An Advanced Spectral–Spatial Classification Framework for Hyperspectral Imagery Based on DeepLab v3+
Abstract
1. Introduction
- (a)
- The spectral bands of hyperspectral images have high dimensionality, which increases the computational complexity. Moreover, according to the Hughes phenomenon, as the dimension number increases, the classification accuracy first increases and then decreases.
- (b)
- The spatial resolution of hyperspectral images is very low, and one pixel typically represents a distance of tens of meters. Therefore, the number of pictures in each dataset is very small, and extracting spatial features is more difficult.
- (1)
- We utilize DeepLab v3+ as the neural network structure to extract spatial domain features and merge them with spectral domain features. DeepLab v3+ is one of the fourth-generation DeepLab series of semantic segmentation networks developed by Google and has the best comprehensive performance so far. We are the first to apply the latest version of the DeepLab network to the hyperspectral image classification task.
- (2)
- We use the PCA method to reduce the dimensionality of the original hyperspectral image. In the spatial feature extraction stage, we select the first three principal components and the first principal component as training data and labels [16] to solve the dimensionality problem of the hyperspectral imagery.
- (3)
- We select different classifiers such as SVM, KNN, etc., to complete the task of hyperspectral image classification. We test and compare the classification accuracy under different conditions.
2. DeepLab v3+ and Proposed Framework
2.1. DeepLab v3+
