Winter Wheat Take-All Disease Index Estimation Model Based on Hyperspectral Data
Abstract
:1. Introduction
2. Materials and Methods
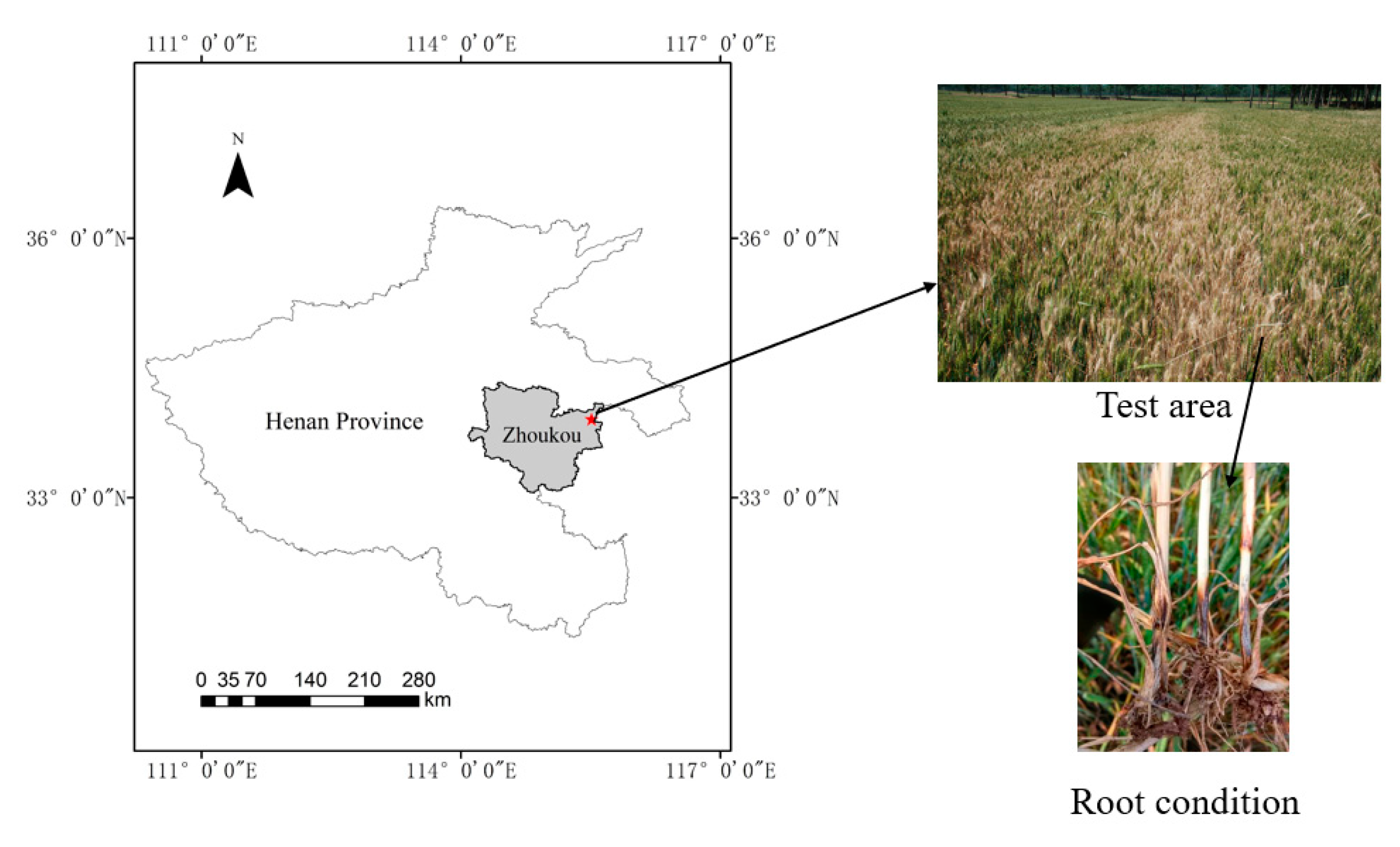
2.1. Test Area
2.2. Data Acquisition and Processing
Disease Investigation and Spectral Collection
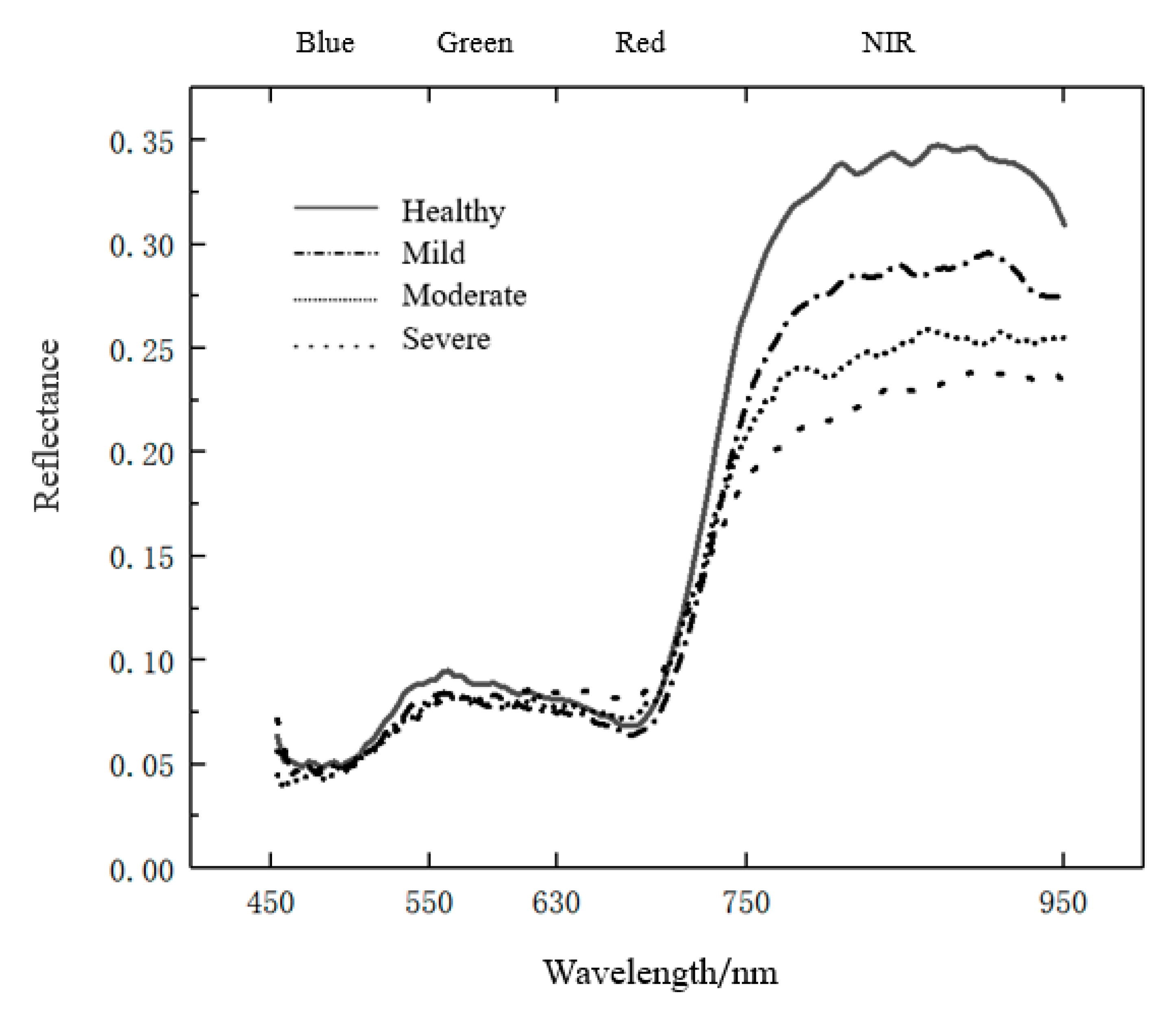
2.3. Spectral Index Construction
2.4. Modeling Method
2.5. Accuracy Evaluation Method
2.6. Data Analysis
3. Results
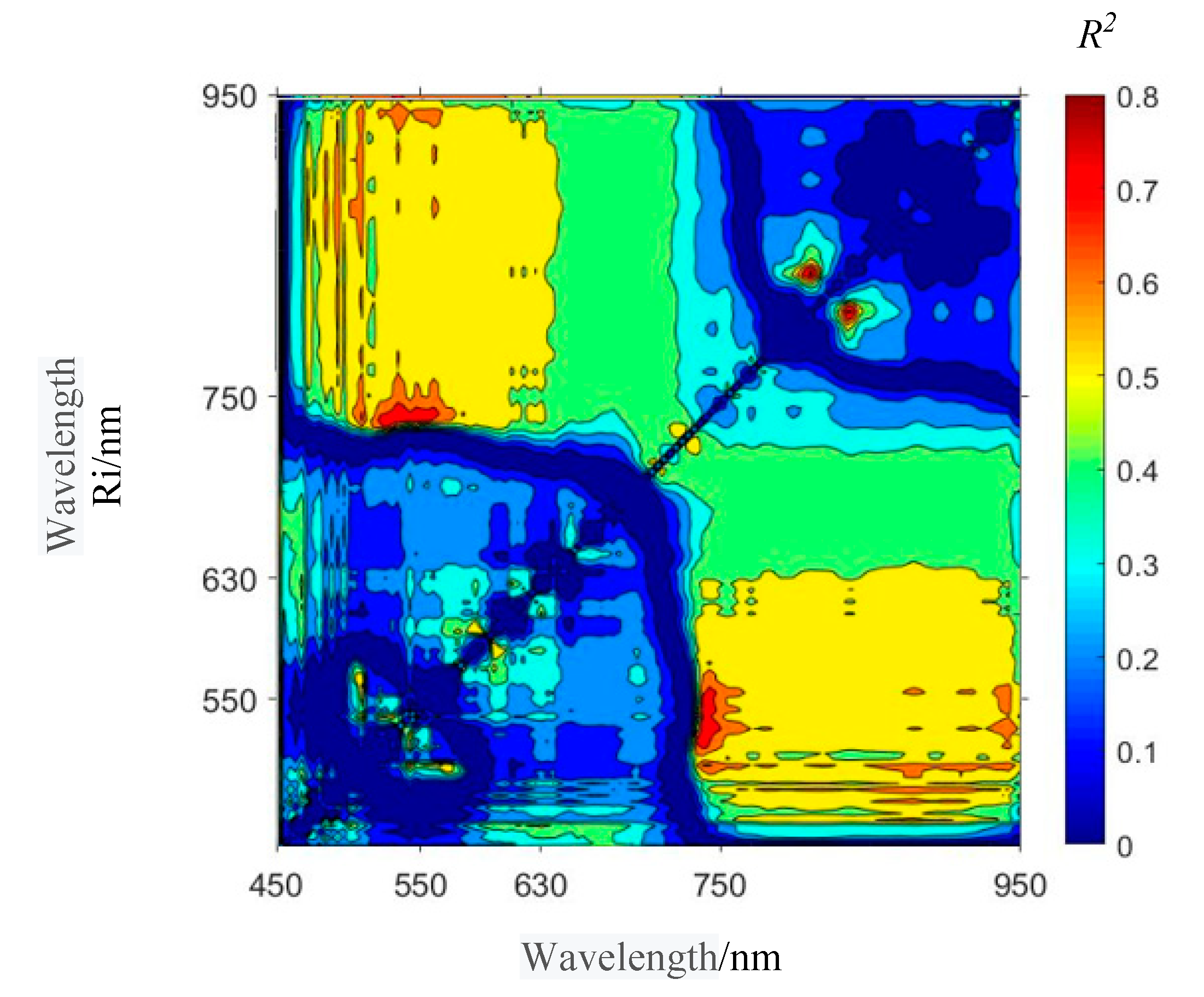
3.1. Selection of Spectral Index Combinations of Winter Wheat Take-All
3.2. Wheat Take-All Disease Model Based on Spectral Index
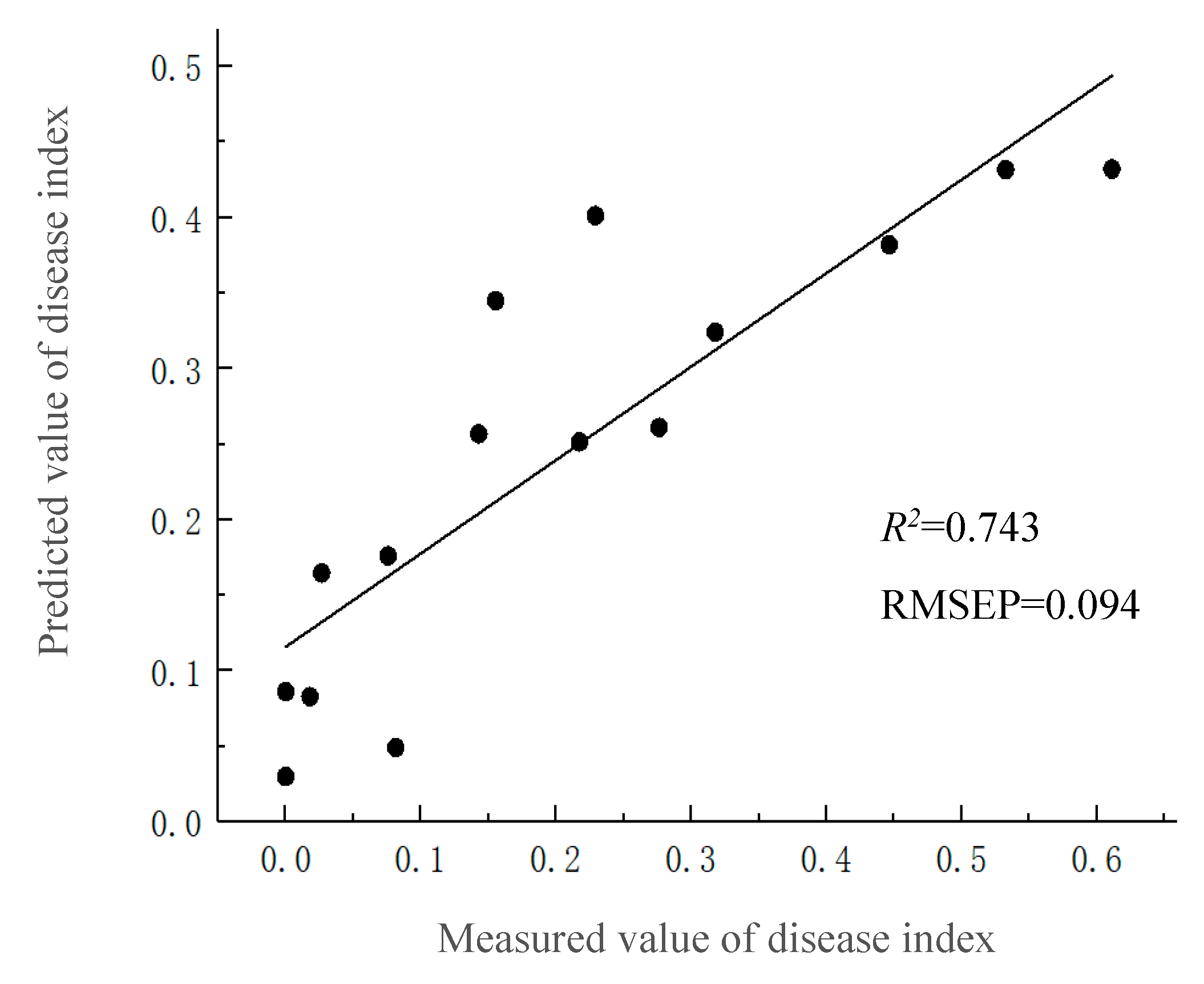
3.3. Validation of Inversion Model of Winter Wheat Take-All Disease Index
4. Discussion
Author Contributions
Funding
Conflicts of Interest
References
- Qiao, H.; Jiao, H.; Wu, X.; Si, H.; Shi, L.; Guo, W.; Ma, X.; Zhou, Y. Remote sensing monitoring of winter wheat take-all damage level based on support vector machine model. J. Triticeae Crop. 2014, 34, 1694–1698. [Google Scholar]
- Zhang, L.; Zhao, J.; Jia, K.; Li, X. Plant Spectral Discrimination Based on Phenological Features. Spectrosc. Spectr. Anal. 2015, 35, 2836–2840. [Google Scholar]
- Li, W. Advances in application of hyperspectral remote sensing technology in monitoring agricultural pests and diseases. Mod. Agric. Sci. Technol. 2019, 14, 126–128. [Google Scholar]
- Tan, C.; Wang, J.; Huang, W.; Liu, L.; Huang, Y.; Yan, W. Application of hyperspectral remote sensing in physicochemical information extraction of vegetation. J. Northwest Agric. For. Univ. 2005, 6, 151–156. [Google Scholar]
