Microbiota “Fingerprint” of Greek Feta Cheese through Ripening
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Chemical Analysis
2.3. Microbiological Analysis
Identification of Bacteria with MALDI-TOF MS Biotyper
2.4. Statistical Analysis
3. Results
3.1. Chemical Characteristics of Feta Cheese
3.2. Microbiological Analysis of Feta Cheese
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- 1829/2002. Regulation (EC) no 1829/2002 of the European Parliament and of the Council of 14 October 2002 Amending the Annex to Regulation (EC) No 1107/96 with Regard to the Name ‘Feta’; European Parliament: Brussels, Belgium, 2002.
- Mantis, A.I. Hygiene and Technology of Milk and Dairy Products; Kyriakidis Bros: Thessaloniki, Greece, 1993. [Google Scholar]
- Ledina, T.G.M.; Djordjević, J.; Magas, V.; Colovic, S.; Bulajic, S. MALDI-TOF mass spectrometry for the identification of Serbian artisanal cheeses microbiota. J. Consum. Prot. Food Saf. 2018, 13, 309–314. [Google Scholar] [CrossRef]
- Sánchez-Juanes, F.; Teixeira-Martín, V.; González-Buitrago, J.M.; Velázquez, E.; Flores-Félix, J.D. Identification of species and subspecies of lactic acid bacteria present in Spanish cheeses type “Torta” by MALDI-TOF MS and pheS gene analyses. Microorganisms 2020, 8, 301. [Google Scholar] [CrossRef]
- Fox, A. Mass spectrometry for species or strain identification after culture or without culture: Past, present, and future. J. Clin. Microbiol. 2006, 44, 2677–2680. [Google Scholar] [CrossRef] [PubMed]
- Gantzias, C.; Lappa, I.K.; Aerts, M.; Georgalaki, M.; Manolopoulou, E.; Papadimitriou, K.; De Brandt, E.; Tsakalidou, E.; Vandamme, P. MALDI-TOF MS profiling of non-starter lactic acid bacteria from artisanal cheeses of the Greek island of Naxos. Int. J. Food Microbiol. 2020, 323, 108586. [Google Scholar] [CrossRef] [PubMed]
- Albesharat, R.; Ehrmann, M.A.; Korakli, M.; Yazaji, S.; Vogel, R.F. Phenotypic and genotypic analyses of lactic acid bacteria in local fermented food, breast milk and faeces of mothers and their babies. Syst. Appl. Microbiol. 2011, 34, 148–155. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, O.S.; Argyri, A.A.; Varzakis, E.E.; Tassou, C.C.; Chorianopoulos, N.G. Greek functional Feta cheese: Enhancing quality and safety using a Lactobacillus plantarum strain with probiotic potential. Food Microbiol. 2018, 74, 21–33. [Google Scholar] [CrossRef]
- Montel, M.-C.; Buchin, S.; Mallet, A.; Delbes-Paus, C.; Vuitton, D.A.; Desmasures, N.; Berthier, F. Traditional cheeses: Rich and diverse microbiota with associated benefits. Int. J. Food Microbiol. 2014, 177, 136–154. [Google Scholar] [CrossRef]
- Hayaloglu, A.A. Chapter 39—Cheese Varieties Ripened Under Brine. In Cheese, 4th ed.; McSweeney, P.L.H., Fox, P.F., Cotter, P.D., Everett, D.W., Eds.; Academic Press: Cambridge, UK, 2017; pp. 997–1040. [Google Scholar]
- Choi, J.; In Lee, S.; Rackerby, B.; Frojen, R.; Goddik, L.; Ha, S.D.; Park, S.H. Assessment of overall microbial community shift during Cheddar cheese production from raw milk to aging. Appl. Microbiol. Biotechnol. 2020, 104, 6249–6260. [Google Scholar] [CrossRef]
- Rantsiou, K.; Urso, R.; Dolci, P.; Comi, G.; Cocolin, L. Microflora of Feta cheese from four Greek manufacturers. Int. J. Food Microbiol. 2008, 126, 36–42. [Google Scholar] [CrossRef]
- Bozoudi, D.; Torriani, S.; Zdragas, A.; Litopoulou-Tzanetaki, E. Assessment of microbial diversity of the dominant microbiota in fresh and mature PDO Feta cheese made at three mountainous areas of Greece. LWT Food Sci. Technol. 2016, 72, 525–533. [Google Scholar] [CrossRef]
- Spyrelli, E.; Stamatiou, A.; Tassou, C.; Nychas, G.J.; Doulgeraki, A. Microbiological and metagenomic analysis to assess the effect of container material on the microbiota of feta cheese during ripening. Fermentation 2020, 6, 12. [Google Scholar] [CrossRef]
- Møller, K.; Rattray, F.; Bredie, W.; Høier, E.; Ardö, Y. Physicochemical and sensory characterization of Cheddar cheese with variable NaCl levels and equal moisture content. Int. J. Dairy Sci. 2013, 96, 1953–1971. [Google Scholar] [CrossRef]
- Karaduman, A.; Ozaslan, M.O.; Kilic, I.H.; Bayil-Oguzkan, S.; Kurt, B.S.; Erdogan, N. Identification by using MALDI-TOF mass spectrometry of lactic acid bacteria isolated from non-commercial yogurts in Southern Anatolia, Turkey. J. Int. Microbiol. 2017, 20, 25–30. [Google Scholar]
- 178/2002. Regulation (EC) No 178/2002 of the European Parliament and of the Council of 28 January 2002 Laying down the General Principles and Requirements of Food Law, Establishing the European Food Safety Authority and Laying down Procedures in Matters of Food Safety; European Parliament: Brussels, Belgium, 2002.
