Abstract
Various tasks in natural language processing (NLP) suffer from lack of labelled training data, which deep neural networks are hungry for. In this paper, we relied upon features learned to generate relation triples from the open information extraction (OIE) task. First, we studied how transferable these features are from one OIE domain to another, such as from a news domain to a bio-medical domain. Second, we analyzed their transferability to a semantically related NLP task, namely, relation extraction (RE). We thereby contribute to answering the question: can OIE help us achieve adequate NLP performance without labelled data? Our results showed comparable performance when using inductive transfer learning in both experiments by relying on a very small amount of the target data, wherein promising results were achieved. When transferring to the OIE bio-medical domain, we achieved an F-measure of 78.0%, only 1% lower when compared to traditional learning. Additionally, transferring to RE using an inductive approach scored an F-measure of 67.2%, which was 3.8% lower than training and testing on the same task. Hereby, our analysis shows that OIE can act as a reliable source task.
1. Introduction
In deep learning for natural language processing (NLP), the collection of labelled data necessary for training and building models is expensive. This has further highlighted the urgency towards transfer learning research. The aim of transfer learning is to benefit from information gathered from previous training data in directly making predictions in the target task by utilizing the extracted information. Deep learning approaches in NLP did not start until the early 2000s [1]. Recently, there has been an exponential increase in the number of scientific publications in neural networks in various NLP tasks [1].
Open information extraction (OIE) is a challenging task of extracting relation tuples from an unstructured corpus. Its main objective is to generate structured information from unstructured data in the form of a relation triple, <Argument 1> <Relation> <Argument 2>, without the need of predefining the relation between the two arguments. The extracted tuples can be binary, ternary, or n-ary, where the relationship is expressed between more than two entities such as the Person–Location–BornIn–BornOn relation (Jack Adams, Michigan, California, 1975).
Relation extraction (RE)—also classified as a category of information extraction—is the processes of identifying semantic relationships between entities. Contrary to OIE, RE requires predefining the relation prior to extraction. Similar to OIE, the extracted relation can either be a binary relation, for instance, Located-In (Berlin, Germany), or a higher order relation (n-ary), for instance, a 3-ary relation between Employee–Position–Company (Adam Smith, Marketing Manager, XYZ Company). Examples of both OIE and RE triples can be found in Table 1.

Table 1.
Open information extraction and relation extraction example.
OIE is a crucial NLP task, and thus it was chosen as a source task to transfer to other NLP tasks due to its various potential applications in information retrieval, information extraction, text summarization, and question answering [2]. While various OIE algorithms have been developed in the past decade, only a small number employ deep learning techniques.
In recent years, researchers have increasingly been showing interest towards model generalization in deep learning due to the lack of labelled data. In this paper, we investigated the ability to transfer OIE to other NLP tasks, ranging from domain–adaptation (news domain to bio-medical) to RE as a semantically related task. RE task was chosen because of the nature of both OIE and RE, and our choice was backed up by the semantic overlap between both tasks. Throughout our research, we also compared and experimented with the use of different word embeddings.
This work aimed to measure how OIE can assist in other NLP tasks. Our primary objective was to conduct a fair comparison of different methods and settings with respect to OIE transfer learning effects to other NLP tasks. Therefore, we did not focus on outperforming state-of-the-art results in the target tasks.
The remainder of the paper is structured as follows. Section 2 presents a brief overview of transfer learning, while Section 3 surveys previous work in both OIE and RE. The neural network architecture is explained in Section 4, and experimental setup is explained in Section 5. Results and evaluation are discussed in Section 6. Finally, Section 7 concludes the paper and discusses future work.
2. Transfer Learning in NLP
Formerly, there was a misconception that a machine learning framework will achieve the desirable results only if the testing data and training data have similar distribution and feature space. Thus, a new framework was required for data with different distribution properties and features, making the collection of labelled training data expensive and difficult. Transfer learning lessens the demand of gathering an immense amount of labelled training data by reemploying the knowledge gained from a different task to tackle new tasks faster and constructively.
Pan and Yang introduced a transfer learning taxonomy [3]. Additionally, they categorized transfer learning into three classes:
Inductive transfer learning: labelled data are accessible in source and target domain.
Transductive transfer learning: labelled data are only available in the source domain.
Unsupervised transfer learning: No labelled data are both source and target domain.
Transfer learning has been implemented in various different machine learning tasks, achieving notable results, for instance, textual summarization [4], named entity recognition [5], question answering [6,7], and text classification [8].
BERT (Bidirectional Encoder Representations from Transformers) [9] was a breakthrough in transfer learning on a range of language-based tasks, not only due to the fact that BERT was pretrained on an immense dataset, but also because it has a substantial number of transformer blocks (encoder layers) and feed-forward networks. Later on, many transfer learning models built on BERT were introduced, for example ULMFiT [10] and OpenAI transformer [11]. This novel development also affected the way words are encoded, with more elaboration being found in Section 4.2.
As shown in Figure 1, in our work, two transductive transfer learning experiments were carried out. The first one transfers knowledge learned from the OIE news domain to the OIE bio-medical domain—this is referred to as domain adaptation. In contrast to transfer learning, domain adaption entails adapting a model trained on one domain to other different domains on the same task. The default process of supervised domain adaptation for neural models involves pre-training the network on data from the source domain followed by fine-tuning hyperparameters on data from the target domain. The second experiment transfers information from the OIE news domain to the RE news domain. Moreover, a small percentage of OIE bio-medical data were added to OIE news data to experiment with inductive transfer learning. Similarly, a small amount of RE training data were inputted to the neural model along with OIE news corpus, with both experiments being referred to as multi-task learning.