2.2. Proposed Framework
3. Experimental Results
3.1. Comparison Between Different Classifiers
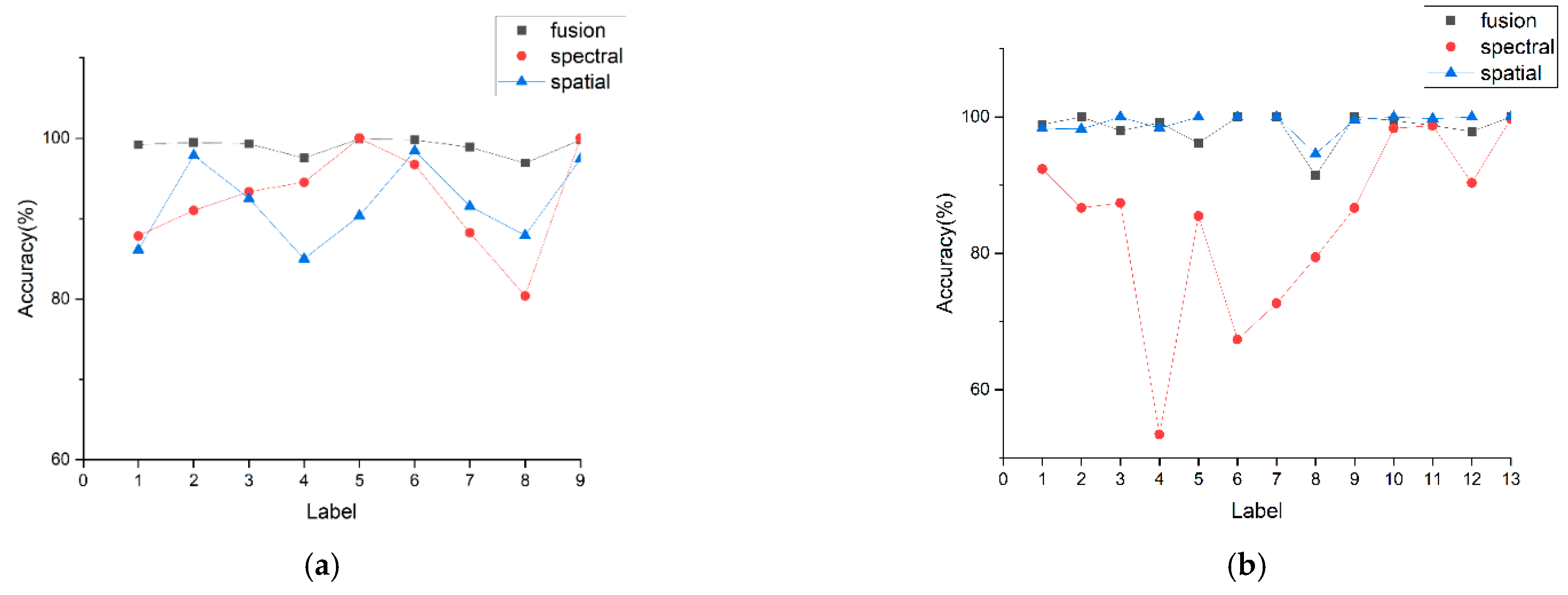
3.2. Comparison of Spectral Features, Spatial Features and Fusion Features
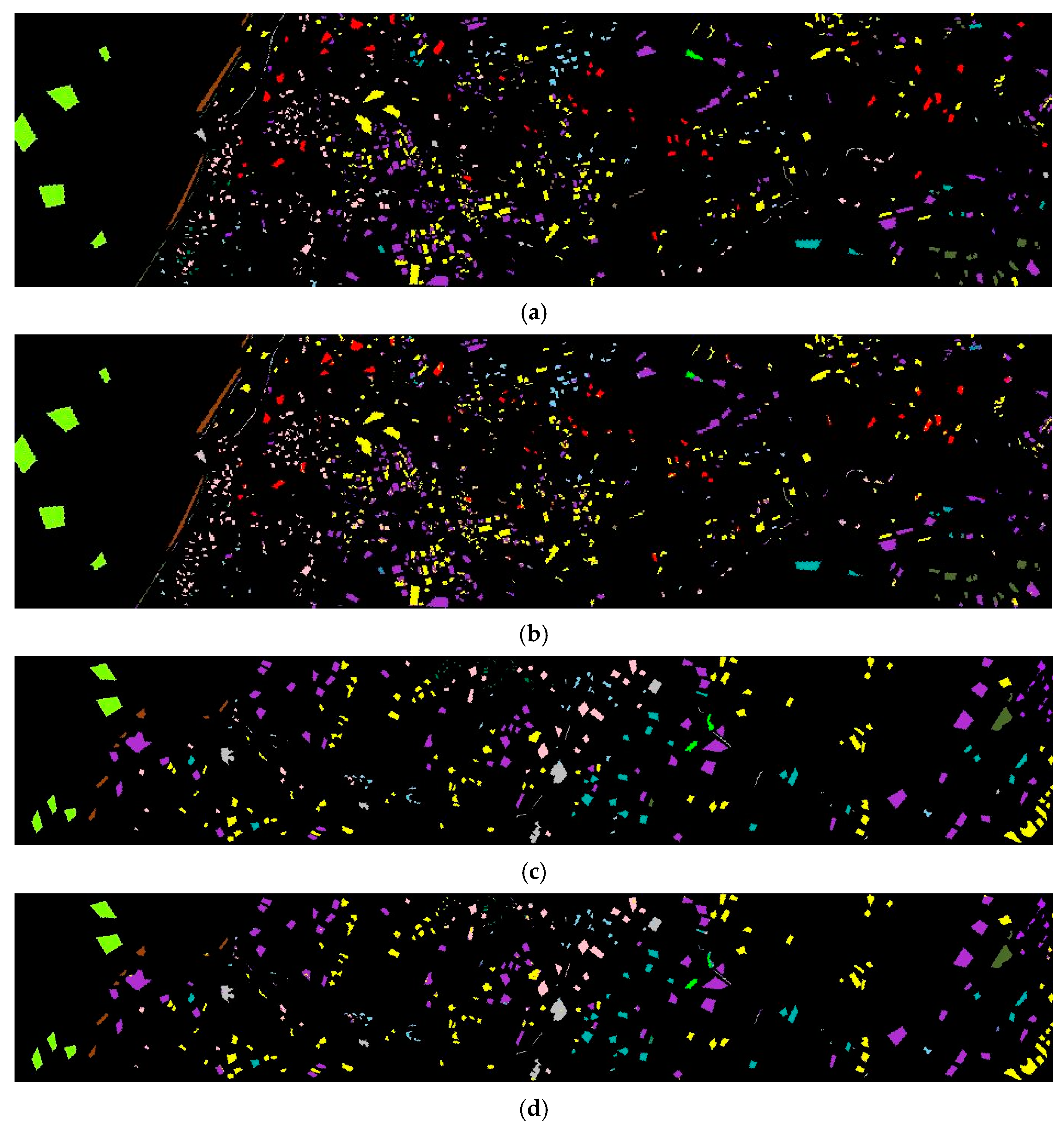
3.3. Generalization Verification
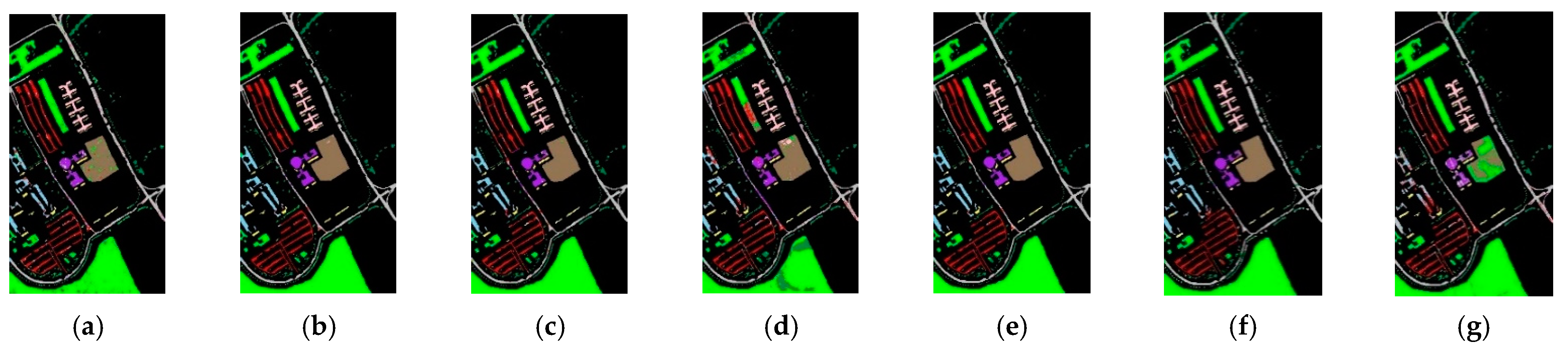
3.4. Visualization
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harsanyi, J.C.; Chang, C.I. Hyperspectral image classification and dimensionality reduction: An orthogonal subspace projection approach. IEEE Trans. Geosci. Remote Sens. 1994, 32, 779–785. [Google Scholar] [CrossRef]
- Licciardi, G.; Marpu, P.R.; Chanussot, J.; Benediktsson, J.A. Linear versus nonlinear PCA for the classification of hyperspectral data based on the extended morphological profiles. IEEE Geosci. Remote Sens. Lett. 2011, 9, 447–451. [Google Scholar] [CrossRef]
- Green, A.A.; Craig, M.D.; Shi, C. The application of the minimum noise fraction transform to the compression and cleaning of hyper-spectral remote sensing data. Int. Geosci. Remote Sens. Symp. IEEE 1988, 3, 1807. [Google Scholar]
- Balasubramanian, M.; Schwartz, E.L.; Tenenbaum, J.B.; de Silva, V.; Langford, J.C. The lsomap algorithm and topological stability. Science 2002, 295, 7. [Google Scholar] [CrossRef] [PubMed]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. Adv. Neural Inf. Process. Syst. 2001, 14, 585–591. [Google Scholar]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep feature extraction and classification of hyperspectral images based on convolutional neural networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef]
