Plant-Growth-Promoting Bacteria Mitigating Soil Salinity Stress in Plants
Abstract
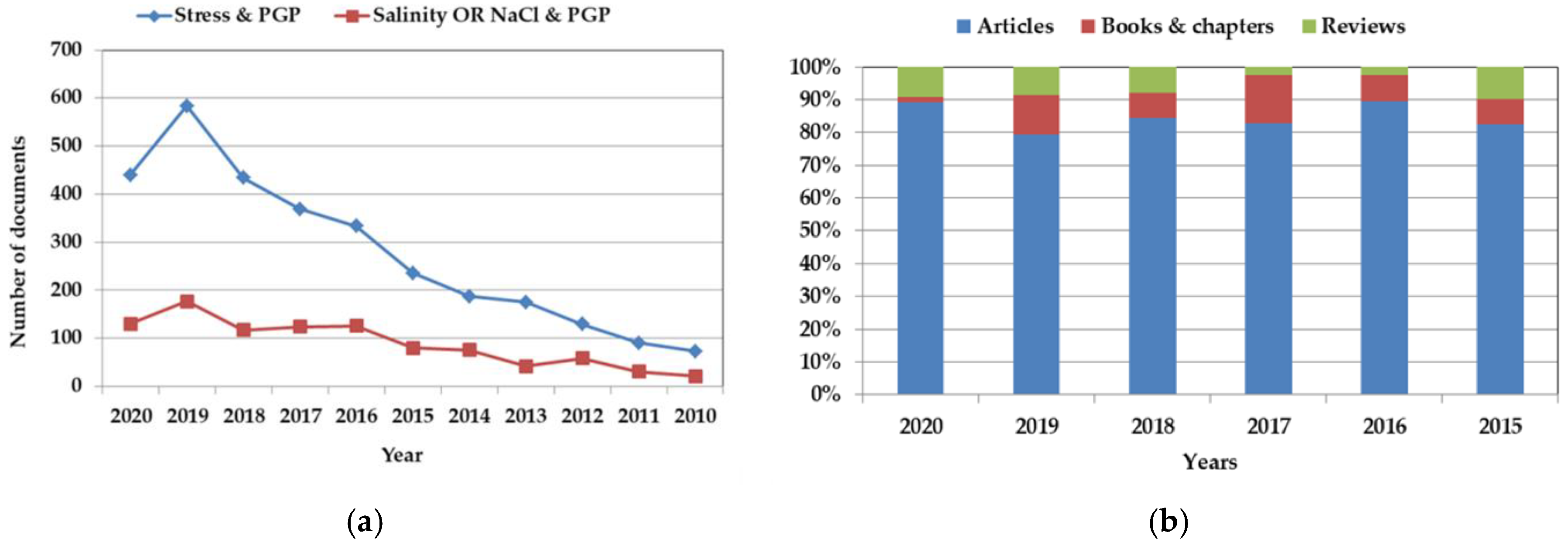
1. Introduction
2. Soil Salinity-Induced Harmful Effects on Plants
3. Plant Response to Salinity Stress
4. Bacteria and Salinity Stress
5. Role of PGPB in Alleviating Salinity Stress to Plants
5.1. Phytohormone Signals
5.1.1. Auxins
5.1.2. Cytokinins, Gibberellic Acid, and Abscisic Acid
5.1.3. Modulation of Plant Ethylene Levels
5.2. PGPB Support Nutrient Acquisition in Plants
5.2.1. Nitrogen Fixation
5.2.2. Phosphate Solubilization
5.2.3. Iron Mobilization
5.3. Contribution to Osmolyte Accumulation in Plants
5.4. Plant Ion Homeostasis
5.5. Extracellular Polymeric Substances (EPSs)
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agricultural Organization. Available online: http://www.fao.org/global-soil-partnership/resources/highlights/detail/en/c/1208623/ (accessed on 2 August 2020).
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Ayers, R.S.; Westcot, D.W. Water Quality for Agriculture; FAO of UN: Rome, Italy, 1985. [Google Scholar]
- Shilev, S.; Sancho, E.D.; Benlloch-Gonzalez, M. Rhizospheric bacteria alleviate salt-produced stress in sunflower. J. Environ. Manag. 2012, 95, S37–S41. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Glick, B.R. Halotolerant plant growth–promoting bacteria: Prospects for alleviating salinity stress in plants. Environ. Exp. Bot. 2020, 178, 104124. [Google Scholar] [CrossRef]
- Guttman, D.; McHardy, A.C.; Schulze-Lefert, P. Microbial genome-enabled insights into plantmicroorganism interactions. Nat. Rev. Genet. 2014, 15, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R.; Liu, C.; Ghosh, S.; Dumbrof, E.B. Early development of canola seedlings in the presence of the plant growth-promoting rhizobacterium Pseudomonas putida GR12-2. Soil Biol. Biochem. 1997, 29, 1233–1239. [Google Scholar] [CrossRef]
- Kang, H.L.; Rae, H.K.; Song, H.G. Enhancement of growth and yield of tomato by Rhodopseudomonas sp. under greenhouse conditions. J. Microbiol. 2008, 46, 641–646. [Google Scholar]
- Etesami, H.; Noori, F. Soil salinity as a challenge for sustainable agriculture and bacterial-mediated alleviation of salinity stress in crop plants. In Saline Soil-Based Agriculture by Halotolerant Microorganisms; Springer: Singapore, 2019; pp. 1–22. [Google Scholar]
- Reich, M.; Aghajanzadeh, T.; Helm, J.; Parmar, S.; Hawkesford, M.J.; De Kok, L.J. Chloride and sulfate salinity differently affect biomass, mineral nutrient composition and expression of sulfate transport and assimilation genes in Brassica rapa. Plant Soil 2017, 411, 319–332. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 2018, 9, 148. [Google Scholar] [CrossRef]
- Lamattina, L.; García-Mata, C.; Graziano, M.; Pagnussat, G. Nitric oxide: The versatility of an extensive signal molecule. Annu. Rev. Plant Biol. 2003, 54, 109–136. [Google Scholar] [CrossRef]
- Nakashima, K.; Tran, L.S.P.; Van Nguyen, D.; Fujita, M.; Maruyama, K.; Todaka, D.; Ito, Y.; Hayashi, N.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 2007, 51, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems BiologyPerspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef]
- Klein, W.; Weber, M.H.W.; Marahiel, M.A. Cold shock response of Bacillus subtilis: Isoleucine-dependent switch in the fatty acid branching pattern for membrane adaptation to low temperatures. J. Bacteriol. 1999, 181, 5341–5349. [Google Scholar] [CrossRef]
- Hidri, R.; Metoui-Ben Mahmoud, O.; Debez, A.; Abdelly, C.; Barea, J.-M.; Azcon, R. Modulation of C:N:P stoichiometry is involved in the effectiveness of a PGPR and AM fungus in increasing salt stress tolerance of Sulla carnosa Tunisian provenances. Appl. Soil Ecol. 2019, 143, 161–172. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-producing plant growth-promoting rhizobacteria under salinity condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Richards, L.A. (Ed.) Diagnosis and improvements of saline and alkali soils. In Agriculture Handbook; USDA: Washintong, DC, USA, 1954; Volume 60, p. 160. [Google Scholar]
- Rengasamy, P. World salinization with emphasis on Australia. J. Exp. Bot. 2006, 57, 1017–1023. [Google Scholar] [CrossRef]
- Rengasamy, P. Transient salinity and subsoil constraints to dryland farming in Australian sodic soils: An overview. Aust. J. Exp. Agric. 2002, 42, 351–361. [Google Scholar] [CrossRef]
- Abrol, I.P.; Yadav, J.S.P.; Massoud, F.I. Salt Affected Soils and Their Management; FAO Soils. Bulletin 39; Food and Agriculture Organization of the United Nations: Rome, Italy, 1988.
