Abstract
The aim of this study was the identification of the causative agent of the basal glume rot of wheat Pseudomonas syringae pv. atrofaciens from the affected weeds in wheat crops, and determination of its virulent properties. Isolation of P. syringae pv. atrofaciens from weeds of wheat crops was carried out by classical microbiological methods. To identify isolated bacteria, their morphological, cultural, biochemical, and serological properties as well as fatty acids and Random Amplification of Polymorphic DNA (RAPD)-PCR (Polymerase chain reaction) profiles with the OPA-13 primer were studied. Pathogenic properties were investigated by artificial inoculation of wheat plants and weed plants, from which bacteria were isolated. For the first time, bacteria that are virulent both for weeds and wheat were isolated from weeds growing in wheat crops. It was shown that the fatty acids profiles of the bacteria isolated from the weeds contained typical for P. syringae pv. atrofaciens fatty acids, in particular, hydroxy acids: 3-hydroxydecanoic, 2-hydroxydodecanoic, and 3-hydroxydodecanoic. RAPD-PCR profiles of the newly isolated strains were identical to those of the collection strains P. syringae pv. atrofaciens UCM B-1011 and P. syringae pv. atrofaciens UCM B-1014 and contained a dominant fragment of 700 bp. The isolated strains, according to their phenotypic and genotypic properties, were identified as P. syringae pv. atrofaciens. It was established that the causative agent of basal glume rot of wheat P. syringae pv. atrofaciens is polyphagous and capable of infecting a wide range of plants. The main control measure for cereals diseases caused by P. syringae pv. Atrofaciens—crop rotations with nonhost species, should be revised, and alternative control methods must be proposed.
1. Introduction
Nowadays, around the world, researchers have noted the increasingly threatening spread and aggressiveness of phytopathogenic bacteria [1,2]. This trend is also observed in Ukraine [3,4]. This also applies to wheat, one of the most valuable and important grain crops and ranks first in the world’s sown area [5]. Bacterial diseases spread data are often not realistic, as they are based as a rule on observing symptoms without isolation of the pathogen and its identification [2]. Pseudomonas syringae pv. atrofaciens has been the main causative agent of bacterial diseases of wheat, particularly basal glume rot of wheat, for many years [6,7,8,9]. The study of this pathogen’s biological properties, the patterns of population formation and the circulation of pathogens, and techniques that will allow influencing these processes are of fundamental and practical importance and will not lose their relevance.
The causative agent of the basal glume rot of wheat belongs to the species of Pseudomonas syringae, which, according to much research, is the most important bacterial plant pathogen in terms of economics and scientific studies [10]. Pseudomonas syringae pv. atrofaciens causes basal glume rot of wheat in Russia, Bulgaria, Italy, Germany, New Zealand, and Iran [8,11]. The scientific literature data confirm the high harmfulness of this pathogen and its ability to cause epiphytotics [12]. This pathogen’s danger is its ability to colonize both the host plant and other plant species of the phyllosphere effectively. Forming epiphytic populations, which is the primary inoculum for plant infection, P. syringae pv. atrofaciens can cause mass plant damage when favorable weather conditions occur [13,14,15].
It is known that for a long time, the causative agent of the basal glume rot of wheat had been found as an epiphyte on non-crops plant species and also on annual and perennial weeds [14]. Weeds’ ability to serve as an ecological niche for agents of bacterial diseases that can affect crops has been discussed [16], and it was proved experimentally that weeds could serve as sources of dangerous epiphytic populations of phytopathogenic bacteria [17]. At the same time, the presence of the pathogens common both for cultivated plants and weeds in crops and the problem of the specialization of the pathogens that affect weeds remain unclear. Therefore, our study aimed to identify phytopathogenic bacteria on affected weeds in wheat crops and to establish their relationship with the causative agent of the basal glume rot of wheat.
2. Material and Methods
Bacterial strains used in research. Strains of Pseudomonas syringae pv. atrofaciens, which are stored in the collection of microorganisms of the Department of phytopathogenic bacteria of the Zabolotny Institute of Microbiology and Virology of the National Academy of Sciences of Ukraine (NASU), Kyiv were used in the study: P. syringae pv. atrofaciens UCM B-1011 (PDDCC 4394), P. syringae pv. atrofaciens UCM B-1014, and P. syringae pv. atrofaciens 9780. Also, we used the type strain P. syringae pv. syringae UCM B-1027 (NCPPB 281).
