Transcriptome Analysis Identified Candidate Genes Involved in Fruit Body Development under Blue Light in Lentinula edodes
Abstract
1. Introduction
2. Materials and Methods
2.1. Fungal Materials
2.2. RNA Sequencing Analysis
2.3. Functional Analysis of Differentially Expressed Genes
2.4. DEG Expression Profiles and Quantitative RT-PCR Validation
2.5. Statistical Analysis
3. Results and Discussion
3.1. Blue Light-Induced Mushroom Phenotypes
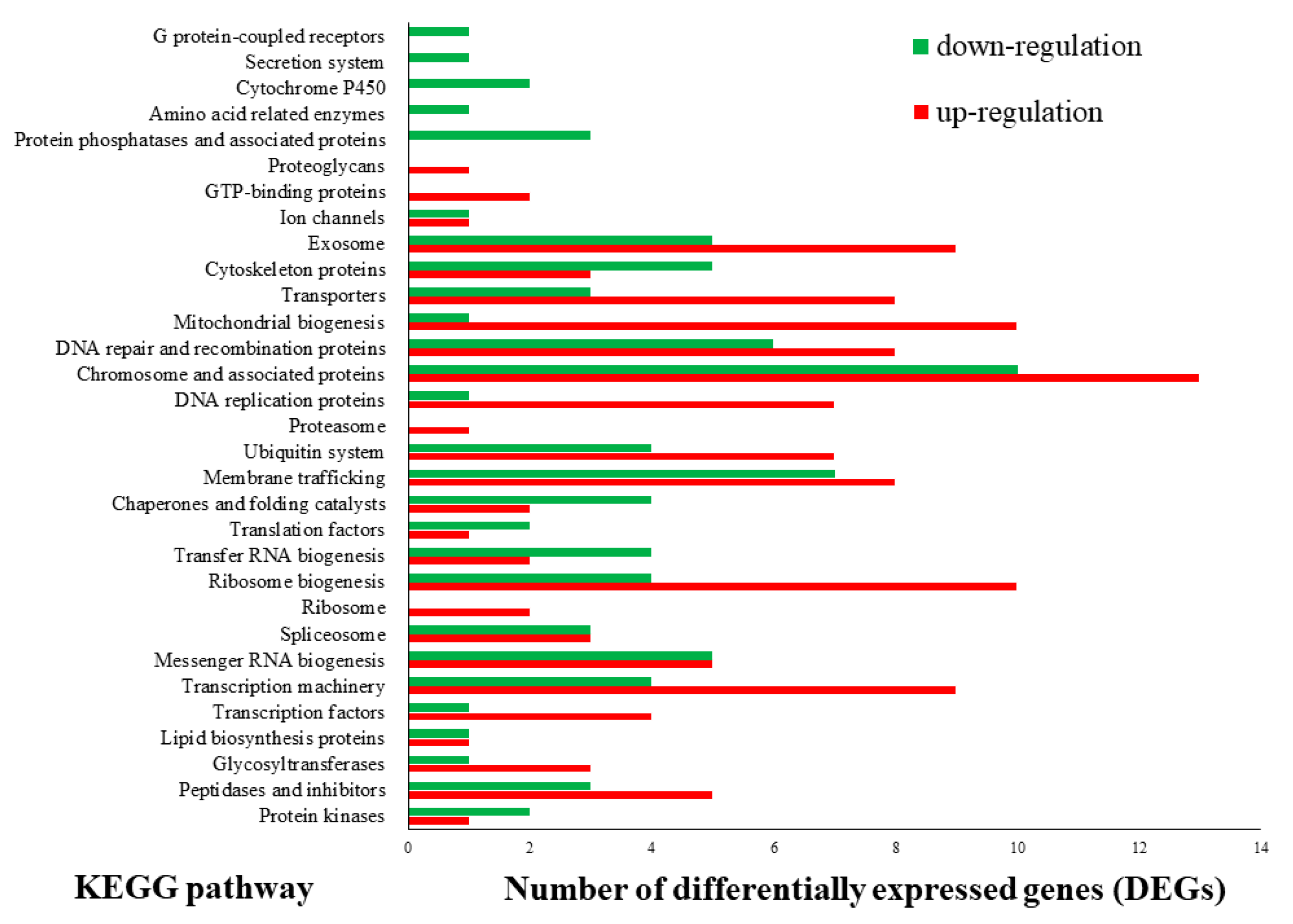
3.2. Transcriptome Sequencing and Functional Annotation of Differentially Expressed Genes (DEGs)
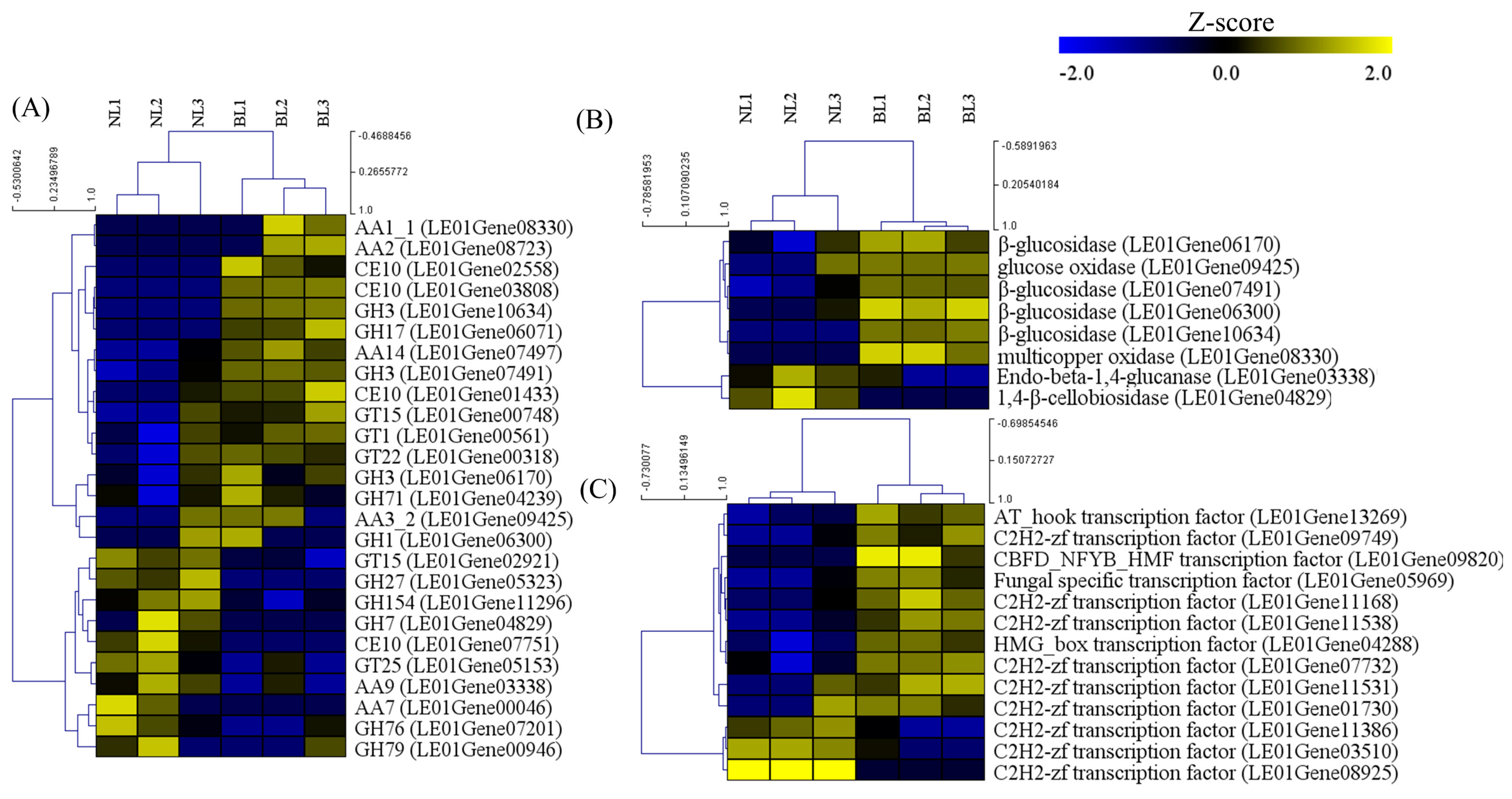
3.3. CAZyme Encoding Genes under Different Light Sources Mapped to the Differentially Expressed Genes (DEGs)
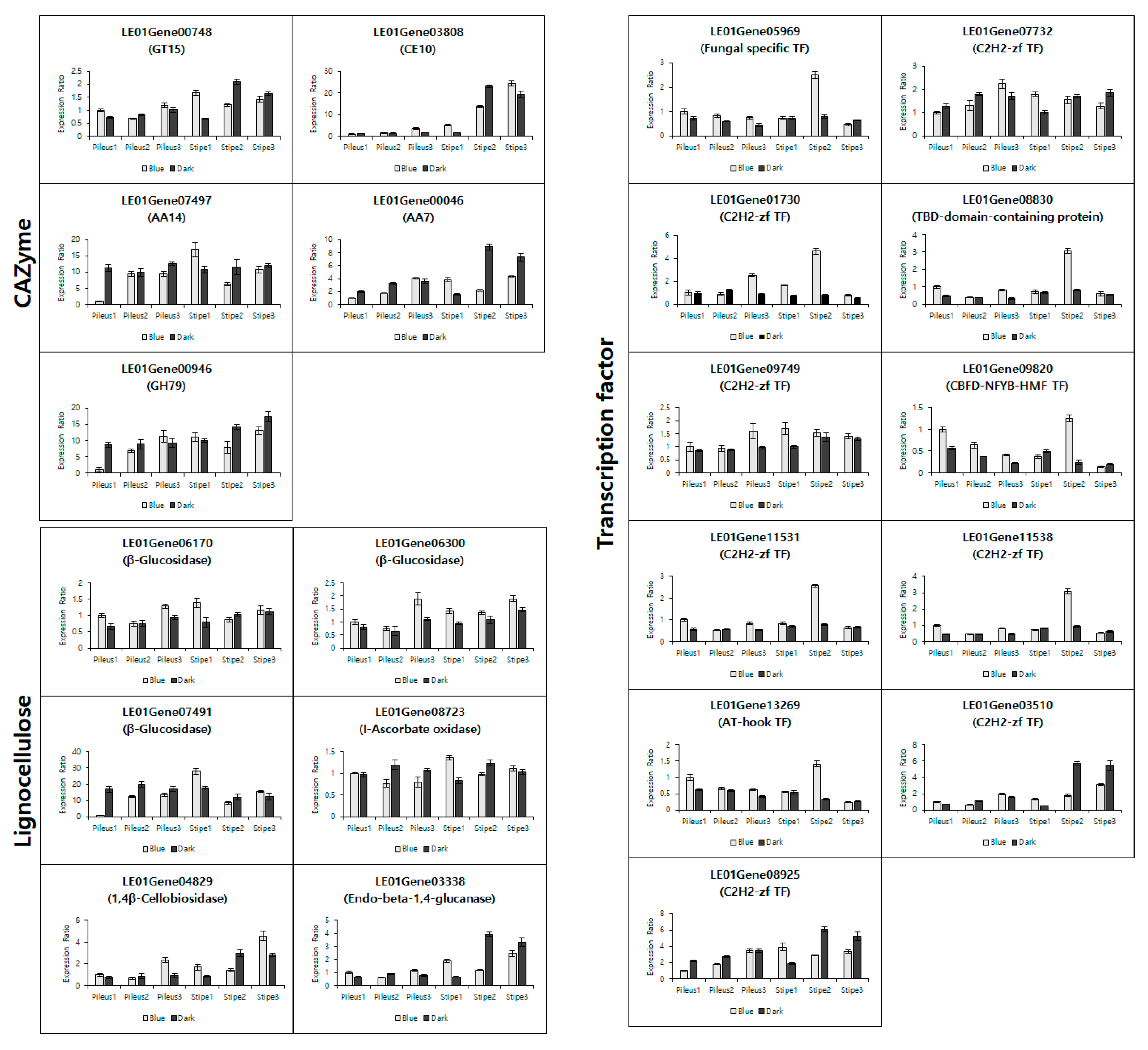
3.4. Validation of the Transcriptome Data by qRT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Mattos-Shipley, K.M.J.; Ford, K.L.; Alberti, F.; Banks, A.M.; Bailey, A.M.; Foster, G.D. The good, the bad, and the tasty: The many roles of mushrooms. Stud. Mycol. 2016, 85, 125–157. [Google Scholar] [CrossRef]
- Yoo, Y.B.; Kong, W.S.; Oh, S.J.; Cheong, J.C.; Jang, K.Y.; Jhune, C.S. Trends of mushroom science and mushroom industry. J. Mushrooms 2015, 3, 1–23. [Google Scholar]
- Han, S.R.; Kim, M.J.; Oh, T.J. Antioxidant activities and antimicrobial effects of solvent extracts from Lentinus edodes. J. Korean Soc. Food Sci. Nutr. 2015, 44, 1144–1149. [Google Scholar] [CrossRef]
- Tayer, D.L. Myco-heterotroph-fungus marriages-is fidelity over-rated? New Phytol. 2004, 163, 217–221. [Google Scholar]