- 852/2004. Regulation (EC) No 852/2004 of the European Parliament and of the Council of 29 April 2004 on the Hygiene of Foodstuffs; European Parliament: Brussels, Belgium, 2004.
- 854/2004. Regulation (EC) No 854/2004 of the European Parliament and of the Council of 29 April 2004 Laying down Specific Rules for the Organization of Official Controls on Products of Animal Origin Intended for Human Consumption; European Parliament: Brussels, Belgium, 2004.
- 2073/2005. Regulation (EC) No 2073/2005 of the European Parliament and of the Council of 15 November 2005 on Microbiological Criteria for Foodstuffs; European Parliament: Brussels, Belgium, 2005.
- 1441/2007. Regulation (EC) No 1441/2007 of the European Parliament and of the Council of 5 December 2007 Amending Regulation (EC) No 2073/2005 on Microbiological Criteria for Foodstuffs; European Parliament: Brussels, Belgium, 2007.
- Acuña, L.; Corbalan, N.; Quintela-Baluja, M.; Barros-Velázquez, J.; Bellomio, A. Expression of the hybrid bacteriocin Ent35-MccV in Lactococcus lactis and its use for controlling Listeria monocytogenes and Escherichia coli in milk. Int. Dairy J. 2020, 104, 104650. [Google Scholar] [CrossRef]
- Chen, J.-Q.; Regan, P.; Laksanalamai, P.; Healey, S.; Hu, Z. Prevalence and methodologies for detection, characterization and subtyping of Listeria monocytogenes and L. ivanovii in foods and environmental sources. Food Sci. Hum. Well. 2017, 6, 97–120. [Google Scholar] [CrossRef]
- Naghili, H.; Tajik, H.; Mardani, K.; Rouhani, S.M.R.; Ehsani, A.; Zare, P. Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. In Veterinary Research Forum; Faculty of Veterinary Medicine, Urmia University: Urmia, Iran, 2013; p. 179. [Google Scholar]
- Boultifat, L.; Guendouze, A.; Hamidechi, M. Application of MALDI-TOF MS technology to the identification and characterization of pathogens in dairy industry. Int. J. Curr. Adv. Res. 2018, 6, 185–192. [Google Scholar] [CrossRef]
- CXS 243-2003. Codex Standard for Fermented Milks; Codex Alimentarius Commission: Rome, Italy, 2003. [Google Scholar]