Isolation of P. syringae pv. atrofaciens. In order to detect the strains of P. syringae pv. atrofaciens, wheat fields in the Kyiv region, Ukraine (50°05′16.3″ N 30°02′56.8″ E) were surveyed 2019 and selected weed plants of Sonchus arvensis L. and Papaver argemone L. with symptoms of bacterial infection were collected. Bacteria isolation was carried out by plating the pieces of plants, pounded with 0.1 mL of sterile tap water, on potato agar. For the bacteria isolation, the parts of plants were selected on the border of healthy and damaged tissues. The morphology and structure of bacterial colonies were studied after 72 h growing on potato agar in Petri dishes [18]. Cell morphology and motility were determined in Gram-stained and “crushed drop” preparations, respectively, under a Sigeta MB-201 microscope using a one-day bacterial culture grown in nutrient broth (NB) [5,18]. For the initial selection of the isolates from weeds, the LOPAT test (Levan production, Oxidase, Protopectinase activity on potato, Arginine dihydrolase, hypersensitive reaction on Tobacco) was used [18].
Physiological and biochemical characterization. For studying the physiological and biochemical properties of the bacterial isolates from weeds, the test system NEFERMtest24 (MikroLaTEST®, ErbaLachema, Czech Republic) was used. Oxidase was detected in the cells of the isolated strains by test strips Bactident Oxidase (Merck, Germany). Fermentative and oxidative glucose metabolism (OF-test) were determined by the breakable microwell plate (OFtest, Erba Lachema). The ability to induce rot on potato was investigated by applying 0.01 mL of bacterial suspension (1 × 107 colony-forming unit (CFU)/mL) on potato discs [18]. Potato discs were incubated for 24 h at 25 °C, and then the presence of rot was registered.
Pathogenicity tests. To study the virulent properties of isolated strains, artificial inoculation of S. arvensis and P. argemone plants, from which bacterial strains were obtained, was carried out under field conditions and of plants of the spring wheat variety Pecherianka in the greenhouse.
Artificial infection of wheat seedlings seven days old was performed by pricking through a drop of bacterial suspension with a concentration of 1 × 107 CFU/mL. The development of visible symptoms of damage evidenced the bacterium virulence. The results of artificial infection were analyzed according to a 5-point scale in 10 days after the inoculation of bacteria [5]. Highly aggressive strains caused the damage from three to four scores, while weakly aggressive—1–2 scores. Aggressiveness was determined as the average score of disease symptoms in ten inoculated plants.
Also, the pathogenic properties of bacteria that were isolated from the weeds were determined by artificial inoculation of the respective plant species from which the strains were isolated. For this purpose, the bacterial suspension containing 1 × 107 CFU/mL was injected into a leaf by a syringe. Sterile tap water was inoculated into the plant as a control. Aggressiveness was calculated as the average score of the results obtained from 6 experiments (during each of them, five plants were artificially infected). The artificial infection of weed plants was evaluated ten days after the inoculation of bacterial suspension due to the five-point scale. The degree of the damage of plants (from zero to five points) was estimated by the size of the necrosis formed: 0—no symptoms of damage; 1—a border around the injection site; 2—small spots of necrosis (up to 5 mm); 3—damage of ½ part of the leaf; 4—damage of up to 2/3 of the leaf and stems and drying leaves.
The ability of P. syringae to induce a hypersensitive reaction on tobacco was determined by the leaf infiltration technique [18]. The suspension of two-day bacterial cultures with a concentration of 1 × 107 CFU/mL was injected under the leaves’ epidermis by a syringe. A cell suspension was prepared in sterile tap water, and it was used as a negative control. The presence of necrosis was observed in a day. The experiment was repeated three times.
Serology. For determining the serological properties of the strains, the method of double diffusion in agar was used. The assignment of the strains of P. syringae pv. atrofaciens to a serological group was determined according to the scheme developed in the Department of phytopathogenic bacteria of the Zabolotny Institute of Microbiology and Virology of NASU [19]. Sera were obtained by the immunization of rabbits with live bacteria. The strains of P. syringae that belonged to serogroups II, IV, V, VI were used for immunization [19].
Fatty acids determination. In order to identify the isolated bacteria, the determination of the fatty acids composition of total cellular lipids was carried out. For this purpose, fatty acid methyl ethers were obtained during the methanolysis of whole bacterial cells in a 5% solution of sulfuric acid in methanol [5]. Chromatographic separation and identification of fatty acids were carried out at the Collective Use Center of the D.K. Zabolotny Institute of Microbiology and Virology NASU. The separation of the fatty acid methyl esters was carried out on chromatographic-mass spectrometric system Agilent 6890N/5973 inert. Peaks were identified by comparing their retention times with retention times of standard Bacterial Acid Methyl Ester Mix (26 components), analytical standard, Supelco®, as well as using an integrated database of NIST 02 mass spectra. The content of individual fatty acids was determined as a percentage of the total peak area.
Analysis of fatty acids profiles of P. syringae pv. atrofaciens isolated from weeds and affected by basal glume rot of wheat plants was performed using RStudio (Version 3.5.1) with the basic functions and library “dendextend” [20]. For the dendrogram building, the data were normalized, the Euclidean distance was calculated, and the Ward clusterization method was used.