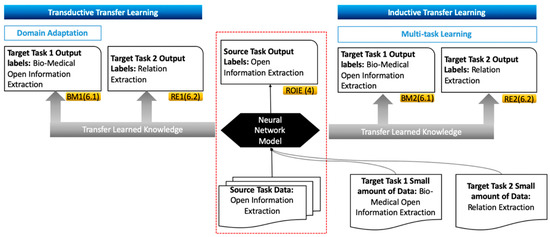
Figure 1.
Open information extraction (OIE) transfer learning assessment. A total of four transfer learning experiments were carried out in our work. Left: the two transductive transfer learning experiments (BM1 and RE1). Right: illustration of the two experiments using inductive approaches (BM2 and RE2). Middle: the red dotted line represents the original model (ROIE), in which we tested our proposed neural model by testing and training on OIE news data, which is discussed in Section 4. The experiments’ ID and the section they are discussed in are encapsulated in the yellow rectangles.
3. Related Work
In this section, we focus on previous works performed on OIE and RE relation extraction in the literature.
3.1. State-of-the-Art Open Information Extraction
OIE can be portrayed in three broad categories [12]: (a) machine learning classifier approaches, (b) hand-crafted rules approaches, and (c) neural network approaches. The first two categories can be further divided into two sub-categories: shallow syntactic analysis and dependency parsing. Below we discuss state-of-the-art work in each of these categories.
3.1.1. Machine Learning Classifiers Approaches
OIE systems that are built on machine learning classifier techniques require automatically generated data to train the classifier. In 2007, Banko et al. introduced the first OIE system based on shallow syntactic analysis, TextRunner [13]. It implements extraction in three main phases. It starts with a self-supervised learner that depends mainly on a conditional random field (CRF) classifier that utilizes unlexicalized features required for relation extraction, followed by a single pass extractor that extracts any potential relation triple and classifies each as either trustworthy or not. Finally, a redundancy-based assessor that re-ranks the extracted relations and assigns a confidence score to each extracted tuple is implemented. Not only did the authors of TextRunner facilitate domain-independent detection of relations from a corpus but their work triggered researchers towards developing OIE systems. For instance, the WOE (Wikipedia-based Open Extractor) [14] system is built on TextRunner, having two modes of operation: WOEPos and WOEParse. The main hypothesis behind WOE is the automated assembly of training samples by heuristically pairing Wikipedia info box values with corresponding texts, hence improving TextRunner’s performance. WOEPos exploits the CRF classifier trained with shallow syntactic proprieties to extract specific words between two noun phrases that represents a relation.
An example of an OIE approach that utilizes dependency parsing is WOEParse; it exploits a rich dictionary of dependency path patterns acquired from Wikipedia extractions. While the OLLIE (Open Language Learning for Information Extraction) approach [15] relies on the bootstrapping concept, it learns semi-lexicalized pattern templates using dependency parses by bootstrapping a plentiful amount of training data that results in surpassing WOE’s performance.
3.1.2. Hand-Crafted Rules Approaches
REVERB, introduced by Fader et al. [16], extracts tuples by singling out relation phrases that satisfy syntactic and lexical constraints; for each relation phrase, a pair of noun phrase arguments are identified. REVERB then uses logistic regression trained on 1000 sentences from the web with shallow syntactic features to assign a confidence score to each extracted relation triple. The R2A2 approach [17] upgrades REVERB by adding ARGLEARNER, an argument identifier that makes use of patterns as features to identify the left and right boundaries of each argument.
KRAKEN [18] is one of the few OIE system that is able to capture N-ary relations. It utilizes hand-crafted patterns to identify relation phrases and their correlated arguments over typed dependency parsers. As a further matter, KRAKEN is able to detect completeness and correctness of the extracted facts, thus increasing the quality of the extracted information. Del Corro and Gemulla proposed ClausIE (Clause-based Open Information Extraction) [19], which locates clauses in input sentences by making use of linguistic information of the English language’s grammar by computing a dependency parse tree of the input phrase to determine its syntactical structure. Each clause is later classified to be compatible with the grammatical function of its constituents. Unlike the aforementioned OIE systems, ClausIE does not exploit any training data.
3.1.3. Neural Network Approaches
Recently, as a result of their successfulness in a diverse NLP tasks [1], deep neural networks paved the way to the OIE task. A recurrent neural network (RNN) encoder–decoder OIE framework was proposed by Cui et al. [20]. A fluctuating length sequence is sent to the network’s encoder as a sole input. The encoder then generates a compressed representation vector to transfer to the decoder in order to produce the output sequence. A three-layer long short-term memory (LSTM) [21] is the internal structure of both the encoder and the decoder. Stanovsky et al. [22] presented a neural OIE paradigm that trains a bidirectional LSTM (bi-LSTM) transducer to label each word, verifying that supervised learning can have a positive effect on OIE performance.
3.2. State-of-the-Art Relation Extraction
RE research falls mainly under one of the following approaches: supervised, semi-supervised, distant supervision, and unsupervised. As always, the main issue of supervised techniques is the necessity of having a large amount of labelled data, which is difficult to gather [23]. Semi-supervised approaches mainly depend on bootstrapping techniques. Distant supervision techniques merge both semi-supervised and unsupervised approaches. However, popularity of unsupervised techniques declined due to the fact that the learner is provided unannotated data, and for that reason, evaluation becomes demanding at a large scale. We limited our discussion to supervised, semi-supervised, and distant supervision approaches. Neural approaches appear as a subclass in all the aforementioned classes.
