Norovirus Detection in Ready-To-Eat Salads by Propidium Monoazide Real Time RT-PCR Assay
Abstract
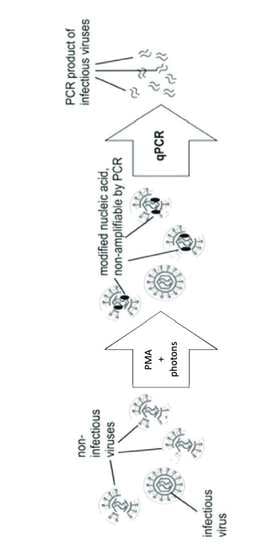
1. Introduction
2. Materials and Methods
2.1. Virus Strains
2.2. PMAxx Real Time RT-PCR Validation
2.2.1. PMAxx Pretreatment on Virus Suspensions
2.2.2. Real Time RT-PCR
2.3. Detection of NoV GI and GII with PMAxx Real Time RT-PCR
2.3.1. Sampling
2.3.2. Virus Concentration and Nucleic Acid Extraction
2.3.3. Mengovirus Real Time RT-PCR
2.4. Statistical Analysis
3. Results
3.1. PMA Real Time RT-PCR Assay Validation
3.2. Detection of NoV GI and GII by Real Time RT-PCR
4. Discussion
5. Conclusions
Future Perspective
Author Contributions
Funding
Conflicts of Interest
References
- Bányai, K.; Estes, M.K.; Martella, V.; Parashar, U.D. Viral gastroenteritis. Lancet 2018, 392, 175–186. [Google Scholar] [CrossRef]
- EFSA and ECDC. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar]
- Berger, C.N.; Sodha, S.V.; Shaw, R.K.; Griffin, P.M.; Pink, D.; Hand, P.; Frankel, G. Fresh fruit and vegetables as vehicles for the transmission of human pathogens. Environ. Microbiol. 2010, 12, 2385–2397. [Google Scholar] [CrossRef]
- Cheong, S.; Lee, C.; Song, S.W.; Choi, W.C.; Lee, C.H.; Kim, S.J. Enteric viruses in raw vegetables and groundwater used for irrigation in South Korea. Appl. Environ. Microbiol. 2009, 75, 7745–7751. [Google Scholar] [CrossRef]
- Kokkinos, P.; Kozyra, I.; Lazic, S.; Bouwknegt, M.; Rutjes, S.; Willems, K.; Moloney, R.; de Roda Husman, A.M.; Kaupke, A.; Legaki, E.; et al. Harmonised investigation of the occurrence of human enteric viruses in the leafy green vegetable supply chain in three European countries. Food Environ. Virol. 2012, 4, 79–191. [Google Scholar] [CrossRef]
- Terio, V.; Bottaro, M.; Pavoni, E.; Losio, M.N.; Serraino, A.; Giacometti, F.; Martella, V.; Mottola, A.; Di Pinto, A.; Tantillo, G. Occurrence of hepatitis A and E and norovirus GI and GII in ready-to-eat vegetables in Italy. Int. J. Food Microbiol. 2017, 249, 61–65. [Google Scholar] [CrossRef]
- Bidawid, S.; Farber, J.M.; Sattar, A. Contamination of foods by food handlers: Experiments onHepatitis a virus transfer to food and its interruption. Appl. Environ. Microbiol. 2000, 66, 2759–2763. [Google Scholar] [CrossRef]
- Mottola, A.; Bonerba, E.; Bozzo, G.; Marchetti, P.; Celano, G.V.; Colao, V.; Terio, V.; Tantillo, G.; Figueras, M.J.; Di Pinto, A. Occurrence of emerging food-borne pathogenic Arcobacter spp. isolated from pre-cut (ready-to-eat) vegetables. Int. J. Food Microbiol. 2016, 236, 33–37. [Google Scholar] [CrossRef]
- European Commission. Commission Regulation (EC) No 2073/2005 of 15 November 2005 on Microbiological Criteria for Foodstuffs; European Commission: Brussels, Belgium, 2005. [Google Scholar]
- EFSA Panel on Biological Hazards. Scientific opinion on the risk posed by pathogens in food of non-animal origin. Part 2 (Salmonella and norovirus in leafy greens eaten raw as salads). EFSA J. 2014, 12, 118. [Google Scholar]
- Haramoto, E.; Kitajima, M.; Hata, A.; Torrey, J.R.; Masago, Y.; Sano, D.; Katayama, H. A review on recent progress in the detection methods and prevalence of human enteric viruses in water. Water Res. 2018, 135, 168–186. [Google Scholar]
