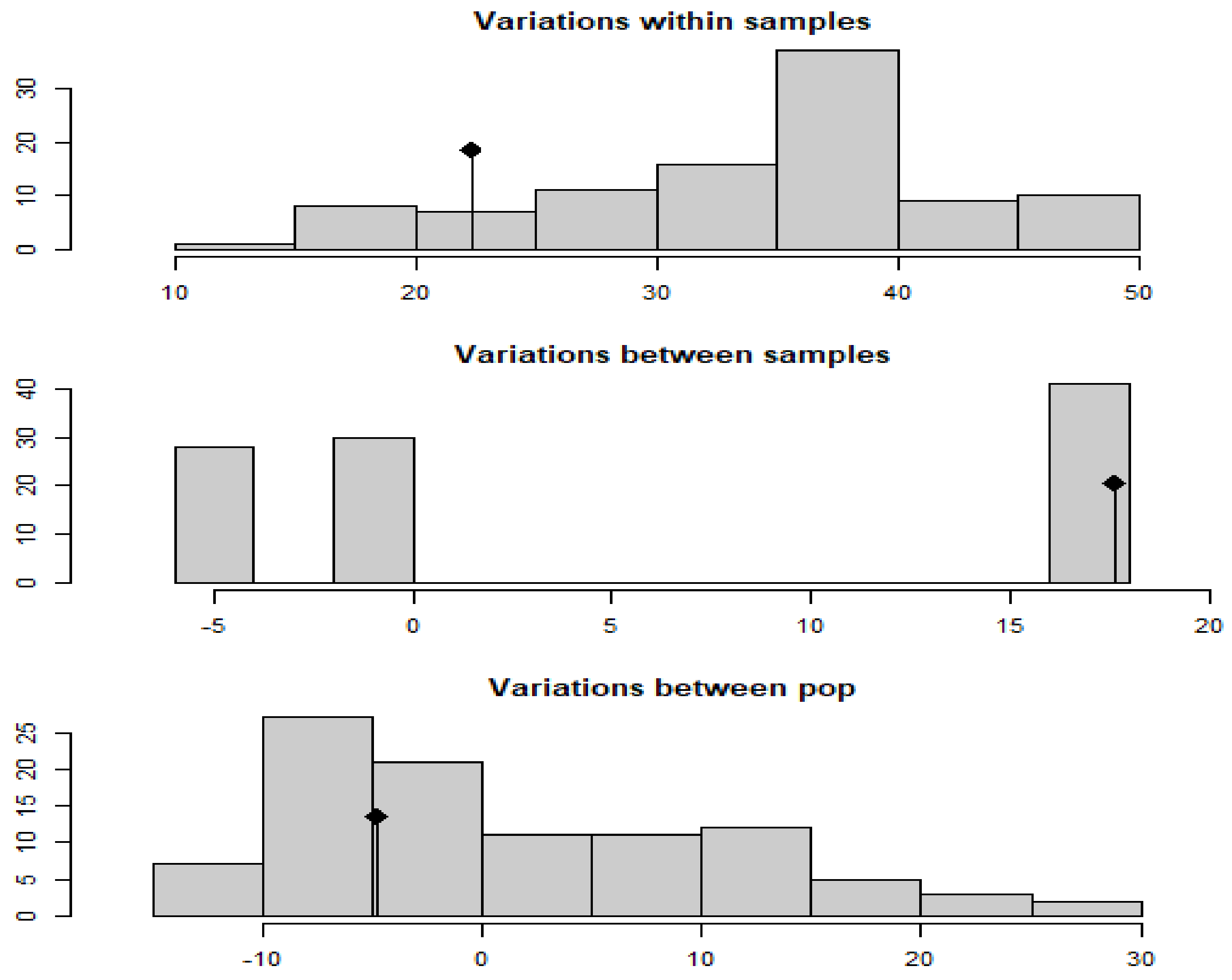
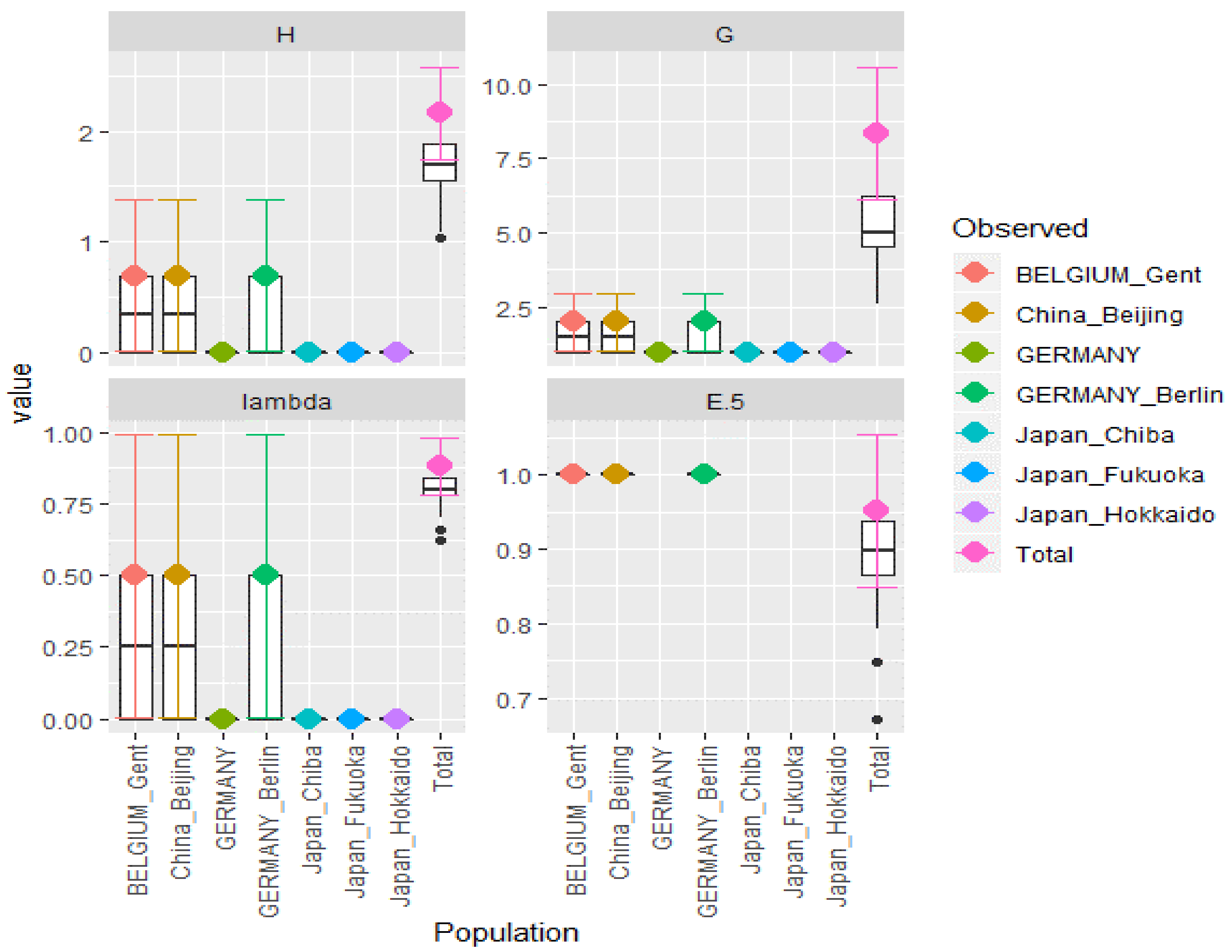
| Test | Obs | Std.Obs | Alter | p-Value |
|---|---|---|---|---|
| Variation within samples | 22.291667 | −1.4080075 | less | 0.13 |
| Variation between samples | 17.637500 | 1.1759021 | greater | 0.01 |
| Variation between sub pop | −4.784459 | −0.6088021 | greater | 0.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kanwal, H.; Di Cerbo, A.; Zulfiqar, F.; Sabia, C.; Nawaz, A.; Siddiqui, F.M.; Aqeel, M.; Ghazanfar, S. Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry. Appl. Sci. 2021, 11, 9586. https://doi.org/10.3390/app11209586
Kanwal H, Di Cerbo A, Zulfiqar F, Sabia C, Nawaz A, Siddiqui FM, Aqeel M, Ghazanfar S. Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry. Applied Sciences. 2021; 11(20):9586. https://doi.org/10.3390/app11209586
Chicago/Turabian StyleKanwal, Hafsa, Alessandro Di Cerbo, Freeha Zulfiqar, Carla Sabia, Amara Nawaz, Fariha Masood Siddiqui, Muhammad Aqeel, and Shakira Ghazanfar. 2021. "Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry" Applied Sciences 11, no. 20: 9586. https://doi.org/10.3390/app11209586
APA StyleKanwal, H., Di Cerbo, A., Zulfiqar, F., Sabia, C., Nawaz, A., Siddiqui, F. M., Aqeel, M., & Ghazanfar, S. (2021). Probiotic Characterization and Population Diversity Analysis of Gut-Associated Pediococcus acidilactici for Its Potential Use in the Dairy Industry. Applied Sciences, 11(20), 9586. https://doi.org/10.3390/app11209586