- Zhao, H.; Yang, C.; Guo, W.; Zhang, L.; Zhang, D. Automatic Estimation of Crop Disease Severity Levels Based on Vegetation Index Normalization. Remote Sens. 2020, 12, 1930. [Google Scholar] [CrossRef]
- Guo, J.; Huang, C.; Wang, H.; Sun, Z.; Ma, Z. Inversion of stripe rust disease index of different wheat varieties based on hyperspectral remote sensing technology. Spectrosc. Spectr. Anal. 2009, 29, 3353–3357. [Google Scholar]
- Lei, Y.; Han, D.; Zeng, Q.; He, D. Grading method of wheat stripe rust disease degree based on hyperspectral imaging technology. Trans. Chin. Soc. Agric. Mach. 2018, 49, 226–232. [Google Scholar]
- Chen, Y.; Jiang, J.; Huang, W.; Wang, Y. Comparative study on severity of stripe rust in winter wheat estimated by principal component Analysis and empirical vegetation index method. Spectrosc. Spectr. Anal. 2009, 29, 2161–2165. [Google Scholar]
- Ashourloo, D.; Mobasheri, M.; Huete, A. Evaluating the Effect of Different Wheat Rust Disease Symptoms on Vegetation Indices Using Hyperspectral Measurements. Remote Sens. 2014, 6, 5107–5123. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Qi, L.; Ma, X.; Zhu, X.; Wang, W. Grading method of rice leaf blast disease based on hyperspectral imaging technology. Trans. Chin. Soc. Agric. Eng. 2013, 29, 138–144. [Google Scholar]
- Qiao, H.; Shi, Y.; Guo, W.; Shi, L.; Ma, X.; Zhou, Y. Monitoring of wheat take-all by near-earth imaging hyperspectral remote sensing. J. Plant Prot. 2015, 42, 475–476. [Google Scholar]
- Guo, W.; Zhu, Y.; Wang, H.; Zhang, J.; Dong, P.; Qiao, H. Study on winter wheat take-all monitoring model based on UAV hyperspectral image. Trans. Chin. Soc. Agric. Mach. 2019, 50, 162–169. [Google Scholar]