- Sano, H.; Narikiyo, T.; Kaneko, S.; Yamazaki, T.; Shishido, K. Sequence analysis and expression of a blue-light photoreceptor gene, Le.phrA from the basidiomycetous mushroom Lentinula edodes. Biosci. Biotechnol. Biochem. 2007, 71, 2206–2213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Baek, I.S.; Lee, Y.H.; Jang, M.J.; Jeoung, Y.K.; Lee, H.B.; Chi, J.H. Effects of cultural characteristics of Lentinula edodes according to LED wavelength with sawdust substrate cultivation. J. Mushrooms 2013, 11, 226–229. [Google Scholar] [CrossRef]
- Shim, D.; Park, S.G.; Kim, K.M.; Bae, W.S.; Lee, G.W.; Ha, B.S.; Ro, H.S.; Kim, M.K.; Ryoo, R.; Rhee, S.K.; et al. Whole genome de novo sequencing and genome annotation of the world popular cultivated edible mushroom, Lentinula edodes. J. Biotechnol. 2016, 223, 24–25. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, Y.; Nakade, K.; Sato, S.; Yoshida, K.; Miyazaki, K.; Natsume, S.; Konno, N. Lentinula edodes genome survey and postharvest transcriptome analysis. Appl. Environ. Microbiol. 2017, 83, e02990-16. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.I.; Lee, H.Y.; Markkandan, K.; Moon, S.; Ahn, Y.J.; Ji, S.; Ko, J.; Kim, S.J.; Ryu, H.; Hong, C.P. Comparative transcriptome analysis identified candidate genes involved in mycelium browning in Lentinula edodes. BMC Genom. 2019, 20, 121. [Google Scholar] [CrossRef] [PubMed]
- Song, H.Y.; Kim, D.H.; Kim, J.M. Comparative transcriptome analysis of dikaryotic mycelia and mature fruiting bodies in the edible mushroom Lentinula edodes. Sci. Rep. 2018, 8, 8983. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kim, D.Y.; Park, Y.J.; Jang, M.J. Transcriptome analysis of the edible mushroom Lentinula edodes in response to blue light. PLoS ONE 2020, 15, e0230680. [Google Scholar]