3.2.1. Supervised Approaches
RE is treated as a multi-class classification task in supervised approaches. Supervised categories can be classified into kernel-based approaches and feature-based approaches. An example of the latter is the work of [24], who merged diverse features of lexical, syntactic, and semantic knowledge by employing a support vector machine (SVM) to extract relations, proving the effectiveness of base phrase chunking information. Authors of [25] introduced a kernel-based RE paradigm that incorporates term generalization techniques—word clustering and latent semantic analysis—with structured kernels to enhance RE results in different domains. Moreover, a neural approach based on adversarial training was proposed by Peng Su and K. Vijay-Shanker [26], aiming to boost RE task performance through various adversarial examples and adding perturbation on all input features of the model. Adversarial learning is built on the basis that similar data instances are assigned the same label.
3.2.2. Semi-Supervised Approaches
The first bootstrapping algorithm was DIPRE (Dual Iterative Pattern Relation Expansion) [27], which employs a pattern-matching model as classifier by using a set of seeds to extract patterns from the dataset in order to extract new candidate relations. The DualRE model [28] was proposed to overcome the problem of semantic drift associated with bootstrapping approaches. The key idea behind DualRE is training a retrieval module along a relation prediction module, hereby mutually improving the quality of one another through labelling data to use as auxiliary training data. In [29], a convolutional neural network (CNN) RE architecture was proposed that employs graph-structured data where label knowledge is smoothed over the graph by means of explicit graph-based regularization.
3.2.3. Distant Supervision Approaches
The traditional distant supervision RE approaches claim that if a sentence consists of two related entities then the same relation lies between those two entities. Nevertheless, Sebastian et al. proposed an RE model that supports a different claim, “if two entities participate in a relation, then at least one sentence that mentions those two entities might express that relation” [30], by utilizing a factor graph to aid in determining if two entities are related or not. Additionally, a learning algorithm is employed to train this graphical framework by structuring distant supervision as an instance of constraint-driven semi-supervision.
A piecewise CNN RE technique was proposed by [31], not only to overcome the noise generated from the feature extraction phase, but also to address the issue of handling distant relation extraction as a multi-instance task, which leads to lack of certainty of instance labels. By designing a convolutional framework with piecewise max pooling as an alternative to feature engineering to automatically learn related features, the authors of [31] were able to overcome the aforementioned problems.
4. ROIE: A Recurrent Neural Network Model for Open Information Extraction
Our recurrent neural network (RNN) model is based on our work in [32] by tackling the OIE task as a sequencing labeling problem resulting in the extraction of multiple, overlapping tuples for each sentence.
4.1. Neural Model Architecture
Throughout the back-propagation process, RNNs are prone to vanishing and exploding gradient descent complications, making RNN training challenging. Thus, LSTMs and gated recurrent units (GRUs) were established to address the issues related to the unstable gradient. When the gradient becomes too big or simply disappears, killing the learning process, LSTMs and GRUs aid by using the relevant gates to allow the gradient to flow backward through time, freely and effectively keeping long-term dependencies [33].
Both LSTMs and GRUs are able to train on long word contexts and connect information using cell states. LSTM has three gates (input, output, and forget), contrary to GRU, which couples input and forget gates in one gate—update gate, in addition to reset gate, which determines how to incorporate previous memory with the current input. As a result, our model employs GRUs instead of LSTMs, since GRUs are less complex with only two gates, and hereby they require less training parameters and utilize less memory, effectively making GRU faster than LSTM.
The default operation in RNN captures context in a single direction, which may lead to comprehending issues; for instance, consider the following two sentences:
“Second place is not as prestigious as first place.”
“Second is the standard international unit of time.”
In these sentences, the word “second” carries different meanings, which traditional RNNs will not be able to comprehend, since it is the first word in the sentence; nevertheless, bidirectional RNNs support learning from both ends. A bidirectional GRU (Bi-GRU) was employed in our model to learn forward and backward lexical semantics of each word in a given sentence. There are two different methods to implement a bidirectional network; either by having two RNNs operating in opposite directions or within the internal architecture of the RNN itself. In our ROIE framework, we implemented the latter approach.
4.2. Word Embeddings
Recently, several types of word embeddings have been introduced; nevertheless, they all serve the same purpose of mapping words to low-dimensional vector representations. The aforementioned OIE and RE deep learning-based approaches in Section 3.1.3 and Section 3.2, respectively, utilized one of the traditional word embeddings, either GloVe [34] or Word2Vec [35].
In our work, we incorporated the novel contextualized word embeddings. Due to their ability to capture complex syntactic and semantic features of a word, deep contextualized word embeddings have proven to be successful in various NLP tasks when compared to the traditional word embeddings. The main concept behind contextualized word embeddings is that a word’s representation varies according to its neighboring words, and thus the same word can have different representations depending on its adjacent words.
Table 2 shows the word embeddings we employed in our experiment, along with the dimensionality of each embedding and the data they are trained on. We picked one traditional non-contextualized embedding, GloVe, and three contextualized embeddings with different dimensionalities: BERT [9], XLNet [36], and XLM-RoBERTa [37]. XLNet is trained on data much larger than Google’s BERT training data, and thus it outperforms BERT on 20 different NLP tasks [36]. Facebook’s XLM-RoBERTa depends on the masked language model objective and is effective in text processing from 100 different languages.

Table 2.
Word embeddings employed in our work.