- Litopoulou-Tzanetaki, E.; Tzanetakis, N. Microbiological characteristics of Greek traditional cheeses. Small Rumin. Res. 2011, 101, 17–32. [Google Scholar] [CrossRef]
- Skoufos, I.; Tzora, A.; Giannenas, I.; Karamoutsios, A.; Tsangaris, G.; Fthenakis, G.C. Milk quality characteristics of Boutsiko, Frisarta and Karagouniko sheep breeds reared in the mountainous and semimountainous areas of Western and Central Greece. Int. J. Dairy Technol. 2017, 70, 345–353. [Google Scholar] [CrossRef]
- Skoufos, I.; Tzora, A.; Karamoutsios, A.; Tsangaris, G.; Giannenas, I.; Fthenakis, G. Milk quality characteristics from Greek indigenous goats. J. Hell. Vet. Med. Soc. 2016, 67, 243–252. [Google Scholar] [CrossRef]
- Freiwald, A.; Sauer, S. Phylogenetic classification and identification of bacteria by mass spectrometry. Nat. Protoc. 2009, 4, 732–742. [Google Scholar] [CrossRef]
- Welker, M.J. Proteomics for routine identification of microorganisms. Proteomics 2011, 11, 3143–3153. [Google Scholar] [CrossRef]
- Krásný, L.; Hynek, R.; Hochel, I. Identification of bacteria using mass spectrometry techniques. Int. J. Mass Spectrom. 2013, 353, 67–79. [Google Scholar] [CrossRef]
- Cui, Z.H.; Zheng, Z.J.; Tang, T.; Zhong, Z.X.; Cui, C.Y.; Lian, X.L.; Fang, L.X.; He, Q.; Wang, X.R.; Chen, C.; et al. Rapid detection of high-Level tigecycline resistance in tet(X)-producing Escherichia coli and Acinetobacter spp. based on MALDI-TOF MS. Front. Cell. Infect. Microbiol. 2020, 10, 583341. [Google Scholar] [CrossRef]
- Garcia, E.F.; Luciano, W.A.; Xavier, D.E.; da Costa, W.C.; de Sousa Oliveira, K.; Franco, O.L.; de Morais Junior, M.A.; Lucena, B.T.; Picao, R.C.; Magnani, M.; et al. Identification of lactic acid bacteria in fruit pulp processing by-products and potential probiotic properties of selected Lactobacillus strains. Front. Microbiol. 2016, 7, 1371. [Google Scholar] [CrossRef] [PubMed]
- Nacef, M.; Chevalier, M.; Chollet, S.; Drider, D.; Flahaut, C. MALDI-TOF mass spectrometry for the identification of lactic acid bacteria isolated from a French cheese: The Maroilles. Int. J. Food Microbiol. 2017, 247, 2–8. [Google Scholar] [CrossRef] [PubMed]