- Zhang, J.; Yuan, L.; Wang, J.; Luo, J.; Du, S.; Huang, W. Advances in remote sensing monitoring of crop diseases and insect pests. Trans. Chin. Soc. Agric. Eng. 2012, 28, 1–11. [Google Scholar]
- Sun, J.; Song, Y.; He, W.; Yang, G. Research on wheat take-all and its pathogens. J. Henan Agric. Sci. 2010, 5, 134–137. [Google Scholar]
- Teke, M.; Deveci, H.S.; Haliloglu, O.; Gurbuz, S.Z.; Sakaray, U. A short survey of hyperspectral remote sensing applications in agriculture. In Proceedings of the International Conference on Recent Advances in Space Technologies, Istanbul, Turkey, 12–14 June 2013. [Google Scholar]
- Zhang, N.; Yang, G.; Zhao, C.; Zhang, J.; Yang, X.; Pan, Y.; Huang, W.; Xu, B.; Li, M. Progress and prospect of hyperspectral remote sensing for crop pests and diseases. J. Remote. Sens. 2021, 25, 403–422. [Google Scholar]
- Fu, K.; Zhang, W.; Cao, H.; Zhu, Y.; Sha, W.; Zhang, W.; Feng, C. Research progress of crop pests and diseases monitoring based on spectral. China Agric. Sci. Technol. Rev. 2014, 16, 90–98. [Google Scholar]
- Yuan, L.; Huang, Y.; Loraamm, R.W.; Nie, C.; Wang, J.; Zhang, J. Spectral analysis of winter wheat leaves for detection and differentiation of diseases and insects. Field Crop. Res. 2014, 156, 199–207. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Niu, Z.; Tang, Q. Modification of leaf photochemical vegetation index (PRI) and its sensitivity analysis. Spectrosc. Spectr. Anal. 2008, 2014, 2018. [Google Scholar]