RAPD-PCR analysis. RAPD-PCR (Random Amplification of Polymorphic DNA—Polymerase chain reaction) typing was carried out with the primer OPA-13 (5′-CAGCACCCAC-3′) [21].
For DNA extraction, the strains of P. syringae pv. atrofaciens were grown on nutrient broth (NB) at 28 °C under shaking conditions (160 rpm) for 24 h. The cells were then precipitated by centrifugation on ELMI CM-50 at 8000× g for 10 min, resuspended in 0.9% NaCl, and again centrifuged at the same speed [5]. According to the manufacturer’s instruction, DNA extraction was carried out with “DNA sorbent B” (AmpliSens, Moscow, Russia) kit. The DNA was stored at −20 °C.
Amplification with RAPD primers was performed in a 25 μL mixture: 200 ng of genomic DNA; 25 pmol of primer; 2.5 Units SynTaq Polymerase; 0.2 mM of each deoxynucleotide triphosphate; 2.5 μL of 10× PCR buffer (Helicon, Moscow, Russia). The amplification was conducted in Gene ATAQ Controller thermocycler (Pharmacia LKB, Uppsala, Sweden). The procedure was as follows: initial denaturation 95 °C—5 min; 45 cycles: 94 °C—1 min, 38 °C—1 min, 74 °C—1 min; terminal elongation 72 °C—7 min [21]. PCR products were run on 1.5% agarose gel for 40 min at 90 V; 200 bp DNA Ladder (O’RangeRuller, Standart, Fermentas) was used as a marker. Gels were stained with Ethidium bromide, visualized in UV light, and photographed by digital camera.
Data were analyzed with RStudio (Version 3.5.1) using the basic functions [20]. The Jaccard distance was calculated for the dendrogram building, and the nearest neighbor clusterization method was used.
3. Results
In the wheat fields examined in the Kyiv region, weeds with the symptoms characteristic of phytopathogenic bacteria were found. Particularly, S. arvensis plants had light brown angular spots and P. argemone plants had a roundish shape with whitish spots on the leaves (Figure 1). From 53 symptomatic plants, 25 bacterial isolates were obtained. Among them, seven isolates (L8, L9, L11, L11a, L13, L15, L16) were found to be aerobic non-spore-forming Gram-negative motile rods. These strains formed gray, translucent, smooth, rounded colonies that were 1.0–2.5 mm in diameter and had slightly wavy edges with a compacted and convex center. These isolates were oxidase-negative and induced a hypersensitivity reaction (HR) on tobacco leaves. According to the results of LOPAT tests (levan production, oxidase, protopectinase activity on potato, arginine dihydrolase, HR on tobacco leaves) (L+, O−, P−, A−, T+), these seven strains were assigned to LOPAT group 1a (P. syringae) (Table 1).

Figure 1.
Symptoms on of Sonchus arvensis L. (A) and Papaver argemone L. (B) found in wheat fields.

Table 1.
Physiological and biochemical properties of bacteria used in this study.
The basic properties of P. syringae strains L8, L9, L11, L11a, L13, L15, L16 were similar to the properties of neopathotype strain P. syringae pv. atrofaciens UCM B-1011 (PDDCC 4394) (Table 1). All the new isolated strains utilized citrate, mannitol, xylose, arabinose, galactose, saccharose, and did not use lactose, trehalose, malonate, maltose, and cellobiose. These strains showed reactions as β-glucosidase, phosphatase, aesculin hydrolysis, and did not demonstrate urease, ornithine decarboxylase, lysine decarboxylase, n-acetyl-β-d-glucosaminidase, α-galactosidase, γ-glutamyl transferase activities. Some newly isolated strains showed weak utilization of acetamide (L11, L11a, L13, L15, L16) and inositol fermentation (L8, L9, L11, L11a, L16).
Strains of bacteria isolated from weeds were different in aggressiveness against wheat and host plants (Table 2). The highly aggressive against wheat strains of P. syringae (L15 and L16) were isolated from the S. arvensis. The score of aggressiveness of these strains was 3 (5-point scale) for wheat. At the same time, the strain L16 was medium aggressive against the host plant—S. arvensis (score of aggressiveness was 3 on a 5-point scale). Strains L8, L9, L11, L11a, and L13 were medium or weak aggressive against weeds and wheat (score of aggressiveness 1–2).

Table 2.
Virulence and serological properties of the bacterial strains used in this study.
In order to study the heterogeneity of P. syringae pv. atrofaciens in Ukraine and to establish the relationship between collection strains of this pathogen and the strains of P. syringae that were isolated from affected weeds in wheat fields, serological properties, fatty acid composition, and RAPD profiles were studied.