- Jang, M.J.; Lee, Y.H.; Ju, Y.C.; Kim, S.M.; Koo, H.M. Effect of color of light emitting diode on development of fruit body in Hypsizygus marmoreus. Mycobiology 2013, 41, 63–66. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Chen, L.; Gong, Y.; Cai, Y.; Liu, W.; Zhou, Y.; Xiao, Y.; Xu, Z.; Liu, Y.; Lei, X.; Wang, G.; et al. Genome sequence of the edible cultivated mushroom Lentinula edodes (Shiitake) reveals insights into lignocellulose degradation. PLoS ONE 2016, 11, e0160336. [Google Scholar]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Aschan-Åberg, K. The production of fruit bodies in Collybia velutipes III. Influence of the quality of light. Physiol. Plant. 1960, 13, 276–279. [Google Scholar] [CrossRef]
- Eger-Hummel, G. Blue-light photomorphogenesis in mushrooms (Basidiomycetes). In The Blue Light Syndrome; Eger-Hummel, G., Ed.; Springer: Berlin/Heidelberg, Germany, 1980; pp. 555–562. [Google Scholar]
- Idnurm, A.; Heitman, J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005, 3, e95. [Google Scholar] [CrossRef]
- Yu, Z.; Fischer, R. Light sensing and responses in fungi. Nat. Rev. Microbiol. 2019, 17, 25–36. [Google Scholar] [CrossRef]
- Du, F.; Zou, Y.; Hu, Q.; Zhang, H.; Ye, D. Comparative transcriptomic analysis reveals molecular processes involved in pileus morphogenesis in Pleurotus eryngii under different light conditions. Genomics 2020, 112, 1707–1715. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.L.; Cox, M.M. Carbohydrates and glycobiology. In Lehninger Principles of Biochemistry, 4th ed.; Lehninger, A.L., Nelson, D.L., Cox, M.M., Eds.; W H Freeman & Co.: New York, NY, USA, 2005; pp. 305–325. [Google Scholar]
- Cao, Y.Y.; Yang, J.F.; Liu, T.Y.; Su, Z.F.; Zhu, F.Y.; Chen, M.X.; Fan, T.; Ye, N.H.; Feng, Z.; Wang, L.J.; et al. A Phylogenetically Informed Comparison of GH1 Hydrolases between Arabidopsis and Rice Response to Stressors. Front. Plant. Sci. 2017, 8, 350. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.W.; Fisher, J.F.; Mobashery, S. Bacterial cell-wall recycling. Ann. N. Y. Acad. Sci. 2014, 1277, 54–75. [Google Scholar] [CrossRef]