- Shannon, M.C.; Grieve, C.M. Tolerance of vegetable crops to salinity. Sci. Hortic. 1998, 78, 5–38. [Google Scholar] [CrossRef]
- Ashraf, M.; McNeilly, T. Salinity tolerance in Brassica oilseeds. Crit. Rev. Plant Sci. 2004, 23, 157–174. [Google Scholar] [CrossRef]
- Meng, R.; Saade, S.; Kurtek, S.; Berger, B.; Brien, C.; Pillen, K.; Tester, M.; Sun, Y. Growth curve registration for evaluating salinity tolerance in barley. Plant Methods 2017, 13, 18. [Google Scholar] [CrossRef]
- Joshi, R.; Mangu, V.R.; Bedre, R.; Sanchez, L.; Pilcher, W.; Zandkarimi, H.; Baisakh, N. Salt adaptation mechanisms of halophytes: Improvement of salt tolerance in crop plants. In Elucidation of Abiotic Stress Signaling in Plants; Springer: New York, NY, USA, 2015; pp. 243–279. [Google Scholar]
- Al-Mutawa, M.M. Effect of salinity on germination and seedling growth of chick pea (Cicer arietinum L.) genotypes. Int. J. Agro. Biol. 2003, 5, 227–229. [Google Scholar]
- Egamberdieva, D. Alleviation of salt stress by plant growth regulators and IAA producing bacteria in wheat. Acta Phys Plant. 2009, 31, 861–864. [Google Scholar] [CrossRef]
- Essa, T.A. Effect of salinity stress on growth and nutrient composition of three soybean (Glycine max L. Merrill) cultivars. J. Agron. Crop Sci. 2002, 188, 86–93. [Google Scholar] [CrossRef]
- Hasegawa, P.M.; Bressan, R.A.; Zhu, J.-K.; Bohnert, H.J. Plant cellular and molecular responses to high salinity. Annu. Rev. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef]
- Prakash, L.; Parthapasenan, G. Interactive effect of NaCl salinity and gibberellic acid on shoot growth, content of abscisic acid and gibberellin like substances and yield of rice (Oruza sativa). Plant Sci. 1990, 100, 173–181. [Google Scholar]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Ali, S.; Ali, B.; Farooq, M.A.; Gill, R.A. Priming-induced antioxidative responses in two wheat cultivars under saline stress. Acta Physiol. Plantarum. 2015, 37, 153. [Google Scholar] [CrossRef]
- Munns, R. Salinity, Growth and Phytohormones; Springer: Berlin, Germany, 2002. [Google Scholar]
- Flowers, T.J.; Colmer, T.D. Salinity tolerance in halophytes. New Phytol. 2008, 179, 945–963. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.; Bhowmik, P.C.; Hossain, M.; Rahman, M.M.; Prasad, M.N.V.; Ozturk, M.; Fujita, M. Potential use of halophytes to remediate saline soils. Biomed. Res. Int. 2014, 2014, 589341. [Google Scholar] [CrossRef]
- Etesami, H.; Maheshwari, D.K. Use of plant growth promoting rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotox. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef]
- Kumar, M.; Etesami, H.; Kumar, V. Saline Soil-Based Agriculture by Halotolerant Microorganisms; Springer Nature: Singapore, 2019. [Google Scholar]
- Ferreira, M.J.; Silva, H.; Cunha, A. Siderophore-producing rhizobacteria as a promising tool for empowering plants to cope with iron limitation in saline soils: A review. Pedosphere 2019, 29, 409–420. [Google Scholar] [CrossRef]
- Sanchez, D.H.; Siahpoosh, M.R.; Roessner, U.; Udvardi, M.; Kopka, J. Plant metabolomics reveals conserved and divergent metabolic responses to salinity. Physiol. Plant. 2008, 132, 209–219. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Kakehi, J.-I. Polyamines: Ubiquitous polycations with unique roles in growth and stress responses. Ann. Bot. 2009, 105, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, salinity and drought stresses: An overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef] [PubMed]
- Shilev, S. Soil rhizobacteria regulating the uptake of nutrients and undesirable elements by plants. In Plant Microbe Symbiosis—Fundamentals and Advance; Arora, N.K., Ed.; Springer: New Delhi, India, 2013; pp. 147–167. [Google Scholar]
- Wang, X.; Chang, L.; Wang, B.; Wang, D.; Li, P.; Wang, L.; Yi, X.; Huang, Q.; Peng, M.; Guo, A. Comparative proteomics of Thellungiella halophila leaves from plants subjected to salinity reveals the importance of chloroplastic starch and soluble sugars in halophyte salt tolerance. Mol. Cell. Proteom. 2013, 12, 2174–2195. [Google Scholar] [CrossRef]
- Sairam, R.K.; Tyagi, A. Physiology and molecular biology of salinity stress tolerance in plants. Curr. Sci. 2004, 86, 407–421. [Google Scholar]
- Stepien, P.; Johnson, G.N. Contrasting responses of photosynthesis to salt stress in the glycophyte Arabidopsis and the halophyte Thellungiella: Role of the plastid terminal oxidase as an alternative electron sink. Plant Physiol. 2009, 149, 1154–1165. [Google Scholar] [CrossRef]