- Manolopoulou, E.; Sarantinopoulos, P.; Zoidou, E.; Aktypis, A.; Moschopoulou, E.; Kandarakis, I.G.; Anifantakis, E.M. Evolution of microbial populations during traditional Feta cheese manufacture and ripening. Int. J. Food Microbiol. 2003, 82, 153–161. [Google Scholar] [CrossRef]
- Levante, A.; De Filippis, F.; La Storia, A.; Gatti, M.; Neviani, E.; Ercolini, D.; Lazzi, C. Metabolic gene-targeted monitoring of non-starter lactic acid bacteria during cheese ripening. Int. J. Food Microbiol. 2017, 257, 276–284. [Google Scholar] [CrossRef]
- Thage, B.V.; Broe, M.L.; Petersen, M.H.; Petersen, M.A.; Bennedsen, M.; Ardö, Y. Aroma development in semi-hard reduced-fat cheese inoculated with Lactobacillus paracasei strains with different aminotransferase profiles. Int. Dairy J. 2005, 15, 795–805. [Google Scholar] [CrossRef]
- Fuquay, J.W.; McSweeney, P.L.; Fox, P.F. Encyclopedia of Dairy Sciences; Academic Press: Cambridge, MA, USA, 2011. [Google Scholar]
- Coton, E.; Desmonts, M.-H.; Leroy, S.; Coton, M.; Jamet, E.; Christieans, S.; Donnio, P.-Y.; Lebert, I.; Talon, R. Biodiversity of coagulase-negative staphylococci in French cheeses, dry fermented sausages, processing environments and clinical samples. Int. J. Food Microbiol. 2010, 137, 221–229. [Google Scholar] [CrossRef]
- McSweeney, P.L. Biochemistry of cheese ripening. Int. J. Dairy Technol. 2004, 57, 127–144. [Google Scholar] [CrossRef]
- Fernández, E.; Alegría, Á.; Delgado, S.; Martín, M.C.; Mayo, B. Comparative phenotypic and molecular genetic profiling of wild Lactococcus lactis subsp. lactis strains of the L. lactis subsp. lactis and L. lactis subsp. cremoris genotypes, isolated from starter-free cheeses made of raw milk. Appl. Environ. Microbiol. 2011, 77, 5324–5335. [Google Scholar] [CrossRef]
- Uymaz, B.; Akçelik, N.; Yüksel, Z. Physicochemical and microbiological characterization of protected designation of origin Ezine cheese: Assessment of non-starter lactic acid bacterial diversity with antimicrobial activity. Food Sci. Anim. Resour. 2019, 39, 804. [Google Scholar] [CrossRef]
