Long-Term Effects of Mercury on Biofilms Grown in Contaminated Microcosms: A Pilot Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Labware Cleaning
2.2. Biofilm Culture in Hg-Enriched Microcosms
2.3. Determination of the Water Quality Variables
2.4. Measurements of Accumulated Hg in Biofilms
2.5. Biofilm Characterization
2.6. Statistics
3. Results and Discussion
3.1. Biofilm Culture Conditions
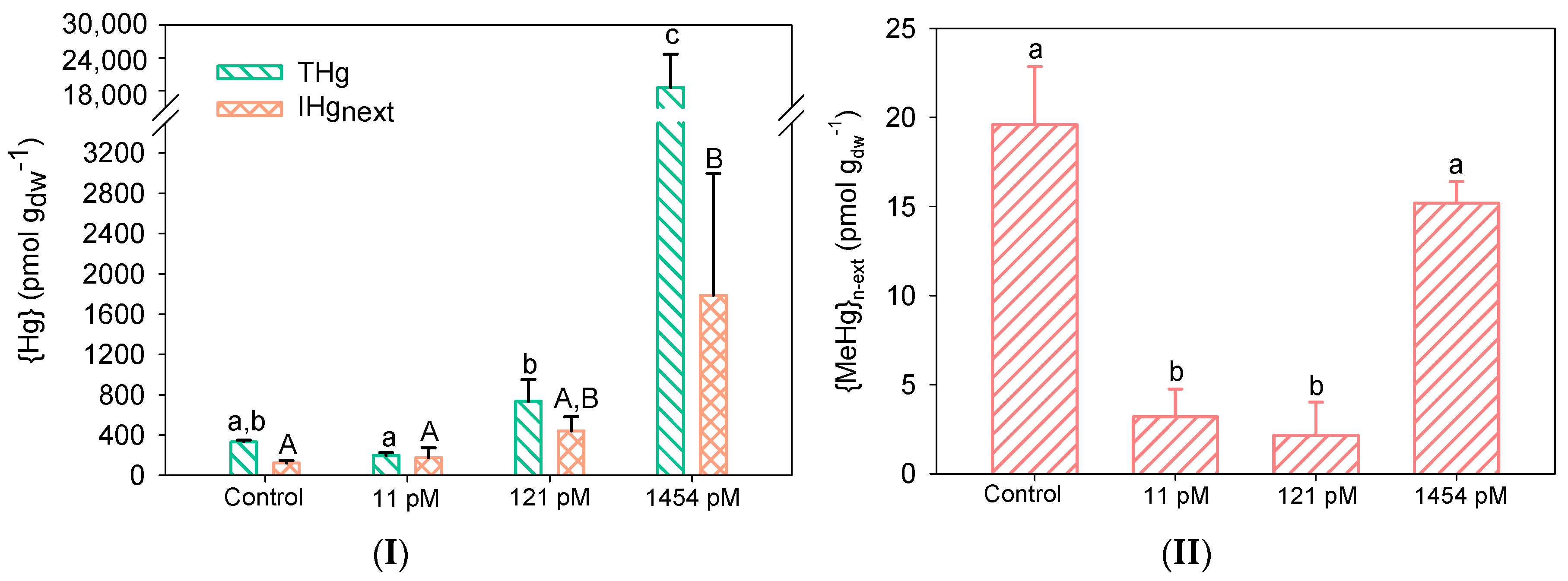
3.2. Hg Accumulation by Biofilms
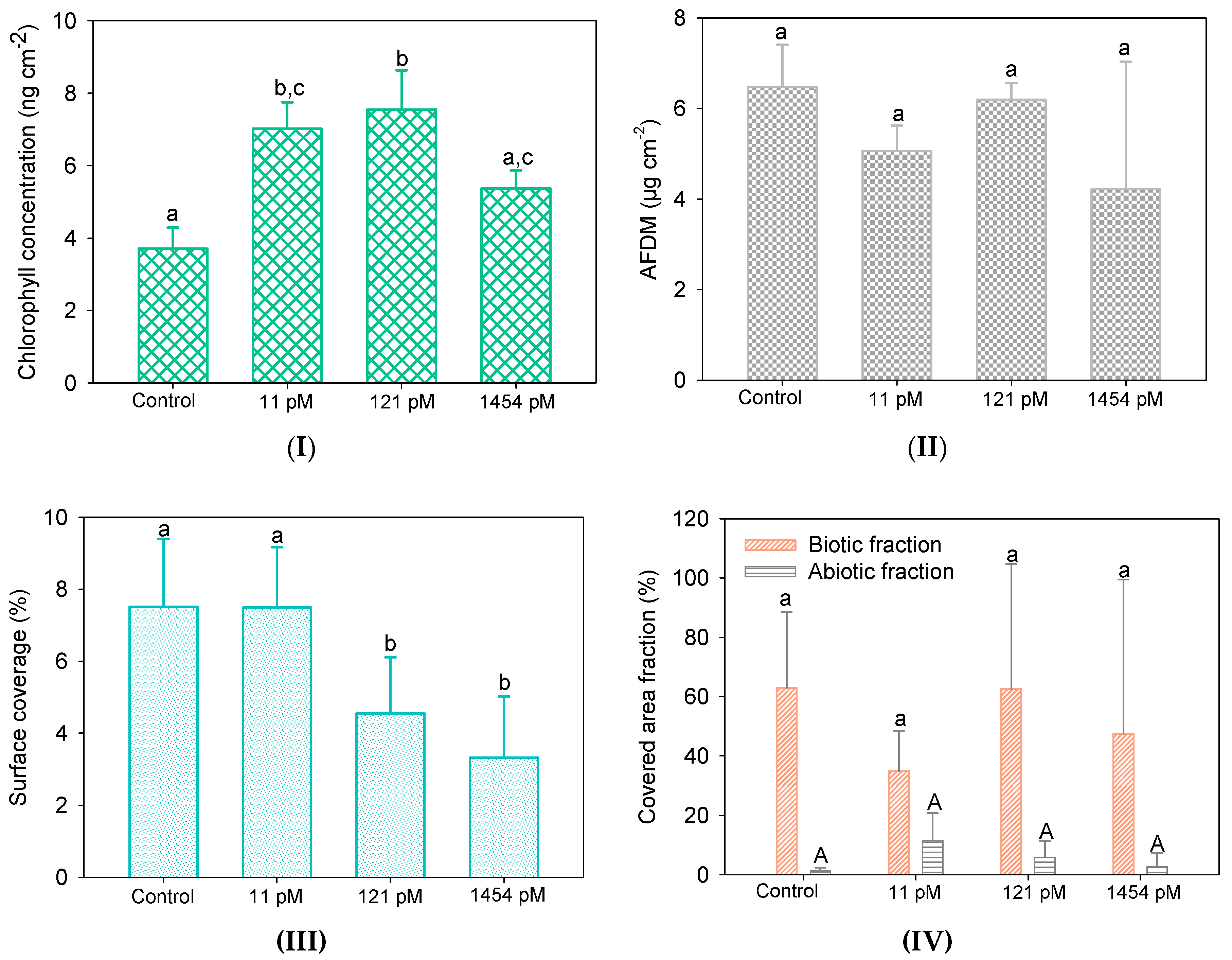
3.3. Hg Impacts on Biofilms
3.3.1. Biofilm Biomass
3.3.2. Bacterial Biofilm Composition
3.3.3. Algal Biofilm Composition
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Battin, T.J.; Besemer, K.; Bengtsson, M.M.; Romani, A.M.; Packmann, A.I. The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 2016, 14, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Vannote, R.L.; Minshall, G.W.; Cummins, K.W.; Sedell, J.R.; Cushing, C.E. River continuum concept. Can. J. Fish. Aquat. Sci. 1980, 37, 130–137. [Google Scholar] [CrossRef]
- Azim, M.E. Photosynthetic periphyton and surfaces. In Encyclopedia of Inland Waters; Likens, G.E., Ed.; Academic Press: Oxford, UK, 2009; pp. 184–191. [Google Scholar]
- Lear, G.; Dopheide, A.; Ancion, P.-Y.; Roberts, K.V.W.; Smith, J.G.; Lewis, G.D. Biofims in freshwater: Their importance for maintenance and monitoring of freshwater health. In Microbial Biofilms: Current Research and Applications; Lear, G., Lewis, G.D., Eds.; Caiser Academic Press: Norfolk, UK, 2012; pp. 129–152. [Google Scholar]
- Sabater, S.; Guasch, H.; Ricart, M.; Romani, A.; Vidal, G.; Kluender, C.; Schmitt-Jansen, M. Monitoring the effect of chemicals on biological communities. The biofilm as an interface. Anal. Bioanal. Chem. 2007, 387, 1425–1434. [Google Scholar] [CrossRef] [PubMed]
- Burns, A.; Ryder, D.S. Potential for biofilms as biological indicators in Australian riverine systems. Ecol. Manag. Restor. 2001, 2, 53–64. [Google Scholar] [CrossRef]