- Lees, K.J.; Artz, R.; Khomik, M.; Clark, J.M.; Ritson, J.; Hancock, M.H.; Cowie, N.R.; Quaife, T. Using Spectral Indices to Estimate Water Content and GPP in Sphagnum Moss and Other Peatland Vegetation. IEEE Trans. Geosci. Remote Sens. 2020, 58, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Xu, C.; Zhu, X.; Pan, Y.; Zhu, W.; Lei, Y. Comparative study of rice Information extraction based on NDII and NDVI. Geogr. Geogr. Inf. Sci. 2008, 5, 44–46. [Google Scholar]
- Zhang, J.; Wang, S. Spatio-Temporal Evolution and Prediction of Tourism Comprehensive Climate Comfort in Henan Province, China. Atmosphere 2021, 12, 1–19. [Google Scholar]
- Yang, K.; Guo, D. Analysis of vegetation hyperspectral characteristics and extraction of disease information. Geogr. Geogr. Inf. Sci. 2006, 13, 31–34. [Google Scholar]
- Wang, N.; Feng, Y.; Du, W.; Wang, Y.; Chen, H. Study on pathogenicity of wheat take-all and resistance of wheat varieties. J. Plant Genet. Resour. 2012, 13, 478–483. [Google Scholar]
- Zhang, L.; Huo, Z.; Wang, L.; Wu, L.; Zhang, G. Dynamic warning of wheat powdery mildew occurrence by meteorological grade in Hebei Province. J. Ecol. 2015, 34, 2489–2497. [Google Scholar]
- Pei, H.; Feng, H.; Li, C.; Jin, X.; Li, Z.; Yang, G. Remote sensing monitoring of winter wheat growth by UAV based on comprehensive indexes. Trans. Chin. Soc. Agric. Eng. 2017, 33, 74–82. [Google Scholar]
- Kristin, T.; Harald, M. Analyzing complex mathematical model behavior by partial least squares regression-based multivariate metamodeling. Wiley Interdiscip. Rev. Comput. Stat. 2014, 6, 440–475. [Google Scholar]
- Krishna, G.; Sahoo, R.N.; Pargal, S.; Gupta, V.K.; Sinha, P.; Bhagat, S.; Saharan, M.S.; Singh, R.; Chattopadhyay, C. Assessing wheat yellow rust disease through hyperspectral remote sensing. ISPRS-International Archives of the Photogrammetry. Remote Sens. Spat. Inf. Sci. 2014, 8, 1413–1416. [Google Scholar]