- Irmler, S.; Bavan, T.; Oberli, A.; Roetschi, A.; Badertscher, R.; Guggenbühl, B.; Berthoud, H. Catabolism of serine by Pediococcus acidilactici and Pediococcus pentosaceus. Appl. Environ. Microbiol. 2013, 79, 1309–1315. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barrangou, R.; Yoon, S.-S.; Breidt, F.; Fleming, H.P.; Klaenhammer, T.R. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 2002, 68, 5452–5458. [Google Scholar] [CrossRef] [PubMed]
- Domingos-Lopes, M.F.P.; Stanton, C.; Ross, P.R.; Dapkevicius, M.L.E.; Silva, C.C.G. Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol. 2017, 63, 178–190. [Google Scholar] [CrossRef] [PubMed]
- Alegría, Á.; Delgado, S.; Flórez, A.B.; Mayo, B. Identification, typing, and functional characterization of Leuconostoc spp. strains from traditional, starter-free cheeses. J. Dairy Sci. 2013, 93, 657–673. [Google Scholar] [CrossRef]
- Psoni, L.; Tzanetakis, N.; Litopoulou-Tzanetaki, E. Characteristics of Batzos cheese made from raw, pasteurized and/or pasteurized standardized goat milk and a native culture. Food Control 2006, 17, 533–539. [Google Scholar] [CrossRef]
- Ambadoyiannis, G.; Hatzikamari, M.; Litopoulou-Tzanetaki, E.; Tzanetakis, N. Probiotic and Technological Properties of Enterococci Isolates from Infants and Cheese. Food Biotechnol. 2004, 18, 307–325. [Google Scholar] [CrossRef]
- Hanchi, H.; Mottawea, W.; Sebei, K.; Hammami, R. The genus Enterococcus: Between probiotic potential and safety concerns—An Update. Front. Microbiol. 2018, 9, 1791. [Google Scholar] [CrossRef]
- Soares, J.C.; Marques, M.R.; Tavaria, F.K.; Pereira, J.O.; Malcata, F.X.; Pintado, M.M. Biodiversity and characterization of Staphylococcus species isolated from a small manufacturing dairy plant in Portugal. Int. J. Food Microbiol. 2011, 146, 123–129. [Google Scholar] [CrossRef]
- Chajecka-Wierzchowska, W.; Zadernowska, A.; Nalepa, B.; Sierpinska, M.; Laniewska-Trokenheim, L. Coagulase-negative staphylococci (CoNS) isolated from ready-to-eat food of animal origin--phenotypic and genotypic antibiotic resistance. Food Microbiol. 2015, 46, 222–226. [Google Scholar] [CrossRef]