- Kushner, D.; Kamekura, M. Physiology of halophilic eubacteria. In Halophilic Bacteria; Rodriguez-Valera, F., Ed.; CRC Press: Boca Raton, FL, USA, 1988; Volume 1, pp. 109–138. [Google Scholar]
- Bowers, K.J.; Mesbah, N.M.; Wiegel, J. Biodiversity of poly-extremophilic bacteria: Does combining the extremes of high salt, alkaline pH and elevated temperature approach a physico-chemical boundary for life? Saline Syst. 2009, 5, 9. [Google Scholar] [CrossRef]
- Wang, S.; Sun, L.; Ling, N.; Zhu, C.; Chi, F.; Li, W.; Hao, X.; Zhang, W.; Bian, J.; Chen, L.; et al. Exploring Soil Factors Determining Composition and Structure of the Bacterial Communities in Saline-Alkali Soils of Songnen Plain. Front. Microbiol. 2020, 10, 2902. [Google Scholar] [CrossRef]
- Nelson, D.R.; Mele, P.M. Subtle changes in the rhizosphere microbial community structure in response to increased boron and sodium chloride concentrations. Soil Biol. Biochem. 2007, 39, 340–351. [Google Scholar] [CrossRef]
- Ruppel, S.; Franken, P.; Witzel, K. Properties of the halophyte microbiome and their implications for plant salt tolerance. Funct. Plant Biol. 2013, 40, 940–951. [Google Scholar] [CrossRef]
- Oren, A. The order Halobacteriales. In The Prokaryotes, 3rd ed.; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: Singapore, 2006; Volume 3, pp. 113–164. [Google Scholar]
- Whatmore, A.M.; Chudek, J.A.; Reed, R.H. The effects of osmotic upshock on the intracellular solute pools of Bacillus subtilis. Microbiology 1990, 136, 2527–2535. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Khan, A.L.; Bilal, S.; Waqas, M.; Kang, S.-M.; Lee, I.-J. Inoculation of abscisic acid-producing endophytic bacteria enhances salinity stress tolerance in Oryza sativa. Environ. Exp. Bot. 2017, 136, 68–77. [Google Scholar] [CrossRef]
- Shilev, S.; Azaizeh, H.; Vassilev, N.; Georgiev, D.; Babrikova, I. Interactions in soil-microbe-plant system: Adaptation to stressed agriculture. In Microbial Interventions in Agriculture and Environment; Singh, D.P., Gupta, V.K., Prabha, R., Eds.; Springer: Singapore, 2019; Volume 1, pp. 131–171. [Google Scholar]
- Kidd, P.; Barceló, J.; Bernal, M.P.; Navari-Izzo, F.; Poschenrieder, C.; Shilev, S.; Clemente, R.; Monteroso, C. Trace element behavior at the root-soil interface: Implications in phytoremediation. J. Environ. Exp. Bot. 2009, 67, 243–259. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, V.K.; Tripathi, V.; Singh, P.P.; Singh, A.K. Plant Growth-promoting Rhizobacteria (PGPR): Perspective in Agriculture under Biotic and Abiotic Stress. In New and Future Developments in Microbial Biotechnology and Bioengineering; Prasad, R., Gill, S.S., Tuteja, N., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 333–342. [Google Scholar]
- Lugtenberg, B.J.; Chin, A.W.T.F.; Bloemberg, G.V. Microbe-plant interactions: Principles and mechanisms. Antonie Van Leeuwenhoek 2002, 81, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Tyagi, S.P.; Kaushik, R.; Saxena, A.K. Culturable diversity and functional annotation of psychrotrophic bacteria from cold desert of Leh Ladakh (India). World J. Microbiol. Biotechnol. 2015, 31, 95–108. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Wirth, S.J.; Alqarawi, A.A.; Abd_Allah, E.F.; Hashem, A. Phytohormones and beneficial microbes: Essential components for plants to balance stress and fitness. Front. Microbiol. 2017, 8, 2104. [Google Scholar] [CrossRef]
- Pilet, P.E.; Saugy, M. Effect of root growth on endogenous and applied IAA and ABA. Plant Physiol. 1987, 83, 33–38. [Google Scholar] [CrossRef]
- Ali, S.; Charles, T.C.; Glick, B.R. Amelioration of high salinity stress damage by plant growth-promoting bacterial endophytes that contain ACC deaminase. Plant Physiol. Biochem. 2014, 80, 160–167. [Google Scholar] [CrossRef]
- Shilev, S.; Naydenov, M.; Sancho Prieto, M.; Sancho, E.D.; Vassilev, N. PGPR as inoculants in management of lands contaminated with trace elements. In Bacteria in Agrobiology: Stress Management; Maheshwari, D.K., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 259–277. [Google Scholar]
- Albacete, A.; Ghanem, M.E.; Martínez-Andújar, C.; Acosta, M.; Sánchez-Bravo, J.; Martínez, V.; Lutts, S.; Dodd, I.C.; Pérez-Alfocea, F. Hormonal changes in relation to biomass partitioning and shoot growth impairment in salinized tomato (Solanum lycopersicum L.) plants. J. Exp. Bot. 2008, 59, 4119–4131. [Google Scholar] [CrossRef]