To study the antigen properties of the strains isolated from weeds that grew in wheat crops, sera specific to II, IV, V, VI serogroups were used, as the bacteria found on wheat plants belong to one of such serogroups. Two strains of P. syringae isolated from P. argemone (L8, L9) were belonged to serogroup II, while strains isolated from S. arvensis assigned to serogroup II (L11, L11a) and serogroup VI (L13, L15, L16) (Table 2).
When studying the composition of fatty acids of total cellular lipids of the isolates L8, L9, L11, L11a, L13, L15, L16, the presence of the following fatty acids was detected: saturated—dodecanoic (C12:0), tetradecanoic (C14:0), hexadecanoic (C16:0), octadecanoic (C18:0); unsaturated—cis-9-hexadecene (C16:1), cis-11-octadecene (C18:1); cyclic—cis-9,10-methylenehexadecanoic (C17:0 cyclo), cis-9,10-methylenoctadecanoic (C19:0 cyclo), hydroxy acids—3-hydroxydecanoic (3-OH C10:0), 2-hydroxydodecanoic (2-OH C12:0), and 3-hydroxydodecanoic (3-OH C12:0) (Figure 2). There were three predominant fatty acids: C16:0, C16:1, and C18:1. The last ones’ content was 25.67–28.89%, 41.63–47.98%, and 12.13–17.12% in the cells of newly isolated strains of P. syringae. This was similar to the content of predominant fatty acids in the cells of collection strains of P. syringae: 28.28–34.25%, 31.16–38.78%, and 18.25–24.49%, respectively. Besides, hydroxy acids have an important role in the taxonomy of P. syringae. Three hydroxy fatty acids were identified in all investigated strains from 0.24 to 4.50%. The fatty acid profiles of all seven isolates were similar to each other and the collection strains of the causative agent of the basal glume rot of wheat, isolated from the infected wheat plants.
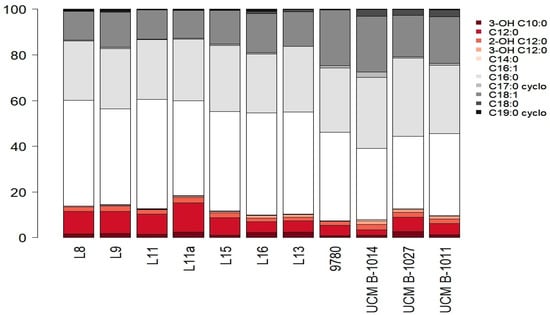
Figure 2.
The fatty acid composition of the total cellular lipids of P. syringae isolates.
We carried out a comparative analysis of the fatty acid composition of the strains isolated from weeds and P. syringae pv. atrofaciens strains, isolated from wheat (2399, 9400, 9417, 9748, 9819, 9771, 9785, P204, and P203) and isolated from rye (7194, 7836, 7959, 8099, 8116, 8274, 8281, 8317, 8462, 8904, and 9010). All these strains were described by us earlier in the works [22,23]. They are stored in the Collection of microorganisms of the Department of phytopathogenic bacteria of the Zabolotny Institute of Microbiology and Virology of NASU. Data on the fatty acid composition of these strains are available in [22]. It was found that bacteria from weeds tended to group into one large cluster together with the strains isolated from wheat. At the same time, strains of P. syringae pv. atrofaciens from rye formed another cluster with some strains from wheat (Figure 3). We found no correlation between the aggressiveness of the strains P. syringae pv. atrofaciens and the composition of fatty acids.
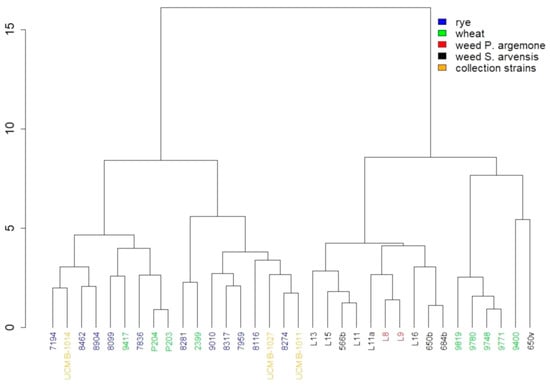
Figure 3.
Cluster analysis of fatty acids composition of Pseudomonas syringae strains (Euclidean distance, Ward method).
RAPD-PCR profiles of new isolated and collection strains of P. syringae are reported in Figure 4. The range of polymorphic loci ranged from 500 to 1300 kb. For all strains of P. syringae pv. atrofaciens, the dominant product was a DNA fragment of about 700 kb in size. It should be noticed that RAPD-profiles with primer OPA-13 of P. syringae strains L8, L9, L11 and L11a were identical to each other, and they were close to the profiles of P. syringae strain L16 and neopathotype strain P. syringae pv. atrofaciens UCM B-1011. At the same time RAPD-profiles of P. syringae strains L13 and L15 were similar to P. syringae pv. atrofaciens UCM B-1011 and P. syringae pv. atrofaciens 9780.