Flair [38] is a simple framework that offers a unified interface for conceptually varying types of word and document embeddings, which we utilized in our experiments.
4.3. Work Flow
The embedded sentence—composed of a fixed-length vector—is sent as an input to our ROIE neural network framework. Specifically, predicates—the part of a sentence or clause containing a verb and stating something about the subject—are regarded as the building blocks of most languages, as they denote significant actions that are deemed extremely efficient in extracting relations of interest. Therefore, in line with the work of [22,32], the predicate in each sentence is presumed to be the relation that links the tuple; consequently, the predicate is inputted to the neural network framework as a feature vector alongside the part of speech (POS) tag of the input sentence obtained using the NLTK toolkit [39], as shown in Figure 2.
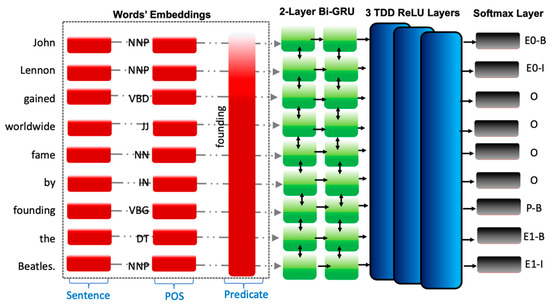
Figure 2.
Our ROIE transferable neural model architecture.
After embedding the three aforementioned inputs, we concatenated them all to form our feature vector of shape (3, length of sentence, embedding size); the feature vector is defined as follows:
Feature Vector = Embedded Word ⊕ Embedded POS ⊕ Embedded Predicate
The generated feature vector is then passed to the two-layer Bi-GRU, which in turn outputs a tensor that is progressed to three-layer time distributed dense (TDD) layers, which is finally passed to the SoftMax layer for label prediction.
4.4. Sequence Labelling
In NLP, sequence labelling is the task of identifying and assigning a label to each word, for instance the POS task, where each word is tagged to a particular POS. Sequencing labelling achieved more promising results when compared to traditional statistical techniques among a diverse array of NLP tasks [22]. In our work, we used BIO tags (Begin-Intermediate-Outside) [40] to indicate the word’s location in the sentence and label it accordingly. The SoftMax output layer assigns the probability score to each word to determine its corresponding label, as shown in Figure 2. Our proposed ROIE paradigm is only able to capture binary relations. If a sentence contains more than one predicate, another instance of that sentence is created to capture any possible relation. However, if a sentence has no relations, only the predicate is labelled “P-B” (Predicate-Begin), “P-I” (Predicate-Intermediate), while label “O” (Outside) is assigned to the remaining words in the sentence, without assigning any “E” (Entity) labels.
4.5. Dataset
To train and test our OIE neural framework, we used the Wikipedia News Corpus (WikiNews) [41]. Our dataset was split into a training set to train the network, a development set for validation purposes and a test set to assess the performance of our ROIE framework on a 60/20/20 ratio. An overview of the dataset is shown in Table 3.

Table 3.
WikiNews dataset overview.
4.6. Hyperparameter Settings
Our ROIE neural framework was implemented using the Keras framework [42] with a TensorFlow backend [43]. Table 4 shows our model’s hyperparameter configurations that achieved the best results when training and testing on OIE. As shown, our framework was trained on 20 epochs and the training dataset was split into 100 batches. For regularization purposes, in order to avoid over-fitting, the dropout rate was set to 0.1. Furthermore, early stopping was utilized to terminate training when the training performance stopped improving. Both bidirectional GRU layers and the three TDD layers had an identical number of units, 128 units. Additionally, rectified linear unit (ReLU) [44] was the chosen activation function in the three TDD layers, while the Adam optimizer [45] was utilized to train our framework.

Table 4.
Hyperparameter settings used in ROIE.
4.7. Results of our ROIE Model
It should be emphasized that our ROIE neural model outperformed other state-of-the-art neural OIE approaches, as documented in [32], while using ELMo word embeddings [46], also a deep contextualized word embedding that models both complex syntactic and semantic features of a word.
Better results were attained after XLNet was substituted for ELMo [46] when compared to our results in [32]; the results are reported in Table 5. An exhaustive grid search was performed to single out the best batch–epoch pair for each word embedding. Our batches and epochs ranged from 20 to 120 and 1 to 50, respectively, both with increments of 5. GloVe achieved an F-measure of 56.1%, while BERT and XLM-RoBERTa achieved a F-measure of 61.1% and 61.5%, respectively. Nevertheless, XLNet surpassed all the other embeddings—including ELMo’s 59% F-measure—and achieved 65%.

Table 5.
Results of the ROIE model using different word embeddings. Both training and testing were done on the OIE WikiNews dataset. Recall (R), precision (P), and F-measure (F) were used as evaluation metrics.
5. Materials and Methods
In this section, we explain the experiments carried out and dataset utilized in our two main tasks, transferring to OIE bio-medical domain and transferring to RE task. In the source task, the aforementioned WikiNews training set [41] was utilized.
5.1. Transfering to OIE: Bio-Medical Domain
A classifier trained on a news corpus would observe an altered distribution if employed to classify bio-medical data. Therefore, domain adaptation methods are deployed in transfer learning in such scenarios. In the transductive learning task, specifically domain adaptation, we handle our pretrained model as a feature extractor; in our case, the pretrained model was trained on the news domain, where there is a characteristic shift in distribution of the data between source and target domains that necessitates adjustments to effectively transfer knowledge.