- Malik, Y.S.; Verma, A.K.; Kumar, N.; Touil, N.; Karthik, K.; Tiwari, R.; Bora, D.P.; Dhama, K.; Ghosh, S.; Hemida, M.G.; et al. Advances in diagnostic approaches for viral etiologies of diarrhea: From the lab to the field. Front. Microbiol. 2019, 10, 1957. [Google Scholar] [CrossRef] [PubMed]
- ISO/TS 15216-2. Microbiology of Food and Animal Feed—Horizontal Method for Determination of Hepatitis A Virus and Norovirus in Food Using Real-Time RT-PCR. Part 2: Method for Qualitative Detection; International Organization for Standardization: Geneva, Switzerland, 2017. [Google Scholar]
- Parshionikar, S.; Laseke, I.; Fout, G.S. Use of Propidium Monoazide in reverse transcriptase PCR To distinguish between infectious and noninfectious enteric viruses in water samples. Appl. Environ. Microbiol. 2010, 76, 4318–4326. [Google Scholar] [CrossRef] [PubMed]
- Hamza, I.A.; Jurzik, L.; Überla, K.; Wilhelm, M. Methods to detect infectious human enteric viruses in environmental water samples. Int. J. Hyg. Environ. Health 2011, 214, 424–436. [Google Scholar] [CrossRef]
- Rodriguez, A.R.; Pepper, I.L.; Gerba, C.P. Application of PCR-Based methods to assess the infectivity of enteric viruses in environmental samples. Appl. Environ. Microbiol. 2009, 75, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Randazzo, W.; López-Gálvez, F.; Allende, A.; Aznar, R.; Sánchez, G. Evaluation of viability PCR performance for assessing norovirus infectivity in fresh-cut vegetables and irrigation water. Int. J. Food Microbiol. 2016, 229, 1–6. [Google Scholar] [CrossRef]
- Fittipaldi, M.; Nocker, A.; Codony, F. Progress in understanding preferential detection of live cells using viability dyes in combination with DNA amplification. J. Microbiol. Methods 2012, 91, 276–289. [Google Scholar] [CrossRef]
- Fuster, N.; Pintó, R.M.; Fuentes, C.; Beguiristain, N.; Bosch, A.; Guix, S. Propidium monoazide RTqPCR assays for the assessment of hepatitis A inactivation and for a better estimation of the health risk of contaminated waters. Water Res. 2016, 101, 226–232. [Google Scholar] [CrossRef]
- Moreno, L.; Aznar, R.; Sánchez, G. Application of viability PCR to discriminate the infectivity of hepatitis A virus in food samples. Int. J. Food Microbiol. 2015, 201, 1–6. [Google Scholar] [CrossRef]
- Yezli, S.; Otter, J.A. Minimum infective dose of the major human respiratory and enteric viruses transmitted through food and the environment. Food Environ. Virol. 2011, 3, 1–30. [Google Scholar] [CrossRef]
- Randazzo, W.; Khezric, M.; Ollivier, J.; Le Guyaderd, F.S.; Díaz, J.R.; Aznara, R.; Sánchez, G. Optimization of PMAxx pretreatment to distinguish between human norovirus with intact and altered capsids in shellfish and sewage samples. Front. Microbiol. 2018, 9, 1973. [Google Scholar] [CrossRef]