- Li, X.; Sun, P.; Zhang, Y.; Jin, C.; Guan, C. A novel PGPR strain Kocuria rhizophila Y1 enhances salt stress tolerance in maize by regulating phytohormone levels, nutrient acquisition, redox potential, ion homeostasis, photosynthetic capacity and stress-responsive genes expression. Environ. Exp. Bot. 2020, 174, 104023. [Google Scholar] [CrossRef]
- Cardinale, M.; Ratering, S.; Suarez, C.; Zapata Montoya, A.M.; Geissler-Plaum, R.; Schnell, S. Paradox of plant growth promotion potential of rhizobacteria and their actual promotion effect on growth of barley (Hordeum vulgare L.) under salt stress. Microbiol. Res. 2015, 181, 22–32. [Google Scholar] [CrossRef] [PubMed]
- Shilev, S.; Babrikova, I.; Babrikov, T. Consortium of plant growth-promoting bacteria improves spinach (Spinacea oleracea L.) growth under heavy metal stress conditions. J. Chem. Technol. Biotechnol. 2020, 95, 932–939. [Google Scholar] [CrossRef]
- Paulucci, N.S.; Gallarato, L.A.; Reguera, Y.B.; Vicario, J.C.; Cesari, A.B.; García de Lema, M.B.; Dardanelli, M.S. Arachis hypogaea PGPR isolated from Argentine soil modifies its lipidscomponents in response to temperature and salinity. Microbiol. Res. 2015, 173, 1–9. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alamri, S.A.; Ali, H.M.; Alayafi, A.A. Bacillus firmus (SW5) augments salt tolerance in soybean (Glycine max L.) by modulating root system architecture, antioxidant defense systems and stress responsive genes expression. Plant Physiol. Biochem. 2018, 132, 375–384. [Google Scholar] [CrossRef] [PubMed]
- Tanimoto, E. Regulation of root growth by plant hormones-roles for auxin and gibberellin. Crit. Rev. Plant Sci. 2005, 24, 249–265. [Google Scholar] [CrossRef]
- Pertry, I.; Vereecke, D. Identification of Rhodococcus fascians cytokinins and their modus operandi to reshape the plant. Proc. Natl. Acad. Sci. USA 2009, 106, 929–934. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Plant growth-promoting Bacteria: Mechanisms and applications. Scientifica 2012, 2012, 963401. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef]
- Noshin, I.; Roomina, M.; Humaira, Y.; Wajiha, K.; Sumera, I.; El Enshasy, H.; Dailin, D.J. Rhizobacteria Isolated from Saline Soil Induce Systemic Tolerance in Wheat (Triticum aestivum L.) against Salinity Stress. Agronomy 2020, 10, 989. [Google Scholar]
- Cherif-Silini, H.; Thissera, B.; Bouket, A.C.; Saadaoui, N.; Silini, A.; Eshelli, M.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Yahiaoui, B.; et al. Durum wheat stress tolerance induced by endophyte Pantoea agglomerans with genes contributing to plant functions and secondary metabolite arsenal. Int. J. Mol. Sci. 2019, 20, 3989. [Google Scholar] [CrossRef] [PubMed]
- El-Esawi, M.A.; Alaraidh, I.A.; Alsahli, A.A.; Alzahrani, S.M.; Ali, H.M.; Alayafi, A.A.; Ahmad, M. Serratia liquefaciens KM4 improves salt stress tolerance in maize by regulating redox potential, ion homeostasis, leaf gas exchange and stress-related gene expression. Int. J. Mol. Sci. 2018, 19, 3310. [Google Scholar] [CrossRef] [PubMed]
- Safdarian, M.; Askari, H.; Nematzadeh, G.; Sofo, A. Halophile plant growth-promoting rhizobacteria induce salt tolerance traits in wheat seedlings (Triticum aestivum L.). Pedosphere 2020, 30, 684–693. [Google Scholar] [CrossRef]
- Kang, M.; Radhakrishnan, R.; Khan, A.L.; Kim, M.-J.; Park, J.-M.; Kim, B.-R.; Shin, D.H.; Lee, I.-J. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 2014, 84, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Swain, T. Ethylene in Plant Biology; Academic Press: New York, NY, USA, 1974. [Google Scholar]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef]
- Glick, B.R.; Penrose, D.M.; Li, J. A model for the lowering of plant ethylene concentrations by plant growth promoting bacteria. J. Theor. Biol. 1998, 190, 63–68. [Google Scholar] [CrossRef]
- Glick, B.R.; Todorovic, B.; Czarny, J.; Cheng, Z.; Duan, J.; McConkey, B. Promotion of plant growth by bacterial ACC deaminase. Crit. Rev. Plant Sci. 2007, 26, 227–242. [Google Scholar] [CrossRef]
- Misra, S.; Chauhan, P.S. ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech 2020, 10, 119. [Google Scholar] [CrossRef]
- Nautiyal, C.S.; Srivastava, S.; Chauhan, P.S.; Seem, K.; Mishra, A.; Sopory, S.K. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant Physiol. Biochem. 2013, 66, 1–9. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Khan Shinwari, Z.; Khan, A.L.; Khan, A.; AL-Harrasi, A. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Kausar, R.; Shahzad, S.M. Effect of ACC-deaminase containing rhizobacteria on growth promotion of maize under salinity stress. J. Agric. Soc. Sci. 2006, 2, 216–218. [Google Scholar]