- Romain, B.; Alexis, C.; Rémi, M.; Vincent, L.; Philippe, V.; Benjamin, D.; Benoit, M. In-field proximal sensing of septoria tritici blotch, stripe rust and brown rust in winter wheat by means of reflectance and textural features from multispectral imagery. Biosyst. Eng. 2020, 197, 257–269. [Google Scholar]
- Liu, L.; Dong, Y.; Huang, W.; Du, X.; Ren, B.; Huang, L.; Zheng, Q.; Ma, H. A Disease Index for Efficiently Detecting Wheat Fusarium Head Blight Using Sentinel-2 Multispectral Imagery. IEEE Access 2020, 8, 52181–52191. [Google Scholar] [CrossRef]
- Guo, A.; Huang, W.; Ye, H.; Dong, Y.; Ma, H.; Ren, Y.; Ruan, C. Identification of Wheat Yellow Rust using Spectral and Texture Features of Hyperspectral Images. Remote Sens. 2020, 12, 1419. [Google Scholar] [CrossRef]
- Yang, K.; Zhang, W.; Wang, X.; Sun, T.; Cheng, L. Classification and degree monitoring of copper and lead pollution in maize leaves based on spatial spectrum. Spectrosc. Spectr. Anal. 2018, 38, 2200–2208. [Google Scholar]
- Graeff, S.; Link, J.; Claupein, W. Identification of powdery mildew (Erysiphe graminis sp. tritici) and take-all disease (Gaeumannomyces graminis sp. tritici) in wheat (Triticum aestivum L.) by means of leaf reflectance measurements. Cent. Eur. J. Biol. 2006, 1, 275–288. [Google Scholar] [CrossRef]

| Grade | Grading Standard |
|---|---|
| 0 | Healthy |
| 1 | The Root Disease Area Accounted for 1–5% of the Total Root Area |
| 3 | The Root Disease Area Accounted for 6–20% of the Total Root Area |