- Amos, H.M.; Jacob, D.J.; Streets, D.G.; Sunderland, E.M. Legacy impacts of all-time anthropogenic emissions on the global mercury cycle. Glob. Biogeochem. Cycles 2013, 27, 410–421. [Google Scholar] [CrossRef]
- Lamborg, C.H.; Hammerschmidt, C.R.; Bowman, K.L.; Swarr, G.J.; Munson, K.M.; Ohnemus, D.C.; Lam, P.J.; Heimburger, L.E.; Rijkenberg, M.J.A.; Saito, M.A. A global ocean inventory of anthropogenic mercury based on water column measurements. Nature 2014, 512, 65–67. [Google Scholar] [CrossRef] [PubMed]
- Dranguet, P.; Le Faucheur, S.; Slaveykova, V.I. Mercury bioavailability, transformations, and effects on freshwater biofilms. Environ. Toxicol. Chem. 2017, 36, 3194–3205. [Google Scholar] [CrossRef] [PubMed]
- Dominique, Y.; Maury-Brachet, R.; Muresan, B.; Vigouroux, R.; Richard, S.; Cossa, D.; Mariotti, A.; Boudou, A. Biofilm and mercury availability as key factors for mercury accumulation in fish (Curimata cyprinoides) from a disturbed Amazonian freshwater system. Environ. Toxicol. Chem. 2007, 26, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Cremona, F.; Hamelin, S.; Planas, D.; Lucotte, M. Sources of organic matter and methylmercury in littoral macroinvertebrates: A stable isotope approach. Biogeochemistry 2009, 94, 81–94. [Google Scholar] [CrossRef]
- Cheng, J.P.; Zhao, W.C.; Liu, Y.Y.; Wu, C.; Liu, C.; Wang, W.H. Adsorption properties and gaseous mercury transformation rate of natural biofilm. Bull. Environ. Contam. Toxicol. 2008, 81, 516–520. [Google Scholar] [CrossRef] [PubMed]
- Hamelin, S.; Amyot, M.; Barkay, T.; Wang, Y.P.; Planas, D. Methanogens: Principal methylators of mercury in lake periphyton. Environ. Sci. Technol. 2011, 45, 7693–7700. [Google Scholar] [CrossRef] [PubMed]
- Correia, R.R.S.; Miranda, M.R.; Guimaraes, J.R.D. Mercury methylation and the microbial consortium in periphyton of tropical macrophytes: Effect of different inhibitors. Environ. Res. 2012, 112, 86–91. [Google Scholar] [CrossRef] [PubMed]