- Bockelmann, W. Development of defined surface starter cultures for the ripening of smear cheeses. Int. Dairy J. 2002, 12, 123–131. [Google Scholar] [CrossRef]
- Voidarou, C.; Antoniadou, Μ.; Rozos, G.; Tzora, A.; Skoufos, I.; Varzakas, T.; Lagiou, A.; Bezirtzoglou, E. Fermentative Foods: Microbiology, biochemistry, potential human health benefits and public health issues. Foods 2021, 10, 69. [Google Scholar] [CrossRef]
- Iyer, R.; Tomar, S.K.; Uma Maheswari, T.; Singh, R. Streptococcus thermophilus strains: Multifunctional lactic acid bacteria. Int. Dairy J. 2010, 20, 133–141. [Google Scholar] [CrossRef]
- Pompilio, A.; Di Bonaventura, G.; Gherardi, G. An overview on Streptococcus bovis/Streptococcus equinus complex isolates: Identification to the species/subspecies level and antibiotic resistance. Int. J. Mol. Sci. 2019, 20, 480. [Google Scholar] [CrossRef]
- Mounier, J.; Gelsomino, R.; Goerges, S.; Vancanneyt, M.; Vandemeulebroecke, K.; Hoste, B.; Scherer, S.; Swings, J.; Fitzgerald, G.F.; Cogan, T.M. Surface microflora of four smear-ripened cheeses. Appl. Environ. Microbiol. 2005, 71, 6489–6500. [Google Scholar] [CrossRef] [PubMed]
- Tirloni, E.; Ghelardi, E.; Celandroni, F.; Bernardi, C.; Stella, S. Effect of dairy product environment on the growth of Bacillus cereus. J. Dairy Sci. 2017, 100, 7026–7034. [Google Scholar] [CrossRef] [PubMed]
- Aponte, M.; Pepe, O.; Blaiotta, G. Identification and technological characterization of yeast strains isolated from samples of water buffalo Mozzarella cheese. J. Dairy Sci. 2010, 93, 2358–2361. [Google Scholar] [CrossRef] [PubMed]
- Facchin, S.; Barbosa, A.C.; Carmo, L.S.; Silva, M.C.C.; Oliveira, A.L.; Morais, P.B.; Rosa, C.A. Yeasts and hygienic-sanitary microbial indicators in water buffalo mozzarella produced and commercialized in Minas Gerais, Brazil. Braz. J. Microbiol. 2013, 44, 701–707. [Google Scholar] [CrossRef][Green Version]