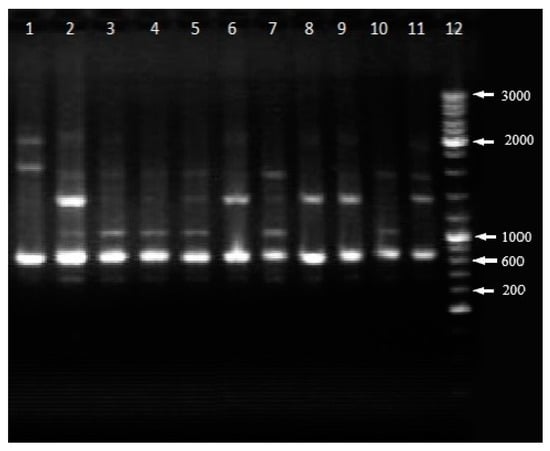
Figure 4.
Random Amplification of Polymorphic DNA (RAPD)-PCR with primer OPA-13: 1—P. syringae pv. syringae UCM B-1027; 2—P. syringae pv. atrofaciens UCM B-1011; 3—P. syringae L8; 4—P. syringae L9; 5—P. syringae L11; 6—P. syringae pv. atrofaciens 9780; 7—P. syringae L11a; 8—P. syringae L13; 9—P. syringae L15; 10—P. syringae L16; 11—P. syringae pv. atrofaciens UCM B-1014; 12—marker.
According to RAPD-PCR analysis results with the OPA-13 primer, the strains isolated from the weeds possessed a high level of similarity with the causative agent of the basal glume rot of wheat P. syringae pv. atrofaciens. Strains of P. syringae isolated from weeds formed a common cluster with three collection strains: P. syringae pv. atrofaciens UCM B-1011, P. syringae pv. atrofaciens UCM B-1014 and P. syringae pv. atrofaciens 9780. Whereas P. syringae pv. atrofaciens UCM B-1027 distinguished into a separate cluster (Figure 5).
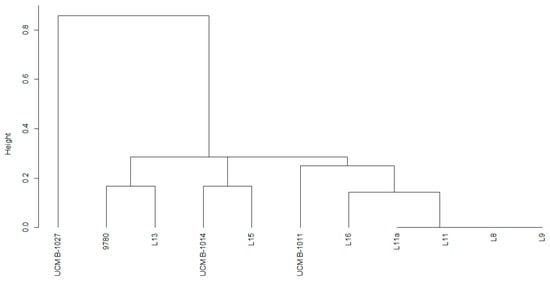
Figure 5.
Cluster analysis of RAPD patterns obtained with primer OPA-13 of Pseudomonas syringae strains (Jaccard distance, the nearest neighbor clusterization method).
4. Discussion
Considering the significant number of weeds in the Ukrainian wheat fields and the ability of weeds to serve as an ecological niche for bacterial pathogens, weeds in wheat crops were collected from symptomatic samples. As a result of the work, seven isolates of P. syringae from S. arvensis and P. argemone grew in wheat crops and were isolated. Further identification of the isolates with the determination of their affiliation to the particular pathovar was difficult due to the high affinity of P. syringae patovars for biochemical, physiological, and even genetic features [10,24,25,26]. The pathovar of the species P. syringae is usually determined by the host plant. It should be noted that the status of such a taxonomic unit as pathovar within the species P. syringae is actively discussed in the scientific literature [24,27,28]. Some researchers consider the division into pathovars unjustified [10,24,27,29] and claim that P. syringae is a universal pathogen. However, other studies have shown the possibility of differentiating pathovar for pathogenic properties and some other biological characteristics [24,30,31,32].
When verifying the virulence properties, these seven isolates’ ability to cause disease to both weeds (S. arvensis and P. argemone) and wheat was confirmed. After artificial infection of wheat, light brown spots with a dark border were formed, which eventually increased in size and covered the stem, leading to a darkening of the stem core. Such symptoms are characteristic of the causative agent of the basal glume rot of wheat [6].
Analyzing the results of serological studies of the causative agent of the basal glume rot of wheat, we concluded that the strains of P. syringae isolated from different crops and weeds were similar in antigenic composition and belonged to serogroups II, IV, V, or VI [19]. The host plant determines the prevalence of specific serogroups and, in our opinion, is possibly associated with the best survival of the strains of these serogroups in various conditions. This hypothesis is supported by the evidence that many strains usually represent these serogroups. The fact that most of the strains isolated from weeds were assigned to the serogroups II and IV according to the results of agar precipitation indicates the predominance of these serogroups among pathogens isolated from wheat also among pathogens isolated from other plant species.