- Martínez-Martínez, M.; Diez-Valcarce, M.; Hernández, M.; Rodríguez-Lázaro, D. Design and application of nucleic acid standards for quantitative detection of enteric viruses by real-time PCR. Food Environ. Virol. 2011, 3, 92–98. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.K.; Le Saux, J.C.; Parnaudeau, S.; Pommepuy, M.; Elimelech, M.; Le Guyader, F.S. Evaluation of removal of noroviruses during wastewater treatment, using real-time reverse transcription-PCR: Different behaviors of genogroups I and II. Appl. Environ. Microbiol. 2007, 73, 7891–7897. [Google Scholar] [CrossRef] [PubMed]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Rzezutka, A.; Cook, N. Survival of human enteric viruses in the environment and food. FEMS Microbiol. Rev. 2004, 28, 441–453. [Google Scholar] [CrossRef]
- La Bella, G.; Martella, V.; Basanisi, M.G.; Nobili, G.; Terio, V.; La Salandra, G. Food-Borne viruses in shellfish: Investigation on norovirus and HAV presence in Apulia (SE Italy). Food Environ. Virol. 2017, 9, 179–186. [Google Scholar] [CrossRef]
- Terio, V.; Di Pinto, A.; Di Pinto, P.; Martella, V.; Tantillo, G. RNA extraction method for the PCR detection of hepatitis A virus in shellfish. Int. J. Food Microbiol. 2010, 142, 198–201. [Google Scholar] [CrossRef]
- Loutreul, J.; Cazeaux, C.; Levert, D.; Nicolas, A.; Vautier, S.; Le Sauvage, A.L.; Perelle, S.; Morin, T. Prevalence of human noroviruses in frozen marketed shellfish, red fruits and fresh vegetables. Food Environ. Virol. 2014, 6, 157–168. [Google Scholar] [CrossRef]
- Terio, V.; Bottaro, M.; Di Pinto, A.; Catella, C.; Chironna, M.; Bozzo, G.; Kingsley, D.H.; Bonerba, E.; Morea, A.; Martella, V. Outbreak of hepatitis A in Italy associated with frozen red currants imported from Poland: A case study. Food Environ. Virol. 2015, 7, 305–308. [Google Scholar] [CrossRef]
- Chiapponi, C.; Pavoni, E.; Bertasi, B.; Baioni, L.; Scaltriti, E.; Chiesa, E.; Cianti, L.; Losio, M.N.; Pongolini, S. Isolation and genomic sequence of Hepatitis A virus from mixed frozen berries in Italy. Food Environ. Virol. 2014, 6, 202–206. [Google Scholar] [CrossRef][Green Version]
- Severi, E.; Verhoef, L.; Thornton, L.; Guzman-Herrador, B.R.; Faber, M.; Sundqvist, L.; Rimhanen-Finne, R.; Roque-Afonso, A.M.; Ngui, S.L.; Allerberger, F.; et al. Large and prolonged food-borne multistate hepatitis A outbreak in Europe associated with consumption of frozen berries, 2013 to 2014. Eurosurveillance 2015, 20, 21192. [Google Scholar] [CrossRef]
- Serracca, L.; Rossini, I.; Battistini, R.; Goria, M.; Sant, S.; De Montis, G.; Ercolini, C. Potential risk of norovirus infection due to the consumption of “ready to eat” food. Food Environ. Virol. 2012, 4, 89–92. [Google Scholar] [PubMed]
- Butot, S.; Putallaz, T.; Sánchez, G. Effects of sanitation, freezing and frozen storage on enteric viruses in berries and herbs. Int. J. Food Microbiol. 2008, 126, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Hewitt, J.; Leonard, M.; Greening, G.E.; Lewis, G.D. Influence of wastewater treatment process and the population size on human virus profiles in wastewater. Water Res. 2011, 45, 6267–6276. [Google Scholar] [CrossRef]
- Baert, L.; Uyttendaele, M.; Vermeersch, M.; Van Coillie, E.; Debevere, J. Survival and transfer of murine norovirus 1, a surrogate for human noroviruses, during the production process of deep-frozen onions and spinach. J. Food Prot. 2008, 71, 1590–1597. [Google Scholar] [CrossRef] [PubMed]