- Brennan, N.M.; Brown, G.; Ward, A.C.; Beresford, T.P.; Simpson, P.J.; Fox, P.F.; Cogan, T.M. Corynebacterium mooreparkense sp. nov. and Corynebacterium casei sp. nov., isolated from the surface of a smear-ripened cheese. Int. J. Syst. Evol. Microbiol. 2001, 51, 843–852. [Google Scholar] [CrossRef]
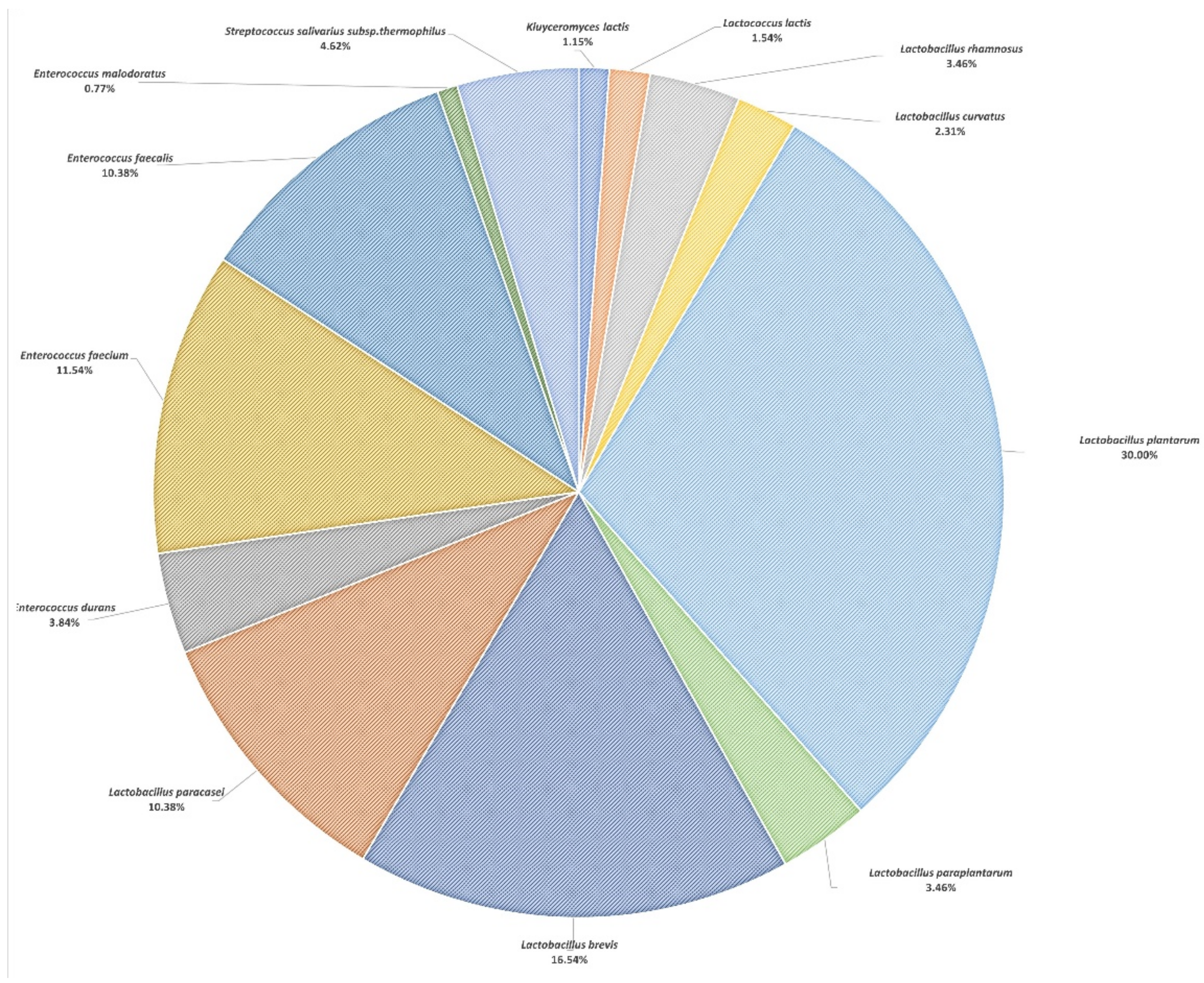
| Ripening Period | Paired Samples t Test | ||||
|---|---|---|---|---|---|
| 3 Months | 6 Months | P (2-Tailed) | Pearson Correlation r | Pearson Correl. Signif. P | |
| Samples N# | 23 | 23 | |||
| Moisture (%) | 51.62 ± 2.41 | 51.86 ± 2.90 | 0.684 | - | - |
| Total solids (%) | 48.55 a ± 2.20 | 47.57 b ± 2.69 | 0.043 | 0.617 | 0.002 |
| Protein (%) | 18.30 a ± 1.96 | 17.30 b ± 2.19 | 0.008 | 0.683 | <0.001 |
| Fat (%) | 25.42 ± 2.15 | 24.60 ± 2.40 | 0.086 | - | - |
| Salt (%) | 1.87 ± 0.06 | 1.87 ± 0.08 | 0.927 | - | - |
| pH | 4.60 a ± 0.09 | 4.50 b ± 0.09 | <0.001 | 0.780 | <0.001 |
| Ripening Period | Paired Samples t Test | ||||
|---|---|---|---|---|---|
| 3 Months | 6 Months | P (2-Tailed) | Pearson Correlation r | Pearson Correl. Signif. P | |
| Samples N# | 23 | 23 | |||
| Total Viable Counts (TVC) (Log10 cfu/g) | 7.316 ± 0.882 | 7.044 ± 0.656 | 0.193 | - | - |
| Total Lactic Acid Bacteria (Log10 cfu/g) | 7.408 ± 0.906 | 7.554 ± 1.142 | 0.497 | - | - |
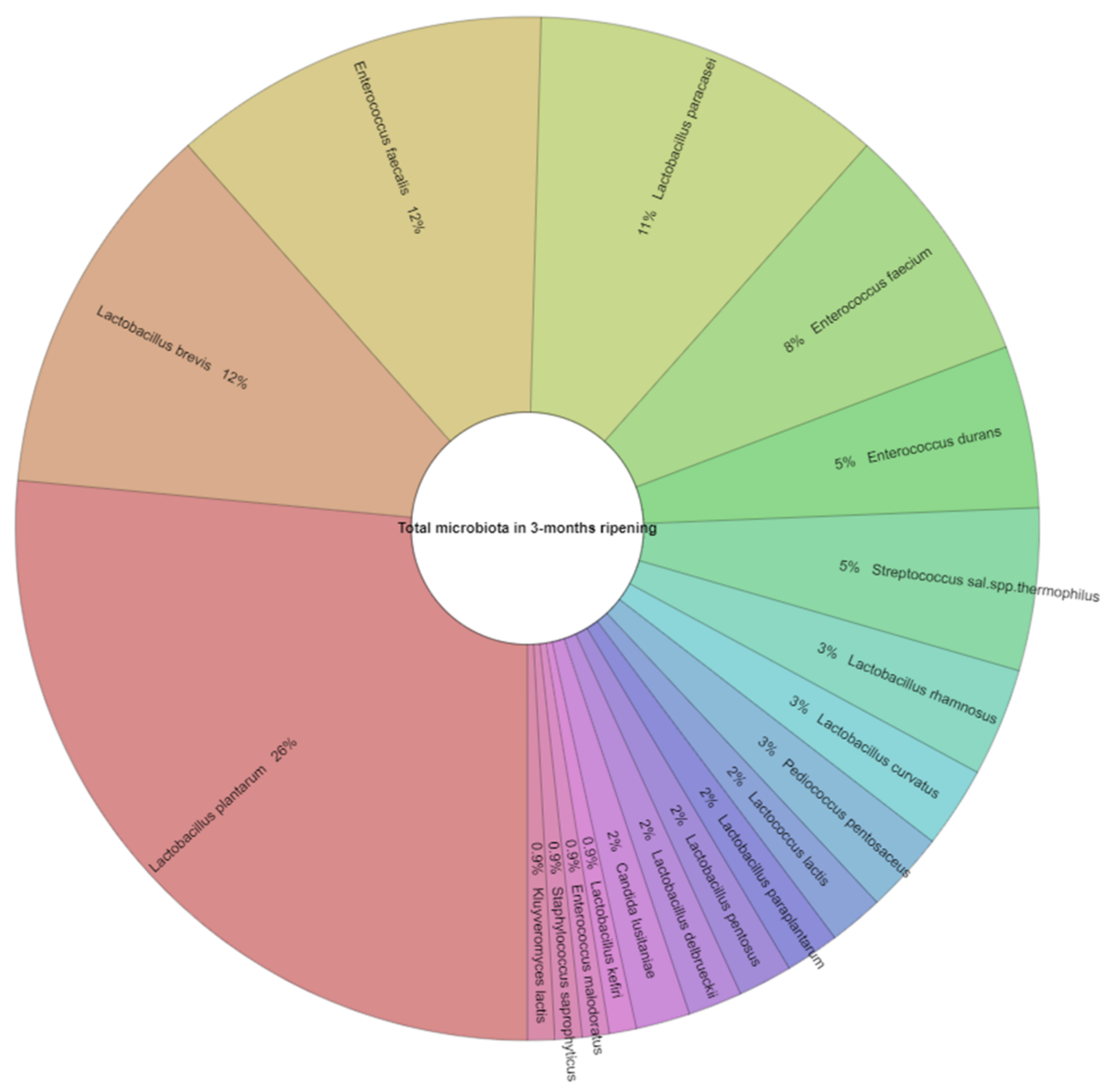
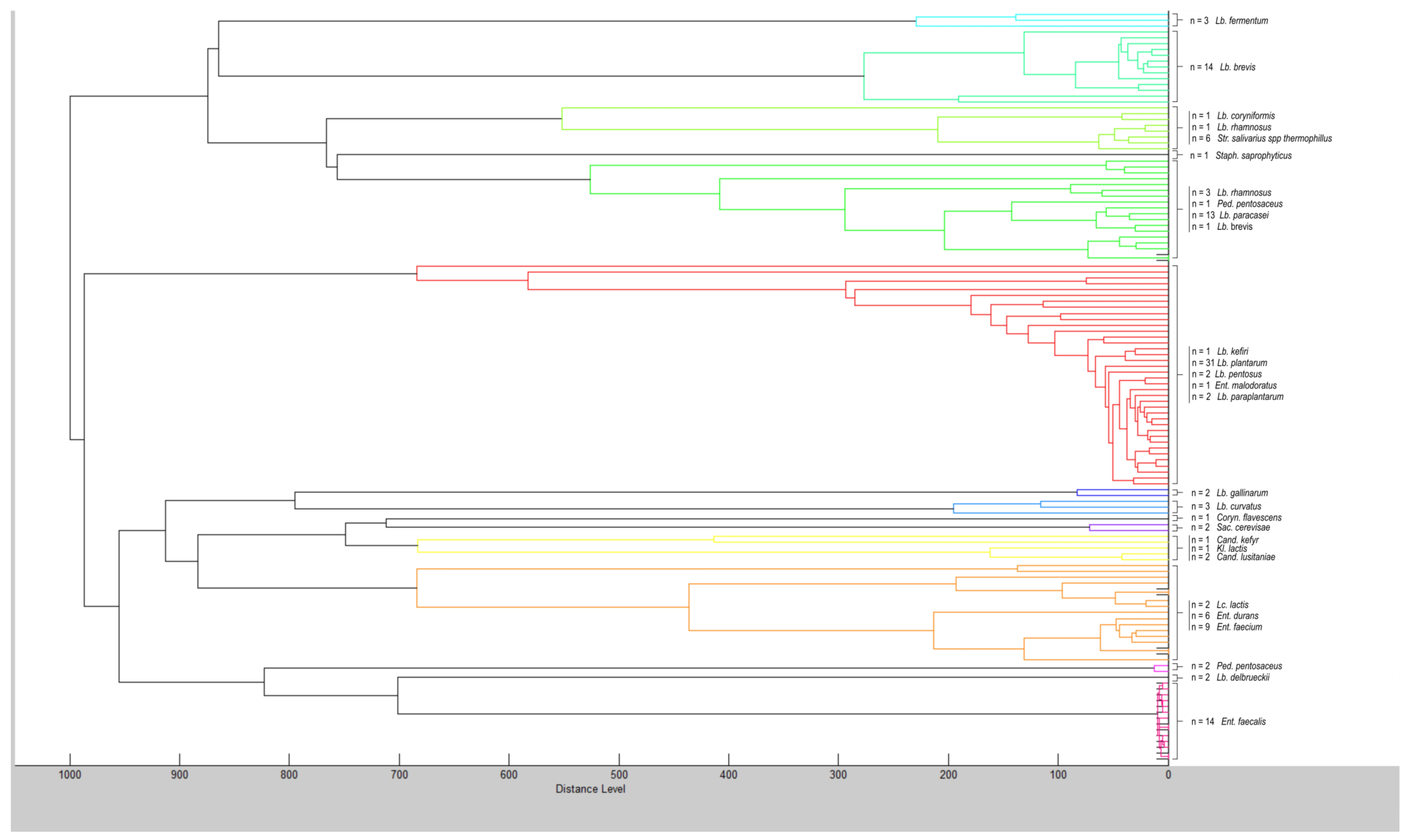
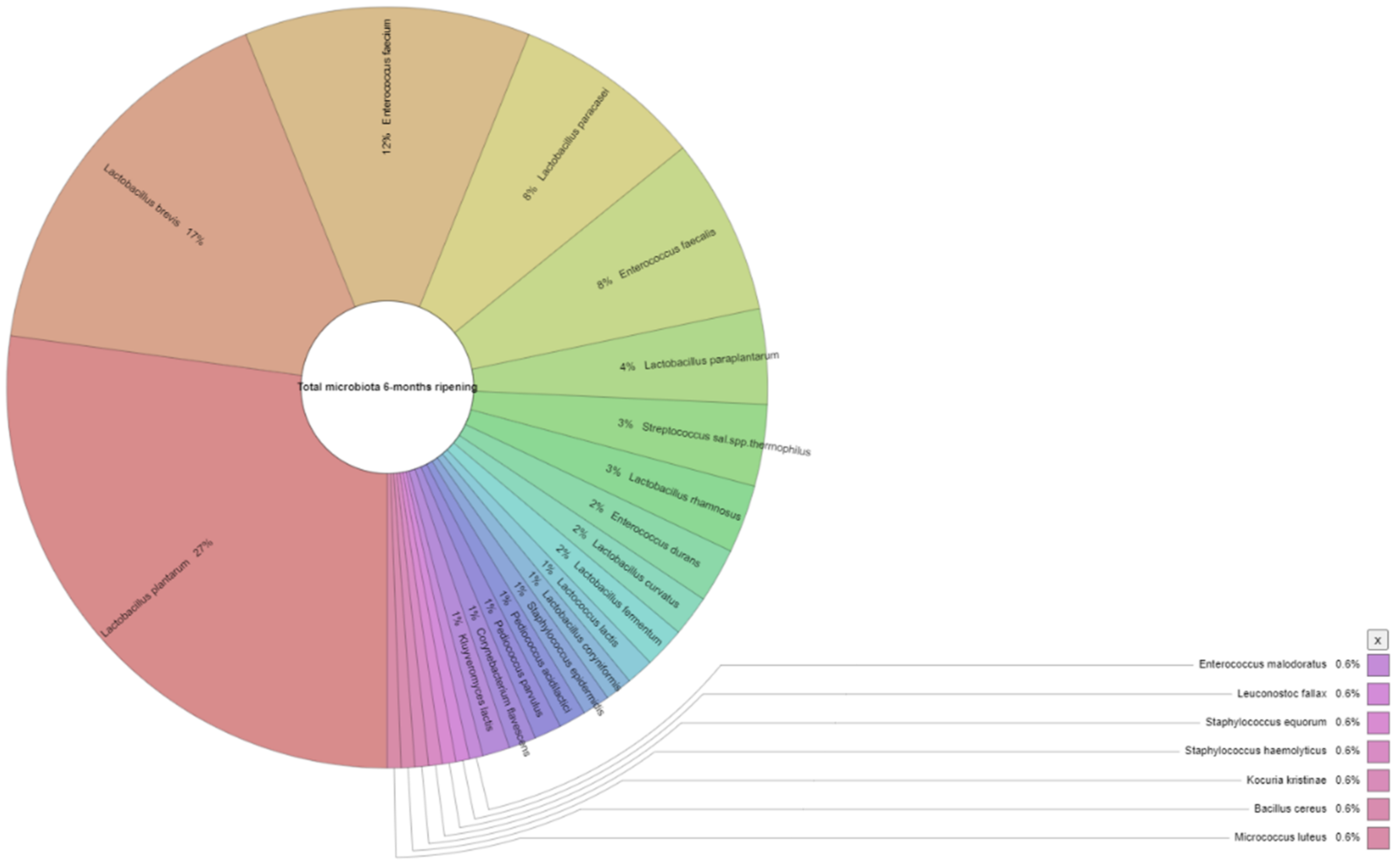
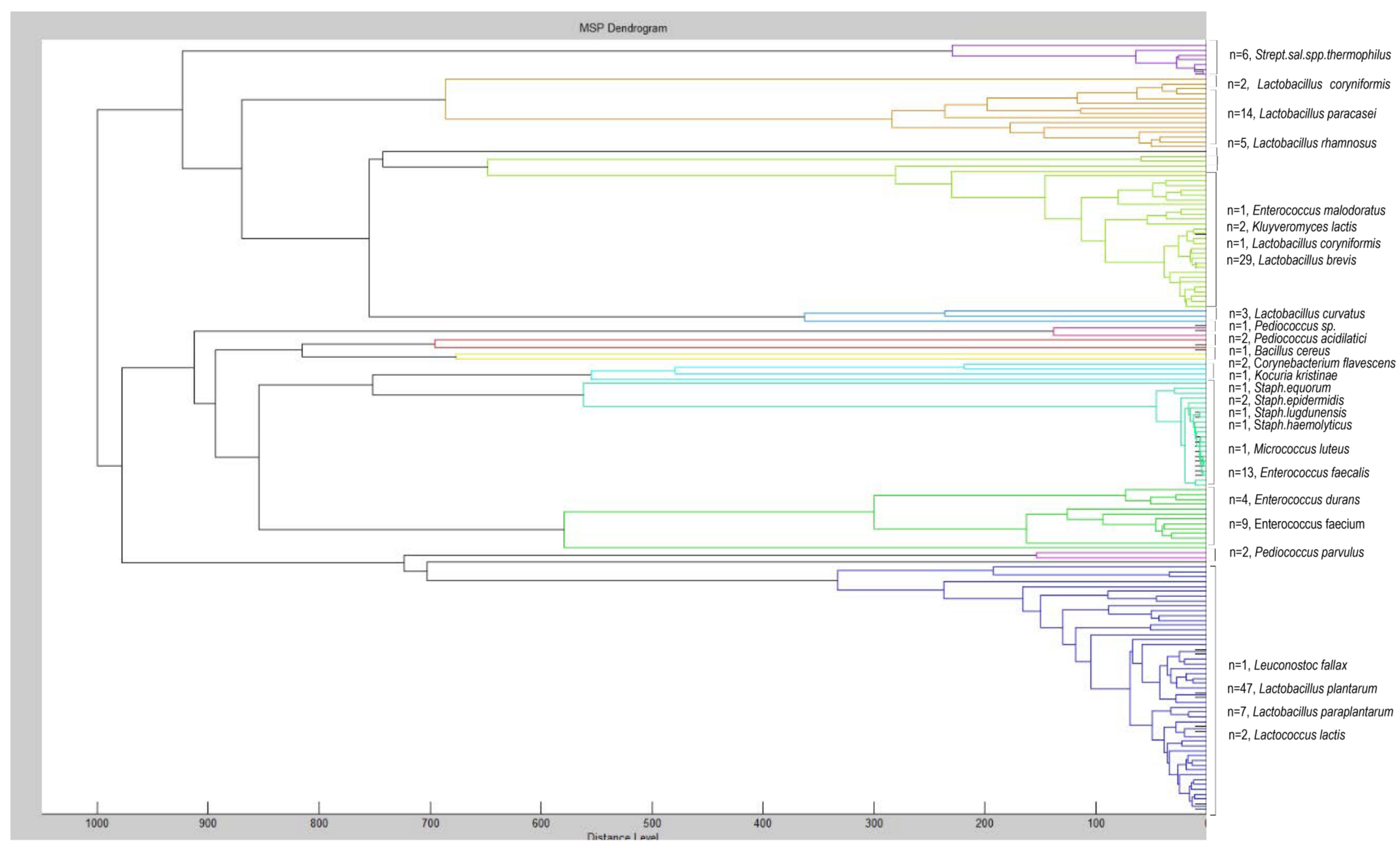
| Appearance in Samples (%) 1 | Number of Isolates 2 | |||
|---|---|---|---|---|
| 3 Months | 6 Months | 3 Months | 6 Months | |
| Lactococcus lactis | 13.0 | 8.7 | 2 | 2 |
| Lactobacillus rhamnosus | 13.0 | 17.4 | 4 | 5 |
| Lactobacillus curvatus | 8.7 | 8.7 | 3 | 3 |
| Lactobacillus plantarum | 100.0 | 87.0 | 31 | 47 |
| Lactobacillus paraplantarum | 4.3 | 26.1 | 2 | 7 |
| Lactobacillus brevis | 56.5 | 73.9 | 14 | 29 |
| Lactobacillus pentosus | 4.3 | 0.0 | 2 | ND |
| Lactobacillus fermentum | 8.7 | 0.0 | 3 | ND |
| Lactobacillus paracasei | 56.5 | 39.1 | 13 | 14 |
| Lactobacillus coryniformis | 0.0 | 4.3 | ND | 2 |
| Lactobacillus delbrueckii | 4.3 | 0.0 | 2 | ND |