All studied bacterial strains, regardless of the source of their isolation, are similar in the qualitative composition of fatty acids, and all strains contain hydroxy fatty acids, which are important for confirming their belonging to P. syringae [33]. The presence of differences in the spectrum of fatty acids is a feature of each strain and does not correlate with differentiation into pathovars [33]. The results of the fatty acid composition of P. syringae cell lipids are consistent with the literature: most of the hydroxy acids in P. syringae are present in the amount at less than 5% of the total peak area. Stead et al. [33] established the possibility of using fatty acid composition analysis to identify phytopathogenic bacteria at the species level.
The RAPD-PCR method with primer OPA-13 (5′-CAGCASSCACC-3′) was successfully used by other researchers for the genetic analysis of P. syringae [25,34] and to analyze the genetic heterogeneity of the wheat pathogen population in Ukraine [23]. RAPD-PCR is a useful technique for studying the genetic relationship between bacterial strains due to specificity and sensitivity. Analysis of literature data [21,25] and our previous studies have shown that OPA-13 is the most optimal primer for identifying P. syringae pv. atrofaciens strains isolated from wheat crops. The implementation of RAPD-PCR analysis with the primer OPA-13 also allowed to establish a high degree of affinity between the causative agent of the basal glume rot of wheat P. syringae pv. atrofaciens and strains of P. syringae that were isolated from the weeds of wheat crops.
Based on RAPD-PCR results, all P. syringae pv. atrofaciens strains, regardless of the geographical region of isolation, the plants from which the bacteria were isolated, their serological and aggressiveness properties, formed a related group. Thereby, it might indicate that P. syringae pv. atrofaciens strains are genetically homogeneous group [23]. It was also shown that P. syringae pv. atrofaciens strains differed from other pathovars of this species, in particular P. syringae pv. phaseolicola, P. syringae pv. coronafaciens, P. syringae pv. lachrymans, P. syringae pv. tabaci, P. syringae pv. tomato, P. corrugata, P. syringae pv. aptata, P. wieringae, and P. syringae pv. syringae. For all P. syringae pv. atrofaciens strains, the dominant product of RAPD amplification with primer OPA-13 is a 700 kb fragment, the presence of which distinguishes the representatives of this pathovar from other pathovars [23]. This method was also useful for differentiating pathovars P. syringae that infect mango, sweet cherry cultivars, and other plants [25,34,35,36].
Therefore, according to the result of morphological, biochemical, and serological studies as well as fatty acids and RAPD-PCR profiles, strains isolated from weeds of wheat crops were identified as P. syringae pv. atrofaciens.
5. Conclusions
For the first time, bacteria that are virulent both for weeds and wheat were isolated from weeds growing in wheat crops. The strains L8, L9, L11, L11a, L13, L15, and L16, isolated from weeds, was identified as a P. syringae through biochemical tests such as the LOPAT test, studying the composition of fatty acids, and determining the serological properties. They were serologically similar to the causative agent of the basal glume rot of wheat P. syringae pv. atrofaciens. Based on the study of their phenotypic properties and RAPD-PCR profiles, they have been identified as P. syringae pv. atrofaciens.
Thus, the causative agent of basal glume rot P. syringae pv. atrofaciens is not a highly specialized plant pathogen. It is capable of infecting various plant species. In wheat crops, the weeds can also be the host plants of P. syringae pv. atrofaciens. The main control measures for cereals diseases caused by phytopathogenic bacteria are crop rotations with nonhost species. Since we have identified a wide range of the hosts of the agent of basal glume rot, the approach to crop rotation should be revised, and alternative control methods should be proposed
Author Contributions
Formal analysis and methodology, L.B., L.P., Y.K., V.P., D.S., M.S. and A.K.; investigation, L.B., L.P. and Y.K.; writing—original draft, L.B., L.P., Y.K., V.P., D.S., M.S. and A.K.; Writing—review and editing, L.B., L.P., V.P. and A.K. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Acknowledgments
The authors would like to appreciate the Collective Use Center of the Zabolotny Institute of Microbiology and Virology NASU for technical support during the analysis of fatty acids and Iuliia Bogdan for the assistance of data analysis with RStudio.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tripathi, D. Bacterial pathogens in plants. J. Bacteriol. Mycol. 2017, 4, 38–39. [Google Scholar] [CrossRef][Green Version]