- Ying, L.; Haokui, Z.; Qiang, S. Spectral–spatial classification of hyperspectral imagery with 3D convolutional neural network. Remote Sens. 2017, 9, 67. [Google Scholar]
- Samaniego, L.; Bárdossy, A.; Schulz, K. Supervised classification of remotely sensed imagery using a modified k-NN technique. IEEE Trans. Geosci. Remote Sens. 2008, 46, 2112–2125. [Google Scholar] [CrossRef]
- Hasanlou, M.; Samadzadegan, F.; Homayouni, S. SVM-based hyperspectral image classification using intrinsic dimension. Arab. J. Geosci. 2015, 8, 477–487. [Google Scholar] [CrossRef]
- Kang, X.; Li, S.; Benediktsson, J.A. Spectral-spatial hyperspectral image classification with Edge-Preserving Filtering. IEEE Trans. Geosci. Remote Sens. 2014, 52, 2666–2677. [Google Scholar] [CrossRef]
- Chen, X.; Wei, Z.; Li, M.; Rocca, P. A review of deep learning approaches for inverse scattering problems (Invited Review). Prog. Electromagn. Res. 2020, 167, 67–81. [Google Scholar] [CrossRef]
- Ma, T.; Lyu, H.; Liu, J.; Xia, Y.; Qian, C.; Evans, J.; Xu, W.; Hu, J.; Hu, S.; He, S. Distinguishing bipolar depression from major depressive disorder using fNIRS and deep neural network. Prog. Electromagn. Res. 2020, 169, 73–86. [Google Scholar] [CrossRef]
- Fajardo, J.E.; Galván, J.; Vericat, F.; Carlevaro, C.M.; Irastorza, R.M. Phaseless microwave imaging of dielectric cylinders: An artificial neural networks-based approach. Prog. Electromagn. Res. 2019, 166, 95–105. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 7, 2094–2107. [Google Scholar] [CrossRef]
- Jonathan, L.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 39, 640–651. [Google Scholar]