- Nadeem, S.M.; Zahir, Z.A.; Naveed, M.; Arshad, M. Rhizobacteria containing ACC-deaminase confer salt tolerance in maize grown on salt-affected fields. Can. J. Microbiol. 2009, 55, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Wu, Z.; He, Y.; Huang, Y.; Li, X.; Ye, B.-C. Plant growth promotion and alleviation of salinity stress in Capsicum annuum L. by Bacillus isolated from saline soil in Xinjiang. Ecotoxicol. Environ. Saf. 2018, 164, 520–529. [Google Scholar] [CrossRef]
- Sandmann, G. Consequences of iron deficiency on photosynthetic and respiratory electron transport in blue-green algae. Photosynth. Res. 1985, 6, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, D.K.; Verma, J.P.; Prakash, S.; Meena, V.S.; Meena, R.S. Potassium as an important plant nutrient in sustainable agriculture: A state of the art. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Meena, V.S., Maurya, B.R., Verma, J.P., Meena, R.S., Eds.; Springer: New Delhi, India, 2016; pp. 21–29. [Google Scholar]
- Noori, F.; Etesami, H.; Najafi Zarini, H.; Khoshkholgh-Sima, N.A.; Hosseini Salekdeh, G.; Alishahi, F. Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotoxicol. Environ. Saf. 2018, 162, 129–138. [Google Scholar] [CrossRef]
- Vimal, S.R.; Singh, J.S.; Arora, N.K.; Singh, S. Soil-plant-microbe interactions in stressed agriculture management: A review. Pedosphere 2017, 27, 177–192. [Google Scholar] [CrossRef]
- Graham, P.H. Principles and Application of Soil Microbiology; Prentice Hall: Upper Saddle River, NJ, USA, 1988. [Google Scholar]
- Gray, E.J.; Smith, D.L. Intracellular and extracellular PGPR: Commonalities and distinctions in the plant-bacterium signaling processes. Soil Biol. Biochem. 2005, 37, 395–412. [Google Scholar] [CrossRef]
- Yadav, A.N.; Sachan, S.G.; Verma, P.; Saxena, A.K. Bioprospecting of Plant Growth Promoting Psychrotrophic Bacilli from the Cold Desert of North Western Indian Himalayas. Ind. J. Exp. Bot. 2016, 54, 142–150. [Google Scholar]
- Dobbelaere, S.; Vanderleyden, J.; Okon, Y. Plant growth-promoting effects of diazotrophs in the rhizosphere. Crit. Rev. Plant Sci. 2003, 22, 107–149. [Google Scholar] [CrossRef]
- Mukhtar, S.; Zareen, M.; Khaliq, Z.; Mehnaz, S.; Malik, K.A. Phylogenetic analysis of halophyte-associated rhizobacteria and effect of halotolerant and halophilic phosphate-solubilizing biofertilizers on maize growth under salinity stress conditions. J. Appl. Microbiol. 2020, 128, 556–573. [Google Scholar] [CrossRef]
- Zahran, H.H. Rhizobium-legume symbiosis and nitrogen fixation under severe conditions and in an arid climate.Microbiol. Mol. Biol. Rev. 1999, 63, 968–989. [Google Scholar] [CrossRef]
- Tilak, K.V.B.; Ranganayaki, N.; Manoharachari, C. Synergistic effects of plant-growth promoting rhizobacteria and Rhizobium on nodulation and nitrogen fixation by pigeonpea (Cajanus cajan). Eur. J. Soil. Sci. 2006, 57, 67–71. [Google Scholar] [CrossRef]
- Egamberdieva, D.; Berg, G.; Lindström, K.; Räsänen, L.A. Alleviation of salt stress of symbiotic Galega officinalis L. (Goat’s Rue) by co-inoculation of rhizobium with root colonising Pseudomonas. Plant Soil. 2013, 369, 453–465. [Google Scholar] [CrossRef]
- Figueiredo, M.V.; Burity, H.A.; Martınez, C.R.; Chanway, C. Alleviation of drought stress in the common bean Phaseolus vulgaris L.) by co-inoculation with Paenibacillus polymyxa and Rhizobium tropici. Appl. Soil Ecol. 2008, 4, 182–188. [Google Scholar] [CrossRef]
- Khalid, R.; Zhang, X.X.; Hayat, R.; Ahmed, M. Molecular characteristics of rhizobia isolated from Arachis hypogaea grown under stress environment. Sustainability 2020, 12, 6259. [Google Scholar] [CrossRef]
- Oldroyd, G.E. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 2013, 11, 252–263. [Google Scholar] [CrossRef]
- Miransari, M.; Smith, D.L. Alleviating salt stress on soybean (Glycine max (L.) Merr.)—Bradyrhizobium japonicum symbiosis, using signal molecule genistein. Eur. J. Soil Biol. 2009, 45, 146–152. [Google Scholar] [CrossRef]
- Rodríguez, H.; Fraga, R.; Gonzalez, T.; Bashan, T. Genetics of phosphate solubilization and its potential applications for improving plant growth-promoting bacteria. Plant Soil 2006, 287, 15–21. [Google Scholar] [CrossRef]
- Anand, K.; Kumari, B.; Mallick, M.A. Phosphate solubilizing microbes: An effective and alternative approach as bio-fertilizers. Int. J. Pharm. Sci. 2016, 8, 37–40. [Google Scholar]