- Lee, R.C.; Hrmova, M.; Burton, R.A.; Lahnstein, J.; Fincher, G.B. Bifunctional Family 3 Glycoside Hydrolases from Barley with α-L-Arabinofuranosidase and β-D-Xylosidase Activity: Characterization, primary structures, and COOH-terminal processing. J. Biol. Chem. 2003, 278, 5377–5387. [Google Scholar] [CrossRef] [PubMed]
- Lashbrook, C.C.; Cai, S. Cell wall remodeling in Arabidopsis stamen abscission zones. Plant Signal. Behav. 2008, 3, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Levasseur, A.; Drula, E.; Lombard, V.; Coutinho, P.M.; Henrissat, B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels 2013, 6, 41. [Google Scholar] [CrossRef] [PubMed]
- Gloster, T.M. Advances in understanding glycosyltransferases from a structural perspective. Curr. Opin. Struct. Biol. 2014, 28, 131–141. [Google Scholar] [CrossRef]
- Latchman, D.S. Transcription factors: An overview. Int. J. Biochem. Cell Biol. 1997, 29, 1305–1312. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger proteins: The classical zinc finger emerges in contemporary plant science. Plant Mol. Biol. 1999, 39, 1073–1078. [Google Scholar] [CrossRef]
- Gamsjaeger, R.; Liew, C.K.; Loughlin, F.E.; Crossley, M.; Mackay, J.P. Sticky fingers: Zinc-fingers as protein-recognition motifs. Trends Biochem. Sci. 2007, 32, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.M.; Xu, J.; Clarkson, B.K.; Martinez-Yamout, M.A.; Dyson, H.J.; Case, D.A.; Wright, P.E. Induced fit and “lock and key” recognition of 5 S RNA by zinc fingers of transcription factor IIIA. J. Mol. Biol. 2006, 357, 275–291. [Google Scholar] [CrossRef]