- Li, J.; Zhao, X.; Li, Y.; Du, Q.; Xi, B.; Hu, J. Classification of hyperspectral imagery using a new fully convolutional neural network. IEEE Geosci. Remote Sens. Lett. 2018, 15, 292–296. [Google Scholar] [CrossRef]
- Chen, L.; Zhu, Y.; Papandreou, G.; Schroff, F.; Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. ECCV 2018, 2018, 801–818. [Google Scholar]
- Yu, F.; Koltun, V. Multi-scale context aggregation by dilated convolutions. In Proceedings of the International Conference on Learning Representations 2016, San Juan, Puerto Rico, 2–4 May 2016; pp. 397–410. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Spatial pyramid pooling in deep convolutional networks for visual recognition. IEEE Trans. Pattern Anal. Mach. Intell. 2014, 37, 1904–1916. [Google Scholar] [CrossRef] [PubMed]
- Niu, Z.; Liu, W.; Zhao, J.; Jiang, G. DeepLab-based spatial feature extraction for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2019, 16, 251–255. [Google Scholar] [CrossRef]
- Pavia University Hyperspectral Satellite Dataset from the Telecommunications and Remote Sensing Laboratory, Ipavia University (Italy). Available online: https://www.kaggle.com/abhijeetgo/paviauniversity (accessed on 21 June 2018).
- Kennedy Space Center Hyperspectral Satellite Dataset from the NASA AVIRIS (Airborne Visible/Infrared Imaging Spectrometer) Instrument. Available online: http://www.ehu.eus/ccwintco/index.php/Hyperspectral_Remote_Sensing_Scenes (accessed on 23 March 1996).
- HyRANK Hyperspectral Satellite Dataset I (Version v001) [Data Set]. Available online: http://doi.org/10.5281/zenodo.1222202 (accessed on 20 April 2008).