- Goldstein, A.H. Involvement of the quinoprotein glucose dehydrogenase in the solubilization of exogenous mineral phosphates by Gram negative bacteria. In Phosphate in Microorganisms: Cellular and Molecular Biology; Torriani-Gorni, A., Yagil, E., Silver, S., Eds.; ASM Press: Washington, DC, USA, 1994; pp. 197–203. [Google Scholar]
- Fernández, L.A.; Zalba, P.; Gómez, M.A.; Sagardoy, M.A. Phosphate-solubilization activity of bacterial strains in soil and their effect on soybean growth under greenhouse conditions. Biol. Fertil. Soils 2007, 43, 805–809. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Wani, P.A. Role of phosphate-solubilizing microorganisms in sustainable agriculture—A review. Agron. Sustain. Dev. 2007, 27, 29–43. [Google Scholar] [CrossRef]
- Babalola, O.O.; Glick, B.R. The use of microbial inoculants in African agriculture: Current practice and future prospects. J. Food Agric. Environ. 2012, 10, 540–549. [Google Scholar]
- Etesami, H. Enhanced phosphorus fertilizer use efficiency with microorganisms. In Nutrient Dynamics for Sustainable Crop Production; Springer: Berlin/Heidelberg, Germany, 2020; pp. 215–245. [Google Scholar]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Tapias, D.; Moreno-Galván, A.; Pardo-Díaz, S.; Obando, M.; Rivera, D.; Bonilla, R. Effect of inoculation with plant growth-promoting bacteria (PGPB) on amelioration of saline stress in maize (Zea mays). Appl. Soil Ecol. 2012, 61, 264–272. [Google Scholar] [CrossRef]
- Hu, X.Y.; Page, M.T.; Sumida, A.; Tanaka, A.; Terry, M.J.; Tanaka, R. The iron-sulfur cluster biosynthesis protein sufb is required for chlorophyll synthesis, but not phytochrome signaling. Plant J. 2017, 89, 1184–1194. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nishizawa, N.K. Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 2012, 63, 131–152. [Google Scholar] [CrossRef]
- Puig, S.; Ramos-Alonso, L.; Romero, A.M.; Martínez-Pastor, M.T. The elemental role of iron in DNA synthesis and repair. Metallomics 2017, 9, 1483–1500. [Google Scholar] [CrossRef]
- Falkowski, P.G.; Lin, H.Z.; Gorbunov, M.Y. What limits photosynthetic energy conversion efficiency in nature? Lessons from the oceans. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160376. [Google Scholar] [CrossRef]
- Abbas, G.; Saqib, M.; Akhtar, J. Interactive effects of salinity and iron deficiency on different rice genotypes. J. Plant Nutr. Soil Sci. 2015, 178, 306–311. [Google Scholar] [CrossRef]
- Gargallo-Garriga, A.; Preece, C.; Sardans, J.; Oravec, M.; Urban, O.; Peñuelas, J. Root exudate metabolomes change under drought and show limited capacity for recovery. Sci. Rep. 2018, 8, 12696. [Google Scholar] [CrossRef]
- Raymond, K.M.; Denz, E. Biochemical and physical properties of siderophores. In Iron Transport in Bacteria; Crosa, J.H., Mey, A.R., Payne, S.M., Eds.; ASM Press: Washington, DC, USA, 2004; pp. 3–17. [Google Scholar]
- Kloepper, J.W.; Leong, J.; Teintze, M.; Schroth, M.N. Enhanced plant growth by siderophores produced by plant growth-promoting rhizobacteria. Nature 1980, 286, 885–886. [Google Scholar] [CrossRef]
- Masalha, J.; Kosegarten, H.; Elmaci, Ö.; Mengel, K. The central role of microbial activity for iron acquisition in maize and sunflower. Biol. Fertil. Soils 2000, 30, 433–439. [Google Scholar] [CrossRef]
- Trapet, P.; Avoscan, L.; Klinguer, A.; Pateyron, S.; Citerne, S.; Chervin, C.; Mazurier, S.; Lemanceau, P.; Wendehenne, D.; Besson-Bard, A. The Pseudomonas fluorescens siderophore pyoverdine weakens Arabidopsis thaliana defense in favor of growth in iron-deficient conditions. Plant Physiol. 2016, 171, 675–693. [Google Scholar] [CrossRef] [PubMed]
- Pii, Y.; Marastoni, L.; Springeth, C.; Fontanella, M.C.; Beone, G.M.; Cesco, S.; Mimmo, T. Modulation of Fe acquisition process by Azospirillum brasilense in cucumber plants. Environ. Exp. Bot. 2016, 130, 216–225. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, P.; Khan, A. Optimization of PGPR and silicon fertilization using response surface methodology for enhanced growth, yield and biochemical parameters of French bean (Phaseolus vulgaris L.) under saline stress. Biocatal. Agric. Biotechnol. 2020, 23, 101463. [Google Scholar] [CrossRef]
- Vaishnav, A.; Singh, J.; Singh, P.; Rajput, R.S.; Singh, H.B.; Sarma, B.K. Sphingobacterium sp. BHU-AV3 Induces Salt Tolerance in Tomato by Enhancing Antioxidant Activities and Energy Metabolism. Front. Microbiol. 2020, 11, 443. [Google Scholar] [CrossRef]