- Bouchet, S.; Goni-Urriza, M.; Monperrus, M.; Guyoneaud, R.; Fernandez, P.; Heredia, C.; Tessier, E.; Gassie, C.; Point, D.; Guedron, S.; et al. Linking microbial activities and low-molecular-weight thiols to Hg methylation in biofilms and periphyton from high-altitude tropical lakes in the Bolivian altiplano. Environ. Sci. Technol. 2018, 52, 9758–9767. [Google Scholar] [CrossRef] [PubMed]
- Peres, E.; Coste, M.; Ribeyre, F.; Ricard, M.; Boudou, A. Effects of methylmercury and inorganic mercury on periphytic diatom communities in freshwater indoor microcosms. J. Appl. Phycol. 1997, 9, 215–227. [Google Scholar] [CrossRef]
- Val, J.; Muniz, S.; Goma, J.; Navarro, E. Influence of global change-related impacts on the mercury toxicity of freshwater algal communities. Sci. Total Environ. 2016, 540, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Kovac Virsek, M.; Hubad, B.; Lapanje, A. Mercury induced community tolerance in microbial biofilms is related to pollution gradients in a long-term polluted river. Aquat. Toxicol. 2013, 144–145C, 208–217. [Google Scholar] [CrossRef] [PubMed]
- Dranguet, P.; Le Faucheur, S.; Cosio, C.; Slaveykova, V.I. Influence of chemical speciation and biofilm composition on mercury accumulation by freshwater biofilms. Environ. Sci. Processes Impacts 2017, 19, 38–49. [Google Scholar] [CrossRef] [PubMed]
- Dranguet, P.; Cosio, C.; Le Faucheur, S.; Hug-Peter, D.; Loizeau, J.L.; Ungureanu, V.G.; Slaveykova, V.I. Biofilm composition in the Olt River (Romania) reservoirs impacted by a chlor-alkali production plant. Environ. Sci. Processes Impacts 2017, 19, 687–695. [Google Scholar] [CrossRef] [PubMed]
- USEPA. Method 1631, Revision E: Mercury in Water by Oxidation, Purge and Trap, and Cold Vapor Atomic Fluorescence Spectrometry; EPA: Washington, DC, USA, 2002; p. 38.
- Dranguet, P.; Slaveykova, V.I.; Le Faucheur, S. Kinetics of mercury accumulation by freshwater biofilms. Environ. Chem. 2018, 14, 458–467. [Google Scholar] [CrossRef]
- Rasband, W.S. ImageJ; U.S. National Institutes of Health: Bethesda, MD, USA, 1997–2018. Available online: https://imagej.nih.gov/ij/ (accessed on 21 February 2019).