- Xiao, C.; Chen, F.; Yu, X.; Lin, C.; Fu, Y.F. Over-expression of an AT-hook gene, AHL22, delays flowering and inhibits the elongation of the hypocotyl in Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Kwon, C.S.; Bezhani, S.; Huvermann, B.; Chen, C.; Peragine, A.; Wagner, D. The N-terminal ATPase AT-hook-containing region of the Arabidopsis chromatin-remodeling protein SPLAYED is sufficient for biological activity. Plant J. 2006, 46, 685–699. [Google Scholar] [CrossRef] [PubMed]
- Lim, P.O.; Kim, Y.; Breeze, E.; Koo, J.C.; Woo, H.R.; Ryu, J.S.; Park, D.H.; Beynon, J.; Tabrett, A.; Buchanan-Wollaston, V.; et al. Overexpression of a chromatin architecture-controlling AT-hook protein extends leaf longevity and increases the post-harvest storage life of plants. Plant J. 2007, 52, 1140–1153. [Google Scholar] [CrossRef]
- Street, I.H.; Shah, P.K.; Smith, A.M.; Avery, N.; Neff, M.M. The AT-hook-containing proteins SOB3/AHL29 and ESC/AHL27 are negative modulators of hypocotyl growth in Arabidopsis. Plant J. 2008, 54, 1–14. [Google Scholar] [CrossRef]
- Xie, C.; Gong, W.; Zhu, Z.; Yan, L.; Hu, Z.; Peng, Y. Comparative transcriptomics of Pleurotus eryngii reveals blue-light regulation of carbohydrate-active enzymes (CAZymes) expression at primordium differentiated into fruiting body stage. Genomics 2018, 110, 201–209. [Google Scholar] [CrossRef]
- Sakamoto, Y.; Sato, S.; Ito, M.; Ando, Y.; Nakahori, K.; Muraguchi, H. Blue light exposure and nutrient conditions influence the expression of genes involved in simultaneous hyphal knot formation in Coprinopsis cinerea. Microbiol. Res. 2018, 217, 81–90. [Google Scholar] [CrossRef]
- Pelkmans, J.F.; Patil, M.B.; Gehrmann, T.; Reinders, M.J.; Wösten, H.A.B.; Lugones, L.G. Transcription factors of Schizophyllum commune involved in mushroom formation and modulation of vegetative growth. Sci. Rep. 2017, 7, 310. [Google Scholar] [CrossRef]