- Costa-Gutierrez, S.B.; Lami, M.J.; Di Santo, M.C.C.; Zenoff, A.M.; Vincent, P.A.; Molina-Henares, M.A.; Espinosa-Urgel, M.; de Cristóbal, R.E. Plant growth promotion by Pseudomonas putida KT2440 under saline stress: Role of eptA. Appl. Microbiol. Biotechnol. 2020, 104, 4577–4592. [Google Scholar] [CrossRef]
- Shultana, R.; Zuan, K.Z.; Yusop, M.R.; Saud, H.M.; Ayanda, A.F. Effect of salt-tolerant bacterial inoculations on rice seedlings differing in salt-tolerance under saline soil conditions. Agronomy 2020, 10, 1030. [Google Scholar] [CrossRef]
- Tchuisseu Tchakounté, G.V.; Berger, B.; Patz, S.; Becker, M.; Turecková, V.; Novák, O.; Tarkowská, D.; Fankem, H.; Silke, R. The response of maize to inoculation with Arthrobacter sp. and Bacillus sp. in phosphorus-deficient, salinity-affected soil. Microorganisms 2020, 8, 1005. [Google Scholar]
- He, A.-L.; Niu, S.-Q.; Zhao, Q.; Li, Y.-S.; Gou, J.-Y.; Gao, H.-J.; Suo, S.-Z.; Zhang, J.-L. Induced salt tolerance of perennial ryegrass by a novel bacterium strain from the rhizosphere of a desert shrub Haloxylon ammodendron. Int. J. Mol. Sci. 2018, 19, 469. [Google Scholar] [CrossRef]
- Yaghoubi Khanghahi, M.; Strafella, S.; Crecchio, C. Changes in photo-protective energy dissipation of photosystem II in response to beneficial bacteria consortium in durum wheat under drought and salinity stresses. Appl. Sci. 2020, 10, 5031. [Google Scholar] [CrossRef]
- Suprasanna, P.; Nikalje, G.C.; Rai, A.N. Osmolyte accumulation and implications in plant abiotic stress tolerance. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Khan, N.A., Eds.; Springer: New Delhi, India, 2016; pp. 1–12. [Google Scholar]
- Norwood, M.; Truesdale, M.R.; Richter, A.; Scott, P. Photosynthetic carbohydrate metabolism in the resurrection plant Craterostigma plantagineum. J. Exp. Bot. 2000, 51, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Ciulla, R.A.; Diaz, M.R.; Taylor, B.F.; Roberts, M.F. Organic osmolytes in aerobic bacteria from mono lake, an alkaline, moderately hypersaline environment. Appl. Environ. Microbiol. 1997, 63, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Marulanda, A.; Azcon, R.; Chaumont, F.; Ruiz-Lozano, J.M.; Aroca, R. Regulation of plasma membrane aquaporins by inoculation with a Bacillus megaterium strain in maize (Zea mays L.) plants under unstressed and salt-stressed conditions. Planta 2010, 232, 533–543. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J.-K. Salt stress signaling and mechanisms of plant salt tolerance. Genet. Eng. 2006, 27, 141–177. [Google Scholar]
- Gupta, J.; Rathour, R.; Singh, R.; Thakur, I.S. Production and characterization of extracellular polymeric substances (EPS) generated by a carbofuran degrading strain Cupriavidus sp. ISTL7. Bioresour. Technol. 2019, 282, 417–424. [Google Scholar] [CrossRef]
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; et al. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 2020. [Google Scholar] [CrossRef]
- Naseem, H.; Bano, A. Role of plant growth-promoting rhizobacteria and their exopolysaccharide in drought tolerance of maize. J. Plant Interact. 2014, 9, 689–701. [Google Scholar] [CrossRef]
- Yang, A.; Akhtar, S.S.; Iqbal, S.; Amjad, M.; Naveed, M.; Zahir, Z.A.; Jacobsen, S.-E. Enhancing salt tolerance in quinoa by halotolerant bacterial inoculation. Funct. Plant Biol. 2016, 43, 632–642. [Google Scholar] [CrossRef]
| Plant | Beneficial Effect(s) | Halotolerant PGPR | PGP-Traits | Conditions | Source |
|---|---|---|---|---|---|
| Phaseolus vulgaris | enhanced growth, yield, and biochemical activity | Bacillus subtilis MTCC 441 and Pseudomonas fluorescence MTCC 103T | - | greenhouse, pots, soil, and vermicompost, 150 mM NaCl | [125] |
| Arachis hypogaea | shoot and root growth promotion | Ochrobactrum intermedium | IAA, siderophores | growth chamber, pots, sterilized nitrogen-free vermiculite | [68] |
| Zea mays | increase of growth, seed germination rate, photosynthetic capacity, antioxidant levels, relative water content, chlorophyll | Kocuria rhizophila Y1 (10% NaCl) | P solubilization, IAA | greenhouse, pots, soil:vermiculite:perlite (2:2:1), 100 and 200 mM NaCl | [66] |
| Sulla carnosa | improved growth, nutrition, and salt tolerance | Bacillus subtilis | IAA, urease, alkaline phosphatase, β-glucosidase and dehydrogenase | greenhouse, pots, soil/sand mixture, 200 mM NaCl | [16] |
| Hordeum vulgare | improved growth | Curtobacterium flaccumfaciens E108 | IAA, phosphate mobilization | greenhouse, tonsubstrat ED 73 substrate, 4.4% or 4.8% NaCl | [66] |