| 5 | The Root Disease Area Accounted for 21–40% of the Total Root Area |
| 7 | The Root Disease Area Accounted for 41–60% of the Total Root Area |
| 9 | The Root Disease Area Accounted for more than 60% of the Total Root Area |
| Range Name | Code | Spectral Index | R2 |
|---|---|---|---|
| Blue | B | NDSI(R510,R542) | 0.649 |
| Green | G | NDSI(R590,R598) | 0.590 |
| Red | R | NDSI(R706,R710) | 0.556 |
| Near Infrared | N | NDSI(R810,R834) | 0.828 |
| Blue-Green | BG | NDSI(R510,R562) | 0.616 |
| Blue-Red | BR | NDSI(R534,R742) | 0.793 |
| Blue—Near Infrared | BN | NDSI(R534,R942) | 0.677 |
| Green—Red | GR | NDSI(R554,R742) | 0.751 |
| Green—Near Infrared | GN | NDSI(R558,R942) | 0.636 |
| Red—Near Infrared | RN | NDSI(R634,R878) | 0.522 |
| ID | Range Combination | R2 | ID | Range Combination | R2 |
|---|---|---|---|---|---|
| 1 * | B-GR | 0.758 | 13 | B-G | 0.298 |
| 2 | B-GN | 0.691 | 14 | B-R | 0.362 |
| 3 | B-RN | 0.643 | 15 | B-N | 0.316 |
| 4 * | G-BR | 0.759 | 16 | G-R | 0.413 |
| 5 | G-BN | 0.731 | 17 | G-N | 0.286 |
| 6 | G-RN | 0.658 | 18 | R-N | 0.372 |
| 7 | R-GB | 0.351 | 19 * | B-GR-N | 0.762 |
| 8 | R-BN | 0.732 | 20 | B-GN-R | 0.692 |
| 9 | R-GN | 0.691 | 21 | B-RN-G | 0.736 |
| 10 | N-BG | 0.300 | 22 * | G-BR-N | 0.760 |
| 11 * | N-BR | 0.755 | 23 | G-BN-R | 0.732 |
| 12 * | N-GR | 0.750 | 24 | R-GB-N | 0.374 |
| Band Combination | Fitting Formula | R2 |
|---|---|---|
| G-BR | Y = 137.507 − 65.786 NDSI(R590,R598) − 228.808 NDSI(R534,R742) | 0.759 |
| N-BR | Y = 133.059 + 32.751 NDSI(R810,R834) − 219.736 NDSI(R534,R742) | 0.755 |
| B-GR | Y = 113.968 + 35.529 NDSI(R590,R598) − 211.716 NDSI(R554,R742) | 0.758 |