- Sundin, G.W.; Castiblanco, L.F.; Yuan, X.; Zeng, Q.; Yang, C.H. Bacterial disease management: Challenges, experience, innovation and future prospects: Challenges in Bacterial Molecular Plant Pathology. Mol. Plant Pathol. 2016, 17, 1506–1518. [Google Scholar] [CrossRef] [PubMed]
- Patyka, V.P. Phytopathogenic bacteria in contemporary agriculture. Microbiolohichnyi Zhurnal 2016, 78, 71–83. [Google Scholar] [CrossRef]
- Kolomiiets, Y.; Grygoryuk, I.; Likhanov, A.; Butsenko, L.; Blume, Y. Induction of Bacterial Canker Resistance in Tomato Plants Using Plant Growth Promoting Rhizobacteria. Open Agric. J. 2019, 13, 215–222. [Google Scholar] [CrossRef]
- Patyka, V.; Pasichnyk, L.; Butsenko, L.; Petrychenko, V.; Zubachev, S.; Dankevych, L.; Gnatiuk, Y.; Huliaiva, H.; Tokovenko, I.; Kalinichenko, A.; et al. Express Diagnostics of Phytopathogenic Bacteria and Phytoplasmas in Agrophytocenosis; Suszanowich, D., Patyka, V., Wyd-wo, I., Eds.; Drukarnia Swietego Krzyza: Opole, Poland, 2019; ISBN 978-83-7342-684-9. [Google Scholar]
- Maraite, H.; Bragard, C.; Duveiller, E. The status of resistance to bacterial diseases of wheat. In Wheat Production in Stressed Environments; Buck, H.T., Nisi, J.E., Salomon, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 37–49. [Google Scholar] [CrossRef]
- Valencia-Botín, A.J.; Cisneros-López, M.E. A Review of the Studies and Interactions of Pseudomonas syringae Pathovars on Wheat. Intern. J. Agron. 2012, 2012, 692350. [Google Scholar] [CrossRef]
- Matveeva, Y.V.; Pekhtereva, E.S.; Polityko, V.A.; Ignatov, A.N.; Nikolaeva, E.V.; Schaad, N.W. Distribution and virulence of Pseudomonas syringae pv. atrofaciens, causal agent of basal glume rot, in Russia. In Pseudomonas Syringae and Related Pathogens; Iacobellis, N.S., Collmer, A., Hutcheson, S.W., Mansfield, J.W., Morris, C.E., Schaad, N.W., Stead, D.E., Surico, G., Ullrich, M.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; pp. 97–105. [Google Scholar] [CrossRef]
- Alexandrova, M.; Zaccardelli, M.; Stefani, E.; Bazzi, C. Testing for Pseudomonas syringae pv. atrofaciens and Xanthomonas campestris pathovars on cereals in Italy. EPPO Bull. 1995, 25, 437–448. [Google Scholar] [CrossRef]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef]
- Kazempour, M.N.; Kheyrgoo, M.; Pedramfar, H.; Rahimian, H. Isolation and identification of bacterial glum blotch and leaf blight on wheat (Triticum aestivum L.) in Iran. Afr. J. Biotechnol. 2010, 9, 2866–2871. Available online: https://www.ajol.info/index.php/ajb/article/view/79941 (accessed on 3 November 2020).
- Sultanov, R.I.; Arapidi, G.P.; Vinogradova, S.V.; Govorun, V.M.; Luster, D.G.; Ignatov, A.N. Comprehensive analysis of draft genomes of two closely related pseudomonas syringae phylogroup 2b strains infecting mono- and dicotyledon host plants. BMC Genom. 2016, 17, 1010. [Google Scholar] [CrossRef]
- Donati, I.; Cellini, A.; Sangiorgio, D.; Vanneste, J.L.; Scortichini, M.; Balesta, G.M.; Spinelli, F. Pseudomonas syringae pv. actinidiae: Ecology, Infection Dynamics and Disease Epidemiology. Microb. Ecol. 2020, 80, 81–102. [Google Scholar] [CrossRef]
- Taghavi, S.M.; Keshavarz, K. Identification of the causal agent of bacterial wheat blight in Fars and Kohgiluyeh Boyrahmad provinces and the reaction of certain wheat cultivars to them. J. Sci. Technol. Agric. Nat. Res. 2003, 6, 171–180. Available online: https://jstnar.iut.ac.ir/browse.php?a_code=A-10-2-146&slc_lang=en&sid=1 (accessed on 3 November 2020).
- Tarkowski, P.; Vereecke, D. Threats and opportunities of plant pathogenic bacteria. Biotechnol. Adv. 2014, 32, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Marques, A.S.D.A.; Samson, R. Population dynamics of Pseudomonas savastanoi pv. phaseolicola in bean, throughout the epiphytic and pathogenic phases. Pesqui. AgropecuáRia Bras. 2016, 51, 623–630. [Google Scholar] [CrossRef]
- Dobrovol’skaya, T.G.; Khusnetdinova, K.A.; Manucharova, N.A.; Golovchenko, A.V. Structure of epiphytic bacterial communities of weeds. Microbiology 2017, 86, 257–263. [Google Scholar] [CrossRef]
- Klement, Z.; Rudolph, K.; Sands, D. Methods in Phytobacteriology; Academiai Kiado: Budapest, Hungary, 1990. [Google Scholar] [CrossRef]
- Pasichnyk, L.A.; Butsenko, L.M. Serological features of bacteria Pseudomonas syringae agroecosystems of cereal. Microbiolohichnyi Zhurnal 2018, 80, 41–54. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. 2018. Available online: https://www.R-project.org/ (accessed on 3 November 2020).