- Onm, R.A.; de Jong, J.F.; de Bekker, C.; Wösten, H.A.B.; Lugones, L.G. Transcription factor genes of Schizophyllum commune involved in regulation of mushroom formation. Mol. Microbiol. 2011, 81, 1433–1445. [Google Scholar]
- Shelest, E. Transcription factors in fungi: TFome dynamics, three major families, and dual-specificity TFs. Front. Genet. 2017, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Gazara, R.K.; Verma, P.K. Transcription factor repertoire of necrotrophic fungal phytopathogen Ascochyta rabiei: Predominance of MYB transcription factors as potential regulators of secretome. Front. Plant Sci. 2017, 8, 1037. [Google Scholar] [CrossRef] [PubMed]


| Light Source | Pileus | Stipe | Pileus Diameter/Stipe Length | ||
|---|---|---|---|---|---|
| Diameter (mm) | Thickness (mm) | Length (mm) | Diameter (mm) | ||
| Fluorescent lamp | 53.25 ± 2.87 b | 19.42 ± 1.19 cd | 42.77 ± 1.00 c | 13.83 ± 1.00 ab | 1.25 ± 0.07 c |
| White LED | 55.68 ± 2.38 ab | 20.07 ± 1.13 bcd | 41.28 ± 0.44 c | 13.45 ± 1.06 abc | 1.35 ± 0.06 bc |
| White–Green LED | 57.27 ± 0.68 ab | 22.63 ± 0.44 ab | 40.73 ± 2.06 c | 11.37 ± 0.45 d | 1.42 ± 0.07 bc |
| White–Blue LED | 59.38 ± 1.95 ab | 24.62 ± 0.79 a | 39.10 ± 1.66 c | 14.03 ± 0.35 a | 1.54 ± 0.10 b |
| Red LED | 46.17 ± 1.68 c | 20.85 ± 0.20 bc | 51.45 ± 1.65 a | 11.93 ± 0.19 bcd | 0.90 ± 0.03 d |
| Green LED | 54.27 ± 1.09 ab | 20.93 ± 0.70 bc | 43.63 ± 1.59 bc | 12.78 ± 0.40 abcd | 1.25 ± 0.04 c |
| Blue LED | 60.13 ± 0.72 a | 22.70 ± 0.19 ab | 32.43 ± 1.05 d | 10.93 ± 0.31 d | 1.87 ± 0.08 a |
| Dark (no light) | 35.63 ± 5.24 d | 17.89 ± 1.76 d | 48.62 ± 5.17 ab | 11.56 ± 0.25 cd | 0.73 ± 0.04 d |
| Sample | Total Reads | Mapped Reads /Rate (%) | Duplication Rate (%) | GC Content (%) | Reads of Genomic Origin | |||
|---|---|---|---|---|---|---|---|---|
| Exonic | Intronic | Intergenic | Intronic/Intergenic Overlapping Exon: | |||||