DDIExtraction 2013 [47] is a bio-medical dataset mainly specialized in the subject of drug–drug interactions. The dataset was structured from the DrugBank database [48] and MEDline abstracts [49] related to drug–drug interactions. We utilized the DDIExtraction as a test set in the following experiments. In our work, the performance of the following three experiments were compared against each other:
Transductive transfer learning: transferring knowledge learnt from the OIE news domain to the OIE bio-medical domain.
Inductive transfer learning: a small amount of bio-medical data also from DDIExtraction is fed to the neural network alongside news data to train the neural network.
Traditional learning: both training and testing on bio-medical data.
5.2. Transfering to Relation Extraction
The OIE and RE tasks are both subclasses of information extraction, making the two tasks similar in semantics. The dataset used in the RE task for training, testing, and validation is Semeval-2010 Task 8 [50]. The nine predefined relations in the dataset are shown in Table 6. The training set consists of 8000 sentences, however, for a fair comparison we trained our neural network on the same number of relation tuples available in the OIE training set; thus, 2906 tuples were randomly selected from the training set. Similarly, the same experiments were compared against each other when transferring from the OIE news domain to RE:

Table 6.
List of predefined relations in the Semeval-2010 corpus and their number of occurrences.
Transductive transfer learning: transferring knowledge learnt from the OIE news domain to the RE news domain.
Inductive transfer learning: a small percentage of the RE corpus is fed into the neural framework along OIE news data to train the neural network.
Traditional learning: both training and testing on the RE news domain.
In all the above-mentioned experiments in both tasks, we used bio-medical OIE and RE, a development set containing 946 tuples composed of the same structure as the source task, for validation purposes.
6. Results and Evaluation
The following measures were used to measure the effect of transferring knowledge learnt from our ROIE framework: Recall (R), Precision (P), and F-measure (F). All the aforementioned evaluation metrics were expressed as percentages throughout the experiments, with the F-measure being the determining performance measure. All hyperparameters—shown previously in Table 4—except for epochs and batches were fixed throughout our experiments. Contextual embeddings were highly sensitive to changes in hyperparameters, specifically with respect to number of epochs and batches. Steep falls and rises were noticed when the number of epochs and batches were changed.
It is worth noting that the dimensionality of the word embeddings refers to the length of the vector; in theory the size of the vector is directly proportional to the information it can store, which allows NLP systems to perform better. However, in practice, there was not much benefit with the embeddings with higher dimensionality when compared with lower dimensionality embeddings.
6.1. Results of Transferring to OIE: Bio-Medical Domain
In order to properly evaluate transfer learning results, we compared it with training and testing on the target task. Detailed results of the experiments can be found in Table 7, indicating the source task (training set) and the target task (testing set). The hyperparameters that achieved the highest scores are the ones reported in Table 7.

Table 7.
Domain adaptation results by transferring from the OIE news domain to the OIE bio-medical domain using four different word embeddings. Bold values indicate the highest achieved F-measure in each of the three experiments (transductive transfer learning, inductive transfer learning, traditional learning).
OIE: Bio-Medical Domain Results Discussion
Our system achieved the highest results using XLM-RoBERTa in all three experiments: transductive transfer learning, inductive transfer learning, and traditional learning, outperforming all other word embeddings.
When our training set was composed entirely of news data, XLM-RoBERTa scored the highest F-measure of 64.4%, with 100 batches and 5 epochs. XLNet and GloVe achieved the same F-measure of 62.9% using the same number of batches and epochs, 100 and 5, respectively. Nevertheless, BERT achieved the lowest F-measure of 60%.
In inductive transfer learning, a small amount of bio-medical data were inputted to the neural framework by sampling a random batch from the DDIExtraction 2013 training data using a 4:1 ratio, with bio-medical data having the lower ratio. A significant increase in the F-measure of 13.6% was attained in inductive transfer learning when comparing to transductive transfer learning. Using both XLM-RoBERTa and XLNet, our inductive transfer approach realized an F-measure of approximately 78%, with XLM-RoBERTa’s precision surpassing XLNet’s by 0.9%. BERT came in third and achieved 75.2%, while GloVe scored an F-measure of 73.7%.
The results scored using traditional learning by training entirely on bio-medical data were only 1% higher than the results achieved using the inductive transfer learning technique. Once again, XLM-RoBERTa outperformed the other embeddings by scoring an F-measure of 79% using 100 batches and 15 epochs. Additionally, BERT achieved roughly the same F-measure as XLM-RoBERTa of 78.9%, using the same number of epochs and batches; however, it achieved a lower precision of 85.9%. It is notable that GloVe achieved a higher F-measure in inductive transfer learning than traditional learning. Our interpretation is that adding news training data to the biomedical tasks resulted in a higher performance with GloVe embeddings. This could correlate with the original training data of the GloVe model used in our experiments. Thus, our results show that using a small percentage from the target task while training our neural network results in a proximate outcome when compared to traditional learning.
6.2. Results of Transfering to Relation Extraction
Equally, in order to establish a fair comparison in the following three experiments, we fixed the training set size to 2906 relation instances. Results of both transductive and inductive transfer learning were compared against the results achieved by traditional learning. Results are reported in Table 8.

Table 8.
Results of transferring from OIE to RE using four different word embeddings. Bold values indicate the highest achieved F-measure in each of the three experiments (transductive transfer learning, inductive transfer learning, traditional learning).