| Triticum aestivum | increased biomass | 11 isolates, 8% NaCl | EPS and auxin production, phosphate solubilization | growth chamber, pots, soil, 1% NaCl | [17] |
| Oryza sativa | increased growth and tolerance to salt | Bacillus amyloliquefaciens NBRISN13, 50–2500 mM NaCl | ACC deaminase, root colonization | hydroponics and soil, 100–300 mM NaCl | [84] |
| Glycine max | plant growth and performance | Bacillus firmus SW5, 500 mM NaCl | IAA, siderophores, phosphate mobilization | hydroponics, 80 mM NaCl | [69] |
| Zea mays | improved plant growth and mineral nutrition | Azotobacter chrooccocum C5 & C9, 5.8% NaCl | N fixation, phosphate solubilization, auxin | greenhouse, pots, soil, 0.58% NaCl | [112] |
| Helianthus annuus | plant biomass, improved K+/Na+ ratio | P. fluorescens biotype F, P. fluorescens CECT 378 T, 50 mM NaCl | IAA, siderophores | growth chamber, pots, soil:peat (1:1), 100 mM NaCl | [4] |
| Lycopersicum esculentum | increased salt tolerance in tomato, enhanced plant biomass, shoot and root length, | Sphingobacterium sp. BHU-AV3, 0.85 M NaCl | IAA, siderophores, phosphate solubilization | growth chamber, pots, soil, 200 mM NaCl | [126] |
| Triticum aestivum | induced salinity tolerance, increased plant biomass and relative water content | Bacillus sp., Ps. stutzeri, Az.brasiliense, Az.lipoferum, 2–15% NaCl | IAA, gibberellic acid, cytokinin, abscisic acid | greenhouse, pots, soil, 150 mM NaCl | [74] |
| Glycine max, Zea mays | improved seed germination, root and stem length | Pseudomonas putida KT2440, 0.5 M NaCl | phosphate solubilization, indoles and siderophores | growth chamber, pots, soil, sand, 80 and 100 mM NaCl | [127] |
| Zea mays | improved growth, induced plant response for defense enzymes, chlorophyll, proline, soluble sugars | B. subtilis, B. subtilis, B. safensis, 1 M NaCl | ACC-deaminase, IAA, P-solubilization, EPS | greenhouse, pots, soil, 100 mM NaCl | [83] |
| Lycopersicum esculentum | improved fresh and dry biomass, chlorophyll contents, and a greater number of flowers and buds | P. fluorescens YsS6 and P. migulae 8R6 | IAA, siderophores, phosphate solubilization, ACC-deaminase | greenhouse, soil, pots, 165 and 185 mM NaCl | [62] |
| Glycine max | length, shoot biomass, chlorophyll content | Pseudomonas putida H-2-3 | gibberellins, abscisic and salicylic acids | greenhouse, pots, peat mixture, 120 mM NaCl | [78] |
| Oryza sativa | dry matter, chlorophyll content, K+/Na+ ratio, | B. tequilensis, B. aryabhattai, Providencia stuartii | glasshouse, pots, soil, 4 g.L−1 NaCl (EC = 8 dS.m−1) | [128] | |
| Zea mays | increase K+/Na+ ratio, plant growth | Arthrobacter sp., Bacillus sp. | auxin, abscisic acid, cytokinins, gibberellins | greenhouse, pots, sand:vermiculite, NaCl (EC = 12 dS.m−1) | [129] |
| Triticum durum | Growth promotion, chlorophyll content, K+/Na+ ratio | Pantoea agglomerans, 1 M NaCl | auxin, ACC-deaminase, phosphate solubilization | growth chamber, pot, soil, 100 and 200 mM NaCl | [75] |
| Zea mays | improving growth and salt stress tolerance, regulating ion homeostasis, redox potential, leaf gas exchange, stress-related genes expression | Serratia liquefaciens KM4, 450 mM NaCl | IAA, siderophores, phosphate solubilization | growth chamber, pots, soil, 160 mM NaCl | [76] |
| Lolium perenne | enhanced growth and salt tolerance and K+/Na+ ratio, chlorophyll, root volume and activity, leaf catalase activity, soluble sugar and proline | Pseudomonas sp. M30-35, | phosphorus solubilization, auxin | greenhouse, pots, soil, 300 mM NaCl | [130] |
| Triticum durum | increasing light absorbed by PSII antenna, PQ ratio and total quenching of chlorophyll fluorescence, increased yield of grains | consortium (A. pittii, A. oleivorans, A. calcoaceticus, Comamonas testosterone) | Phosphate, K, and Zn solubilization, N2 fixation | greenhouse, pots, soil, 150 mM NaCl | [131] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shilev, S. Plant-Growth-Promoting Bacteria Mitigating Soil Salinity Stress in Plants. Appl. Sci. 2020, 10, 7326. https://doi.org/10.3390/app10207326
Shilev S. Plant-Growth-Promoting Bacteria Mitigating Soil Salinity Stress in Plants. Applied Sciences. 2020; 10(20):7326. https://doi.org/10.3390/app10207326
Chicago/Turabian StyleShilev, Stefan. 2020. "Plant-Growth-Promoting Bacteria Mitigating Soil Salinity Stress in Plants" Applied Sciences 10, no. 20: 7326. https://doi.org/10.3390/app10207326
APA StyleShilev, S. (2020). Plant-Growth-Promoting Bacteria Mitigating Soil Salinity Stress in Plants. Applied Sciences, 10(20), 7326. https://doi.org/10.3390/app10207326