- Sazakli, E.; Leotsinidis, M.; Vantarakis, A.; Papapetropoulou, M. Comparative typing of Pseudomonas species isolated from the aquatic environment in Greece by SDS-PAGE and RAPD analysis. J. Appl. Microbiol. 2005, 99, 1191–1203. [Google Scholar] [CrossRef]
- Butsenko, L.M.; Savenko, O.A.; Pasichnyk, L.A.; Shcherbyna, T.M.; Patyka, V.P. Fatty Acid Composition of Cellular Lipids Pseudomonas syringae, Isolated from Cereal Agrophytocenosis. Microbiolohichnyi Zhurnal 2017, 79, 56–64. [Google Scholar] [CrossRef]
- Butsenko, L.M.; Pasichnyk, L.A. Genetic Heterogenicity of Pseudomonas syringae pv. atrofaciens Strains Based on RAPD-PCR Analyze. Microbiolohichnyi Zhurnal 2018, 80, 48–62. [Google Scholar] [CrossRef]
- Young, J.M. Taxonomy of Pseudomonas syringae. J. Plant Pathol. 2010, 92, S1.5–S1.14. [Google Scholar] [CrossRef]
- Gutiérrez-Barranquero, J.A.; Carrión, V.J.; Murillo, J.; Arrebola, E.; Arnold, D.L.; Cazorla, F.M.; De Vicente, A. A Pseudomonas syringae diversity survey reveals a differentiated phylotype of the pathovar syringae associated with the mango host and mangotoxin production. Phytopathology 2013, 103, 1115–1129. [Google Scholar] [CrossRef]
- Barta, T.M.; Willis, D.K. Biological and Molecular Evidence that Pseudomonas syringae pathovars coronafaciens, striafaciens and garcae are likely the same pathovar. J. Phytopathol. 2005, 153, 492–499. [Google Scholar] [CrossRef]
- Lindström, K.; Martínez-Romero, M.E. International Committee on Systematics of Prokaryotes; Subcommittee on the taxonomy of Agrobacterium and Rhizobium. Minutes of the meeting, 26 July 2004, Toulouse, France. Int. J. Syst. Evol. Microbiol. 2005, 55, 1383. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.-H.; Velázquez, E. The current status on the taxonomy of Pseudomonas revisited: An update. Infect. Genet. Evol. 2018, 57, 106–116. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, H.E.; Thakur, S.; Guttman, D.S. Evolution of plant pathogenesis in Pseudomonas syringae: A genomics perspective. Annu. Rev. Phytopathol. 2011, 49, 269–289. [Google Scholar] [CrossRef] [PubMed]
- Sawada, H.; Takeuchi, T.; Matsuda, I. Comparative analysis of Pseudomonas syringae pv. actinidiae and pv. phaseolicola based on phaseolotoxin-resistant ornithine carbamoyltransferase gene (argK) and 16S-23S rRNA intergenic spacer sequences. Appl. Environ. Microbiol. 1997, 63, 282–288. [Google Scholar] [CrossRef]
- Maraite, H.; Weyns, J. Pseudomonas syringae pv. aptata and pv. atrofaciens, Specific Pathovars or Members of pv. syringae? In Pseudomonas Syringae Pathovars and Related Pathogens; Rudolph, K., Burr, T.J., Mansfield, J.W., Stead, D., Vivian, A., von Kietzell, J., Eds.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 515–520. [Google Scholar] [CrossRef]
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328. [Google Scholar] [CrossRef]
- Stead, D.E.; Hennessy, J.; Elphinstone, J.G.; Wilson, J.K. Modern methods for classification of plant pathogenic bacteria including Pseudomonas syringae. Dev. Plant Pathol. 1997, 9, 427–434. [Google Scholar] [CrossRef]
- Iličić, R.; Balaž, J.; Stojšin, V.; Jošić, D. Characterization of Pseudomonas syringae pathovars from different sweet cherry cultivars by RAPD analysis. Genetika 2016, 48, 285–295. [Google Scholar] [CrossRef]
- Butsenko, L.; Pasichnyk, L.; Kolomiiets, Y.; Kalinichenko, T. The Effect of Pesticides on the Tomato Bacterial Speck Disease Pathogen Pseudomonas Syringae pv. Tomato. Appl. Sci. 2020, 10, 3263. [Google Scholar] [CrossRef]
- Kolomiiets, Y.; Grygoryluk, I.; Butsenko, L.; Kalinichenko, A. Biotechnological control methods against phytopathogenic bacteria in tomatoes. Appl. Ecol. Environ. Res. 2019, 17, 3215–3230. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).