Relation Extraction Results Discussion
Firstly, in transductive transfer learning, with 50 batches and 10 epochs, BERT was able to achieve an F-measure of 54.4%. Both XLNet and XLM-RoBERTa scored the same F-measure of 49.1%, which was nearly 4.6% higher than the F-measure achieved using GloVe.
With inductive transfer learning, we found an improvement of 12.8% when compared to transductive learning also using a 4:1 ratio, with the OIE news dataset overtaking the higher ratio. Using XLM-RoBERTa, a 67.2% F-measure was attained when the network was trained on 15 epochs and the training dataset was divided into 100 batches. BERT and XLNet did not fall far behind XLM-RoBERTa, as they achieved F-measures of 66.3% and 65.4%, respectively. GloVe achieved the lowest F-measure of 59.9%.
When employing default learning settings, where we train on our target task, there was a 3.8% enhancement in the F-measure. Once again, BERT outperformed by scoring an F-measure of 71%, only 0.5% higher than XLNet, and 2.6% higher than XLM-RoBERTa. Consistently, GloVe scored the lowest F-measure of 65.9%, hereby proving the notable effect in the model’s performance when using contextualized word embeddings in contrast with traditional word embeddings.
Table 9 summarizes the best results of the three main experiments acquired in our work: ROIE model, transferring to bio-medical domain, and transferring to RE. As seen in Table 9, we could not single out a particular contextualized word embedding to utilize, as the use of word embedding may vary according to the various reasons: type of task (OIE, RE, or sentiment analysis), dataset domain (news, bio-medical data, or financial data), and the computational power available to the user. This is also in agreement with other papers that extensively compared embeddings in various tasks and found that the most suitable one is highly dependent on the task and data nature [51,52].

Table 9.
Summary of the best result obtained in each experiment by different systems described in the paper: original ROIE model, transferring from OIE to bio-medical OIE (transductive transfer learning, inductive transfer learning, traditional learning), and transferring from OIE to (transductive transfer learning, inductive transfer learning, traditional learning).
To further elaborate that the choice of the word embedding is dependent upon the task and nature of data, XLNet outperformed all the other word embeddings when training and testing on the news dataset. However, on bio-Medical data, XLM-RoBERTa performed better in all three experiments: transductive transfer learning, inductive transfer learning, and traditional learning. It is worth noting that XLM-RoBERTa outperformed in four out of a total seven experiments in our work. Thus, we were motivated to compare and experiment with the use of different word embeddings.
7. Conclusions and Future Work
Can we survive without labelled data in NLP? On the basis of our findings: yes! Nevertheless, employing labelled data in NLP tasks still results in better performance. However, the process of collection of labelled data is demanding and, in some cases, inaccessible. In this paper, we utilized training on OIE to diminish the complication of insufficient training data of neural network models in various NLP tasks and encourage model generalization. Since OIE plays a fundamental role in turning massive, unstructured data into factual information that can be used as a foundation to many NLP tasks, we favored OIE as our source task, thereby ensuring our work is useful and beneficial to the NLP community.
In the domain adaptation experiment, we transferred information learnt from one domain to the other on the same task. The neural model was trained on the OIE news domain and tested on the bio-medical domain. Results obtained from the inductive approach indicated that our ROIE neural model can play a fundamental role in domain adaptation.
Moreover, our research also covered the transferability to a semantically related task. Results achieved from transferring from the OIE to RE followed the same pattern as transferring from the OIE news domain to the bio-medical domain. Inductive transfer learning achieved promising and comparable results with traditional learning. Thus, our work demonstrates that OIE can act as a reliable source task, not only in domain adaptation but also when transferring to related tasks.
In the future, we intend to expand our work beyond sequence labelling tasks and experiment with multi-transfer learning thoroughly on several NLP tasks, specifically tasks that are not semantically related to OIE such as sentiment analysis. Additionally, we intend to investigate different transferring mechanisms to study how to leverage knowledge acquired from pre-trained models in varied ways.
Author Contributions
Conception and design of the experiments, I.S. and M.S.; data curation, I.S.; methodology, I.S.; software, I.S.; supervision, M.S.; validation, M.S.; writing—original draft, I.S.; writing—review and editing, M.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was made possible with funding from the European Union’s Horizon 2020 research and innovation program under grant agreement no. 883588 (GEIGER). The opinions expressed and arguments employed herein do not necessarily reflect the official views of the funding body.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Otter, D.W.; Medina, J.R.; Kalita, J. A Survey of the Usages of Deep Learning for Natural Language Processing. arXiv 2019, arXiv:1807.10854. [Google Scholar] [CrossRef] [PubMed]
- Mausam, M. Open Information Extraction Systems and Downstream Applications. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, New York, NY, USA, 15 July 2016. [Google Scholar]
- Yang, Q.; Zhang, Y.; Dai, W.; Pan, S. Foundations of transfer learning. In Transfer Learning; Cambridge University Press: Cambridge, UK, 2020; pp. 1–2. [Google Scholar]
- Keneshloo, Y.; Ramakrishnan, N.; Reddy, C.K. Deep Transfer Reinforcement Learning for Text Summarization. In Proceedings of the 2019 SIAM International Conference on Data Mining, Calgary, AB, Canada, 2–4 May 2019; pp. 675–683. [Google Scholar]
- Bhatia, P.; Arumae, K.; Celikkaya, E.B. Dynamic transfer learning for named entity recognition. In Social Networks: A Framework of Computational Intelligence; Springer: Cham, Switzerland, 2019; pp. 69–81. [Google Scholar]
- Min, S.; Seo, M.; Hajishirzi, H.; Barzilay, R.; Kan, M.Y. Question answering through transfer learning from large fine-grained supervision data. arXiv 2017, arXiv:1702.02171. [Google Scholar]
- Yu, J.; Qiu, M.; Jiang, J.; Huang, J.; Song, S.; Chu, W.; Chen, H. Modelling Domain Relationships for Transfer Learning on Retrieval-based Question Answering Systems in E-commerce. In Proceedings of the Eleventh ACM International Conference on Multimedia—MULTIMEDIA’03, Berkeley, CA, USA, 7 November 2003; pp. 682–690. [Google Scholar]
- Chuong, D.B.; Andrew, N.Y. Transfer learning for text classification. Adv. Neural Inf. Process. Syst. 2006, 299–306. [Google Scholar]
- Devlin, J.; Chang, M.W.; Lee, K.; Toutanova, K. BERT: Pre-training of deep bidirectional transformers for language understanding. arXiv 2018, arXiv:1810.04805. [Google Scholar]
- Howard, J.; Ruder, S. Universal language model fine-tuning for text classification. arXiv 2018, arXiv:1801.06146. [Google Scholar]
- Radford, A.; Karthik, N.; Salimans, T.; Sutskever, I. Improving Language Understanding by Generative Pre-Training. 2018. Available online: https://www.cs.ubc.ca/~amuham01/LING530/papers/radford2018improving.pdf (accessed on 20 August 2020).