| Lactobacillus kefiri | 4.3 | 0.0 | 1 | ND |
| Enterococcus durans | 13.0 | 17.4 | 6 | 4 |
| Enterococcus faecium | 34.8 | 17.4 | 9 | 21 |
| Enterococcus faecalis | 47.8 | 43.5 | 14 | 13 |
| Enterococcus malodoratus | 4.3 | 4.3 | 1 | 1 |
| Leuconostoc fallax | 0.0 | 4.3 | ND | 1 |
| Staphylococcus saprophyticus | 4.3 | 0.0 | 1 | ND |
| Staphylococcus equorum | 0.0 | 4.3 | ND | 1 |
| Staphylococcus haemolyticus | 0.0 | 4.3 | ND | 1 |
| Staphylococcus epidermidis | 0.0 | 13.0 | ND | 2 |
| Streptococcus salivarius subsp. thermophilus | 21.7 | 30.4 | 6 | 6 |
| Pediococcus pentosaceus | 8.7 | 0.0 | 3 | ND |
| Pediococcus acidilactici | 0.0 | 4.3 | ND | 2 |
| Pediococcus parvulus | 0.0 | 4.3 | ND | 2 |
| Candida lusitaniae | 4.3 | 0.0 | 2 | ND |
| Kocuria kristinae | 0.0 | 4.3 | ND | 1 |
| Bacillus cereus | 0.0 | 4.3 | ND | 1 |
| Corynebacterium flavescens | 0.0 | 4.3 | ND | 2 |
| Kluyveromyces lactis | 13.0 | 13.0 | 1 | 2 |
| Micrococcus luteus | 0.0 | 4.3 | ND | 1 |
| TOTAL | - | - | 117 | 173 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tzora, A.; Nelli, A.; Voidarou, C.; Fthenakis, G.; Rozos, G.; Theodorides, G.; Bonos, E.; Skoufos, I. Microbiota “Fingerprint” of Greek Feta Cheese through Ripening. Appl. Sci. 2021, 11, 5631. https://doi.org/10.3390/app11125631
Tzora A, Nelli A, Voidarou C, Fthenakis G, Rozos G, Theodorides G, Bonos E, Skoufos I. Microbiota “Fingerprint” of Greek Feta Cheese through Ripening. Applied Sciences. 2021; 11(12):5631. https://doi.org/10.3390/app11125631
Chicago/Turabian StyleTzora, Athina, Aikaterini Nelli, Chrysoula Voidarou, George Fthenakis, Georgios Rozos, Georgios Theodorides, Eleftherios Bonos, and Ioannis Skoufos. 2021. "Microbiota “Fingerprint” of Greek Feta Cheese through Ripening" Applied Sciences 11, no. 12: 5631. https://doi.org/10.3390/app11125631
APA StyleTzora, A., Nelli, A., Voidarou, C., Fthenakis, G., Rozos, G., Theodorides, G., Bonos, E., & Skoufos, I. (2021). Microbiota “Fingerprint” of Greek Feta Cheese through Ripening. Applied Sciences, 11(12), 5631. https://doi.org/10.3390/app11125631