| Output stride | 16 |
| Learning rate | 0.001 |
| Learning rate scheduler policy | Poly |
| Weight decay | 0.0001 |
| CLASS | KNN | Logistic Regression | Decision Tree | Naive Bayesian Model | SVM |
|---|---|---|---|---|---|
| 1 | 98.72 | 93.79 | 92.16 | 96.17 | 99.19 |
| 2 | 97.81 | 98.34 | 94.98 | 98.04 | 99.48 |
| 3 | 96.69 | 97.05 | 84.67 | 85.92 | 99.30 |
| 4 | 98.57 | 97.07 | 81.25 | 47.66 | 97.53 |
| 5 | 98.00 | 99.92 | 93.48 | 84.52 | 99.92 |
| 6 | 99.39 | 98.43 | 92.06 | 86.60 | 99.79 |
| 7 | 96.63 | 90.20 | 84.68 | 77.17 | 98.90 |
| 8 | 98.01 | 89.37 | 82.19 | 78.53 | 96.91 |
| 9 | 100.00 | 98.04 | 99.22 | 100.00 | 99.78 |
| OA | 98.16 | 96.51 | 91.49 | 85.33 | 99.10 |
| AA | 98.20 | 95.80 | 89.64 | 83.85 | 98.98 |
| Kappa | 97.56 | 95.36 | 88.73 | 81.40 | 98.81 |
| CLASS | KNN | Logistic Regression | Decision Tree | Naive Bayesian Model | SVM |
|---|---|---|---|---|---|
| 1 | 93.89 | 96.65 | 86.07 | 93.47 | 98.89 |
| 2 | 80.07 | 98.65 | 48.60 | 86.59 | 100 |
| 3 | 98.12 | 100 | 76.50 | 95.24 | 97.98 |
| 4 | 97.02 | 100 | 52.36 | 80.75 | 99.13 |
| 5 | 99.32 | 100 | 45.18 | 96.58 | 96.20 |
| 6 | 100 | 100 | 52.97 | 99.43 | 100 |
| 7 | 100 | 93.46 | 40.13 | 100 | 100 |
| 8 | 99.74 | 98.02 | 87.88 | 67.10 | 91.42 |
| 9 | 96.30 | 99.80 | 84.31 | 78.97 | 100 |
| 10 | 99.72 | 100 | 69.65 | 99.68 | 99.47 |
| 11 | 95.38 | 94.75 | 93.16 | 96.21 | 98.74 |
| 12 | 98.09 | 99.33 | 85.58 | 92.10 | 97.88 |
| 13 | 100 | 97.89 | 99.54 | 99.55 | 100 |
| OA | 96.67 | 98.22 | 79.52 | 90.49 | 98.47 |
| AA | 96.74 | 98.35 | 70.92 | 91.21 | 98.44 |
| Kappa | 96.29 | 98.02 | 77.21 | 89.40 | 98.29 |
| CLASS | SRC-T | ELM | SVM-RBF | CNN | FCN | Proposed Framework |
|---|---|---|---|---|---|---|
| 1 | 91.20 | 64.45 | 82.21 | 94.97 | 93.58 | 99.19 |
| 2 | 96.70 | 80.11 | 77.41 | 96.44 | 96.70 | 99.48 |
| 3 | 70.20 | 70.62 | 71.68 | 84.69 | 90.78 | 99.30 |
| 4 | 93.40 | 96.40 | 94.76 | 97.39 | 96.72 | 97.53 |
| 5 | 100.00 | 98.60 | 99.92 | 99.14 | 88.55 | 99.92 |
| 6 | 69.10 | 75.96 | 86.40 | 94.77 | 97.33 | 99.79 |
| 7 | 72.40 | 78.20 | 86.25 | 88.90 | 92.38 | 98.90 |
| 8 | 77.90 | 79.33 | 89.05 | 84.11 | 90.83 | 96.91 |
| 9 | 92.80 | 53.67 | 100.00 | 100.00 | 88.20 | 99.78 |
| OA | 88.70 | 77.76 | 82.49 | 94.35 | 95.11 | 99.10 |
| Kappa | 84.83 | 71.93 | 80.79 | 92.49 | 93.22 | 98.81 |
| CLASS | SAE-LR | Linear SVM | PCA RBF-SVM | RBF-SVM | Proposed Framework |
|---|---|---|---|---|---|
| OA | 96.73 | 95.52 | 95.35 | 96.51 | 98.47 |
| AA | 94.08 | 91.97 | 91.57 | 93.95 | 98.44 |
| Kappa | 96.36 | 95.01 | 94.82 | 96.11 | 98.29 |