- Sarhan, I.; Marco, S. Uncovering algorithmic approaches in open information extraction: A literature review. In Proceedings of the 30th Benelux Conference on Artificial Intelligence, Hertogenbosch, The Netherlands, 8–9 November 2018. [Google Scholar]
- Etzioni, O.; Banko, M.; Soderland, S.; Weld, D. Open information extraction from the web. In Proceedings of the Twentieth International Joint Conference on Artificial Intelligence, Hyderabad, India, 6–12 January 2007; Volume 7, pp. 2670–2676. [Google Scholar]
- Wu, F.; Weld, D.S. Open information extraction using Wikipedia. In Proceedings of the 48th Annual Meeting of the Association for Computational Linguistics, Uppsala, Sweden, 11–16 July 2010; pp. 118–127. [Google Scholar]
- Schmitz, M.; Bart, R.; Soderland, S.; Etzioni, O. Open language learning for information extraction. In Proceedings of the 2012 Joint Conference on Empirical Methods in Natural Language Processing and Computational Natural Language Learning; Association for Computational Linguistics, Jeju Island, Korea, 12–14 July 2012. [Google Scholar]
- Fader, A.; Soderland, S.; Etzioni, O. Identifying relations for open information extraction. In Proceedings of the Conference on Empirical Methods in Natural Language Processing, Association for Computational Linguistics (ACL), Edinburgh, UK, 11 July 2011. [Google Scholar]
- Christensen, J.; Soderland, S.; Etzioni, O. An analysis of open information extraction based on semantic role labeling. In Proceedings of the K-CAP’2011: Knowledge Capture Conference, Banff, AB, Canada, 25–29 June 2011; Volume 11, pp. 3–10. [Google Scholar]
- Akbik, A.; Löser, A. Kraken: N-ary facts in open information extraction. In Proceedings of the Joint Workshop on Automatic Knowledge Base Construction and Web-scale Knowledge Extraction, Montreal, QC, Canada, 7–8 June2012; pp. 52–56. [Google Scholar]
- Del Corro, L.; Gemulla, R. ClausIE: Clause-based open information extraction. In Proceedings of the 22nd International Conference on WWW, Rio de Janeiro, Brazil, 13–17 May 2013; pp. 355–366. [Google Scholar]
- Cui, L.; Wei, F.; Zhou, M. Neural open information extraction. arXiv 2018, arXiv:1805.04270. [Google Scholar]
- Hochreiter, S.; Schmidhuber, J. Long short-term memory. Neural Comput. 1997, 9, 1735–1780. [Google Scholar] [CrossRef]
- Stanovsky, G.; Michael, J.; Zettlemoyer, L.; Dagan, I. Supervised Open Information Extraction. In Proceedings of the 2018 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, New Orleans, LA, USA, 1–6 June 2018; Volume 1. [Google Scholar]
- Sarhan, I.; El-Sonbaty, Y.; El-Nasr, M.A. Arabic relation extraction: A survey. Int. J. Comput. 2016, 5, 430–437. [Google Scholar]
- Guodong, Z.; Jian, S.; Jie, Z.; Min, Z. Exploring various knowledge in relation extraction. In Proceedings of the 43rd Annual Meeting, Ann Harbour, MI, USA, 25–30 June 2005. [Google Scholar]
- Plank, B.; Moschitti, A. Embedding semantic similarity in tree kernels for domain adaptation of relation extraction. In Proceedings of the 51st Annual Meeting of the Association for Computational Linguistics, Sofia, Bulgaria, 4–9 August 2013. [Google Scholar]
- Su, P.; Vijay-Shanker, K. Adversarial learning for supervised and semi-supervised relation extraction in bio-medical literature. arXiv 2020, arXiv:2005.04277. [Google Scholar]
- Brin, S. Extracting patterns and relations from the world wide web. In The World Wide Web and Databases; Springer: Berlin, Germany, 1999; pp. 172–183. [Google Scholar]
- Lin, H.; Yan, J.; Qu, M.; Ren, X. Learning Dual Retrieval Module for Semi-supervised Relation Extraction. In Proceedings of the World Wide Web Conference on—WWW ’19, San Fransisco, CA, USA, 13–17 May 2019; pp. 1073–1083. [Google Scholar]
- Kipf, T.N.; Welling, M. Semi-supervised classification with graph convolutional networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Riedel, S.; Yao, L.; McCallum, A. Modeling Relations and Their Mentions without Labeled Text. In Proceedings of the Joint European Conference on Machine Learning and Knowledge Discovery in Databases, Heidelberg, Germany, 16–20 September 2010. [Google Scholar]
- Zeng, D.; Liu, K.; Chen, Y.; Zhao, J. Distant Supervision for Relation Extraction via Piecewise Convolutional Neural Networks. In Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing, Lisbon, Portugal, 19–23 September 2015. [Google Scholar]