- Goyal, S.M.; Gerba, C.P.; Melnick, J.L. Human enteroviruses in oysters and their overlying waters. Appl. Environ. Microbiol. 1979, 37, 572–581. [Google Scholar] [CrossRef] [PubMed]
- Di Pinto, A.; Forte, V.T.; Tantillo, G.M.; Terio, V.; Buonavoglia, C. Detection of hepatitis A virus in shellfish (Mytilus galloprovincialis) with RT-PCR. J. Food Prot. 2003, 66, 1681–1685. [Google Scholar] [CrossRef]
- Taban, B.M.; Halkman, A.K. Do leafy green vegetables and their ready-to-eat [RTE] salads carry a risk of foodborne pathogens? Anaerobe 2011, 17, 286–287. [Google Scholar] [CrossRef]
- Müller, L.; Rasmussen, L.D.; Jensen, T.; Schultz, A.C.; Kjelsø, C.; Barnadas, C.; Sigsgaard, K.; Larsen, A.R.; Widstrup Jensen, C.; Jeppesen, S.; et al. Series of norovirus outbreaks caused by consumption of green coral lettuce, Denmark, April 2016. PLoS Curr. 2016, 8. [Google Scholar] [CrossRef]
| Viruses | Treatment | Ct Value | Reduction |
|---|---|---|---|
| NoV GI | NT | 28.66 ± 0.23 | - |
| PMAxx [50 μM] | 33.79 ± 0.19 | −5.12 ± 0.37 * | |
| 95 °C | 32.79 ± 0.19 | −3.92 ± 0.18 | |
| 95 °C + PMAxx [50 μM] | 36.35 ± 0.78 | −7.69 ± 0.96 * | |
| NoV GII | NT | 28.59 ± 0.31 | - |
| PMAxx [50 μM] | 32.70 ± 0.18 | −4.10 ± 0.27 * | |
| 95 °C | 33.41 ± 0.32 | −4.82 ± 0.52 | |
| 95 °C + PMAxx [50 μM] | 36.70 ± 0.25 | −8.06 ± 0.27 * |
| Samples | NoV GII Positive | |
|---|---|---|
| Pre-Treatment | Post-Treatment | |
| Mixed salad | 17/21 (80.9%) | 6/21 (28.6%) |
| Carrot | 8/9 (88.8%) | 2/9 (22.2%) |
| Valerian | 27/33 (81.8%) | 9/33 (27.3%) |
| Rocket | 5/9 (55.6%) | 0/9 |
| Spinach | 15/15 (100%) | 6/15 (40%) |
| Iceberg Lettuce | 15/21 (71.4%) | 9/21 (42.9%) |
| Romaine Lettuce | 0/6 | 0/6 |
| Curly Leaf Endive | 8/9 (88.9%) | 3/9 (33.3%) |
| Chicory | 6/9 (66.7%) | 3/9 (33.3%) |
| Trevisano Chicory | 0/3 | 0/3 |
| Total Samples | 101/135 (74.8%) | 38/135 (28.1%) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Terio, V.; Lorusso, P.; Mottola, A.; Buonavoglia, C.; Tantillo, G.; Bonerba, E.; Di Pinto, A. Norovirus Detection in Ready-To-Eat Salads by Propidium Monoazide Real Time RT-PCR Assay. Appl. Sci. 2020, 10, 5176. https://doi.org/10.3390/app10155176
Terio V, Lorusso P, Mottola A, Buonavoglia C, Tantillo G, Bonerba E, Di Pinto A. Norovirus Detection in Ready-To-Eat Salads by Propidium Monoazide Real Time RT-PCR Assay. Applied Sciences. 2020; 10(15):5176. https://doi.org/10.3390/app10155176
Chicago/Turabian StyleTerio, Valentina, Patrizio Lorusso, Anna Mottola, Canio Buonavoglia, Giuseppina Tantillo, Elisabetta Bonerba, and Angela Di Pinto. 2020. "Norovirus Detection in Ready-To-Eat Salads by Propidium Monoazide Real Time RT-PCR Assay" Applied Sciences 10, no. 15: 5176. https://doi.org/10.3390/app10155176
APA StyleTerio, V., Lorusso, P., Mottola, A., Buonavoglia, C., Tantillo, G., Bonerba, E., & Di Pinto, A. (2020). Norovirus Detection in Ready-To-Eat Salads by Propidium Monoazide Real Time RT-PCR Assay. Applied Sciences, 10(15), 5176. https://doi.org/10.3390/app10155176