| Loukia | Dioni | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class | KNN | Logistic Regression | Decision Tree | Naive Bayesian Model | SVM | KNN | Logistic Regression | Decision Tree | Naive Bayesian Model | SVM |
| 1 | 68.06 | 40.15 | 18.88 | 18.03 | 67.82 | 85.66 | 79.19 | 49.04 | 40.40 | 92.81 |
| 2 | 100 | 100 | 50 | 100 | 100 | 93.89 | 94.89 | 31.33 | 100 | 100 |
| 3 | 83.81 | 73.03 | 51.45 | 40.94 | 90.4 | 98.55 | 74.14 | 49.58 | 54.10 | 95.73 |
| 4 | 59.32 | 72.73 | 10.87 | 100 | 100 | 81.44 | 88.89 | 19.37 | 78.26 | 100 |
| 5 | 81.49 | 66.11 | 45.29 | 35.73 | 82.87 | 94.63 | 78.60 | 61.87 | 62.60 | 94.57 |
| 6 | 81.48 | 46.07 | 32.02 | 15.47 | 100 | \ | \ | \ | \ | \ |
| 7 | 77.33 | 66.67 | 39.32 | 40.34 | 94.08 | 97.43 | 92.14 | 81.95 | 63.89 | 100 |
| 8 | 72.98 | 67.16 | 50.46 | 53.84 | 82.19 | \ | \ | \ | \ | \ |
| 9 | 74.22 | 71.08 | 62.85 | 80.10 | 78.02 | 97.26 | 90.82 | 87.79 | 88.45 | 95.34 |
| 10 | 83.06 | 69.7 | 63.53 | 73.31 | 73.2 | 96.78 | 87.90 | 83.64 | 87.34 | 93.84 |
| 11 | 89.66 | 76.69 | 49.78 | 49.40 | 96.25 | 95.42 | 84.01 | 60.58 | 72.53 | 90.79 |
| 12 | 97.31 | 84.54 | 69.60 | 57.93 | 95.51 | 99.55 | 96.84 | 82.33 | 88.99 | 100 |
| 13 | 100 | 99.60 | 95.91 | 100 | 100 | 98.51 | 99.66 | 97.89 | 99.04 | 100 |
| 14 | 100 | 99.01 | 79.33 | 91.82 | 100 | 99.70 | 96.82 | 91.74 | 59.77 | 100 |
| OA | 81.45 | 73.92 | 60.70 | 52.76 | 82.39 | 96.06 | 88.00 | 77.26 | 76.90 | 94.92 |
| AA | 83.48 | 73.75 | 51.38 | 61.21 | 90.03 | 94.90 | 88.66 | 66.43 | 74.61 | 96.92 |
| Kappa | 77.80 | 68.72 | 53.43 | 47.66 | 78.69 | 95.10 | 85.06 | 71.92 | 71.80 | 93.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Si, Y.; Gong, D.; Guo, Y.; Zhu, X.; Huang, Q.; Evans, J.; He, S.; Sun, Y. An Advanced Spectral–Spatial Classification Framework for Hyperspectral Imagery Based on DeepLab v3+. Appl. Sci. 2021, 11, 5703. https://doi.org/10.3390/app11125703
Si Y, Gong D, Guo Y, Zhu X, Huang Q, Evans J, He S, Sun Y. An Advanced Spectral–Spatial Classification Framework for Hyperspectral Imagery Based on DeepLab v3+. Applied Sciences. 2021; 11(12):5703. https://doi.org/10.3390/app11125703
Chicago/Turabian StyleSi, Yifan, Dawei Gong, Yang Guo, Xinhua Zhu, Qiangsheng Huang, Julian Evans, Sailing He, and Yaoran Sun. 2021. "An Advanced Spectral–Spatial Classification Framework for Hyperspectral Imagery Based on DeepLab v3+" Applied Sciences 11, no. 12: 5703. https://doi.org/10.3390/app11125703
APA StyleSi, Y., Gong, D., Guo, Y., Zhu, X., Huang, Q., Evans, J., He, S., & Sun, Y. (2021). An Advanced Spectral–Spatial Classification Framework for Hyperspectral Imagery Based on DeepLab v3+. Applied Sciences, 11(12), 5703. https://doi.org/10.3390/app11125703