- Sarhan, I.; Spruit, M.R. Contextualized Word Embeddings in a Neural Open Information Extraction Model. In Proceedings of the International Conference on Applications of Natural Language to Information Systems, Salford, UK, 26–28 June 2019. [Google Scholar]
- Pascanu, R.; Tomas, M.; Yoshua, B. On the Difficulty of Training Recurrent Neural Networks. In Proceedings of the International Conference on Machine Learning, Atlanta, GA, USA, 16–21 June 2013. [Google Scholar]
- Pennington, J.; Socher, R.; Manning, C. Glove: Global Vectors for Word Representation. In Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP), Doha, Qatar, 19–25 October 2014; pp. 1532–1543. [Google Scholar]
- Mikolov, T.; Sutskever, I.; Chen, K.; Corrado, G.; Dean, J. Distributed representations of words and phrases and their compositionality. arXiv 2013, arXiv:1310.4546. [Google Scholar]
- Yang, Z.; Dai, Z.; Yang, Y.; Carbonell, J.; Salakhutdinov, R.; Le, Q.V. XLNet: Generalized autoregressive pretraining for language understanding. Adv. Neural Inf. Process. Syst. 2019, 32, 5753–5763. [Google Scholar]
- Conneau, A.; Khandelwal, K.; Goyal, N.; Chaudhary, V.; Wenzek, G.; Guzmán, F.; Grave, E.; Ott, M.; Zettlemoyer, L.; Stoyanov, V. Unsupervised cross-lingual representation learning at scale. arXiv 2019, arXiv:1911.02116. [Google Scholar]
- Akbik, A.; Bergmann, T.; Blythe, D.; Rasul, K.; Schweter, S.; Vollgraf, R. FLAIR: An Easy-to-Use Framework for State-of-the-Art NLP. In Proceedings of the 2019 Annual Conference of the North American Chapter of the Association for Computational Linguistics (Demonstrations), NAACL, Princeton, MI, USA, 2–7 June 2019. [Google Scholar]
- Loper, E.; Bird, S. NLTK: The natural language toolkit. arXiv 2002, arXiv:cs/0205028. [Google Scholar]
- Ramshaw, L.; Mitchell, A.; Marcus, P. BIO Labels: Text Chunking Using Transformation-Based Learning. Natural Language Processing Using Very Large Corpora; Springer: Dordrecht, The Netherlands, 1999; pp. 157–176. [Google Scholar]
- Stanovsky, G.; Dagan, I. Creating a Large Benchmark for Open Information Extraction. In Proceedings of the 2016 Conference on Empirical Methods in Natural Language Processing, Austin, TX, USA, 2–6 November 2016; pp. 2300–2305. [Google Scholar]
- Franois, C. Keras. 2015. Available online: https://github.com/fchollet/keras (accessed on 20 March 2020).
- Abadi, M. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th {USENIX} Symposium on Operating Systems Design and Implementation ({OSDI} 16), Savannah, GA, USA, 2–4 November 2016. [Google Scholar]
- Nair, V.; Hinton, G.E. Rectified linear units improve restricted Boltzmann machines. In Proceedings of the 27th International Conference on Machine Learning (ICML-10), Haifah, Isreal, 21–24 June 2010. [Google Scholar]
- Kingma, D.P.; Ba, J. Adam: A method for stochastic optimization. arXiv 2014, arXiv:1412.6980. [Google Scholar]
- Peters, M.; Neumann, M.; Iyyer, M.; Gardner, M.; Clark, C.; Lee, K.; Zettlemoyer, L. Deep contextualized word representations. arXiv 2018, arXiv:1802.05365. [Google Scholar]
- Segura-Bedmar, I.; Martínez, P.; De Pablo-Sánchez, C. Using a shallow linguistic kernel for drug–drug interaction extraction. J. Biomed. Inform. 2011, 44, 789–804. [Google Scholar] [CrossRef]
- Wishart, D.S. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef]
- Bethesda, M.D. National Library of Medicine (US). 2013. Available online: https://medlineplus.gov/ (accessed on 29 March 2020).
- Hendrickx, I.; Kim, S.N.; Kozareva, Z.; Nakov, P.; Séaghdha, D.Ó.; Padó, S.; Pennacchiotti, M.; Romano, L.; Szpakowicz, S. SemEval-2010 Task 8: Multi-way classification of semantic relations between pairs of nominals. arXiv 2019, arXiv:1911.10422. [Google Scholar]
- Tawfik, N.S.; Spruit, M.R. Evaluating sentence representations for biomedical text: Methods and experimental results. J. Biomed. Inform. 2020, 104, 103396. [Google Scholar] [CrossRef]
- Perone, C.S.; Silveira, R.; Paula, T.S. Evaluation of sentence embeddings in downstream and linguistic probing tasks. arXiv 2018, arXiv:1806.06259. [Google Scholar]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).