| N-GR | Y = 111.824 + 11.54 NDSI(R810,R834) − 193.693 NDSI(R554,R742) | 0.750 |
| G-BR-N | Y = 134.577 − 70.301 NDSI(R590,R598) − 223.533 NDSI(R534,R742) + 51.584 NDSI(R810,R834) | 0.760 |
| B-GR-N | Y = 107.966 + 53.344 NDSI(R590,R598) − 206.922 NDSI(R534,R742) + 51.584 NDSI(R810,R834) | 0.762 |
| Band Combination | Spectral Index | R2 | RMSEP |
|---|---|---|---|
| G-BR | NDSI(R590,R598), NDSI(R534,R742) | 0.726 | 0.108 |
| N-BR | NDSI(R810,R834), NDSI(R534,R742) | 0.728 | 0.106 |
| G-BR-N | NDSI(R590,R598), NDSI(R534,R742) NDSI(R810,R834) | 0.743 | 0.094 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, W.; Yang, Y.; Zhao, H.; Song, R.; Dong, P.; Jin, Q.; Baig, M.H.A.; Liu, Z.; Yang, Z. Winter Wheat Take-All Disease Index Estimation Model Based on Hyperspectral Data. Appl. Sci. 2021, 11, 9230. https://doi.org/10.3390/app11199230
Guo W, Yang Y, Zhao H, Song R, Dong P, Jin Q, Baig MHA, Liu Z, Yang Z. Winter Wheat Take-All Disease Index Estimation Model Based on Hyperspectral Data. Applied Sciences. 2021; 11(19):9230. https://doi.org/10.3390/app11199230
Chicago/Turabian StyleGuo, Wei, Yifeng Yang, Hengqian Zhao, Rui Song, Ping Dong, Qian Jin, Muhammad Hasan Ali Baig, Zelong Liu, and Zihan Yang. 2021. "Winter Wheat Take-All Disease Index Estimation Model Based on Hyperspectral Data" Applied Sciences 11, no. 19: 9230. https://doi.org/10.3390/app11199230
APA StyleGuo, W., Yang, Y., Zhao, H., Song, R., Dong, P., Jin, Q., Baig, M. H. A., Liu, Z., & Yang, Z. (2021). Winter Wheat Take-All Disease Index Estimation Model Based on Hyperspectral Data. Applied Sciences, 11(19), 9230. https://doi.org/10.3390/app11199230