| blue light_1 | 21,176,451 | 15,896,792 /75.1 | 44.19 | 47.45 | 6,371,649 | 1,323,094 | 4,725,629 | 1,453,657 |
| blue light_2 | 21,053,355 | 16,067,707 /76.3 | 46.86 | 47.57 | 6,681,571 | 1,380,842 | 4,680,733 | 1,516,080 |
| blue light_3 | 17,564,307 | 13,356,006 /76.0 | 42.66 | 47.52 | 5,656,474 | 1,149,817 | 3,785,002 | 1,271,736 |
| no light_1 | 23,688,201 | 17,545,089 /74.1 | 43.91 | 47.31 | 6,285,984 | 1,276,813 | 5,857,490 | 1,413,930 |
| no light_2 | 25,593,815 | 19,262,298 /75.3 | 46.86 | 47.4 | 7,417,614 | 1,544,413 | 6,028,425 | 1,704,375 |
| no light_3 | 23,973,170 | 18,274,632 /76.2 | 45 | 47.51 | 7,649,221 | 1,525,141 | 5,298,595 | 1,694,124 |
| Average | 22,174,883 | 16,733,754 /75.5 | 44.91 | 47.46 | 6,677,086 | 1,366,687 | 5,062,646 | 1,508,984 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, D.Y.; Jang, M.-J.; Park, Y.-J.; Kim, J.Y. Transcriptome Analysis Identified Candidate Genes Involved in Fruit Body Development under Blue Light in Lentinula edodes. Appl. Sci. 2021, 11, 6997. https://doi.org/10.3390/app11156997
Kim DY, Jang M-J, Park Y-J, Kim JY. Transcriptome Analysis Identified Candidate Genes Involved in Fruit Body Development under Blue Light in Lentinula edodes. Applied Sciences. 2021; 11(15):6997. https://doi.org/10.3390/app11156997
Chicago/Turabian StyleKim, Dae Yeon, Myoung-Jun Jang, Youn-Jin Park, and Jae Yoon Kim. 2021. "Transcriptome Analysis Identified Candidate Genes Involved in Fruit Body Development under Blue Light in Lentinula edodes" Applied Sciences 11, no. 15: 6997. https://doi.org/10.3390/app11156997
APA StyleKim, D. Y., Jang, M.-J., Park, Y.-J., & Kim, J. Y. (2021). Transcriptome Analysis Identified Candidate Genes Involved in Fruit Body Development under Blue Light in Lentinula edodes. Applied Sciences, 11(15), 6997. https://doi.org/10.3390/app11156997