- Zimmermann, J.; Jahn, R.; Gemeinholzer, B. Barcoding diatoms: Evaluation of the V4 subregion on the 18S rRNA gene; including new primers and protocols. Org. Divers. Evol. 2011, 11, 173–192. [Google Scholar] [CrossRef]
- Bricheux, G.; Morin, L.; Le Moal, G.; Coffe, G.; Ballestrino, D.; Charbonnel, N.; Bohatier, J.; Forestier, C. Pyrosequencing assessment of prokaryotic and eukaryotic diversity in biofilm communities from a French river. Microbiologyopen 2013, 2, 402–414. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Crist, T.O.; Chase, J.M.; Vellend, M.; Inouye, B.D.; Freestone, A.L.; Sanders, N.J.; Cornell, H.V.; Comita, L.S.; Davies, K.F.; et al. Navigating the multiple meanings of beta diversity: A roadmap for the practicing ecologist. Ecol. Lett. 2011, 14, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community Ecology Package. R Package Version 2.3-3. 2016. Available online: http://cran.r-project.org/package=vegan (accessed on 1 December 2018).
- Van de Peer Lab, Bioinformatics and Evolutionary Genome, 2018. Available online: http://bioinformatics.psb.ugent.be/beg/software?page=2 (accessed on 1 December 2018).
- R Core-Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015. [Google Scholar]
- CIPEL. Rapports de la Commission Internationale pour la Protection des eaux du Léman Contre la Pollution—Campagne 2013; CIPEL: Nyon, Switzerland, 2014. [Google Scholar]
- Brenda, K.; Lasorsa, G.A.G.; Horvat, M. Analytical methods for measuring mercury in water, sediment and Biota. In Mercury in the Environment; Bank, M.S., Ed.; University of California Press: Berkeley, CA, USA, 2012; pp. 27–54. [Google Scholar]
- Žižek, S.; Milacic, R.; Kovac, N.; Jacimovic, R.; Toman, M.J.; Horvat, M. Periphyton as a bioindicator of mercury pollution in a temperate torrential river ecosystem. Chemosphere 2011, 85, 883–891. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.M.; Bruce, K.D.; Strike, P.; Ritchie, D.A. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 1997, 19, 239–262. [Google Scholar] [CrossRef] [PubMed]
- Dash, H.R.; Das, S. Interaction between mercuric chloride and extracellular polymers of biofilm-forming mercury resistant marine bacterium Bacillus thuringiensis PW-05. RSC Adv. 2016, 6, 109793–109802. [Google Scholar] [CrossRef]
- Hamelin, S.; Planas, D.; Amyot, M. Mercury methylation and demethylation by periphyton biofilms and their host in a fluvial wetland of the St. Lawrence River (QC; Canada) Sci. Total Environ. 2015, 512–513, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Olsen, T.A.; Brandt, C.C.; Brooks, S.C. Periphyton biofilms influence net methylmercury production in an industrially contaminated system. Environ. Sci. Technol. 2016, 50, 10843–10850. [Google Scholar] [CrossRef] [PubMed]
- Murthy, S.D.S.; Bukhov, N.G.; Mohanty, P. Mercury induced alterations of chlorophyll a fluorescence kinetics in Cyanobacteria—Multiple effects of mercury on electron transport. J. Photochem. Photobiol. Biol. 1990, 6, 373–380. [Google Scholar] [CrossRef]
- Kampnielsen, L. Effect of deleterious concentrations of mercury on photosynthesis and growth of Chlorella pyrenoidosa. Physiol. Plant. 1970, 24, 556–561. [Google Scholar] [CrossRef]
- Grégoire, D.S.; Poulain, A.J. A physiological role for Hg-II during phototrophic growth. Nat. Geosci. 2016, 9, 121–127. [Google Scholar] [CrossRef]
- Hiller-Bittrolff, K.; Foreman, K.; Bulseco-McKim, A.N.; Benoit, J.; Bowen, J.L. Effects of mercury addition on microbial community composition and nitrate removal inside permeable reactive barriers. Environ. Pollut. 2018, 242, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, L.D.; Zawadsky, C.; Binnerup, S.J.; Oregaard, G.; Sorensen, S.J.; Kroer, N. Cultivation of hard-to-culture subsurface mercury-resistant bacteria and discovery of new merA gene sequences. Appl. Environ. Microbiol. 2008, 74, 3795–3803. [Google Scholar] [CrossRef] [PubMed]
- Lazaro, W.L.; Guimaraes, J.R.D.; Ignacio, A.R.A.; Da Silva, C.J.; Diez, S. Cyanobacteria enhance methylmercury production: A hypothesis tested in the periphyton of two lakes in the Pantanal floodplain, Brazil. Sci. Total Environ. 2013, 456–457, 231–238. [Google Scholar] [CrossRef] [PubMed]
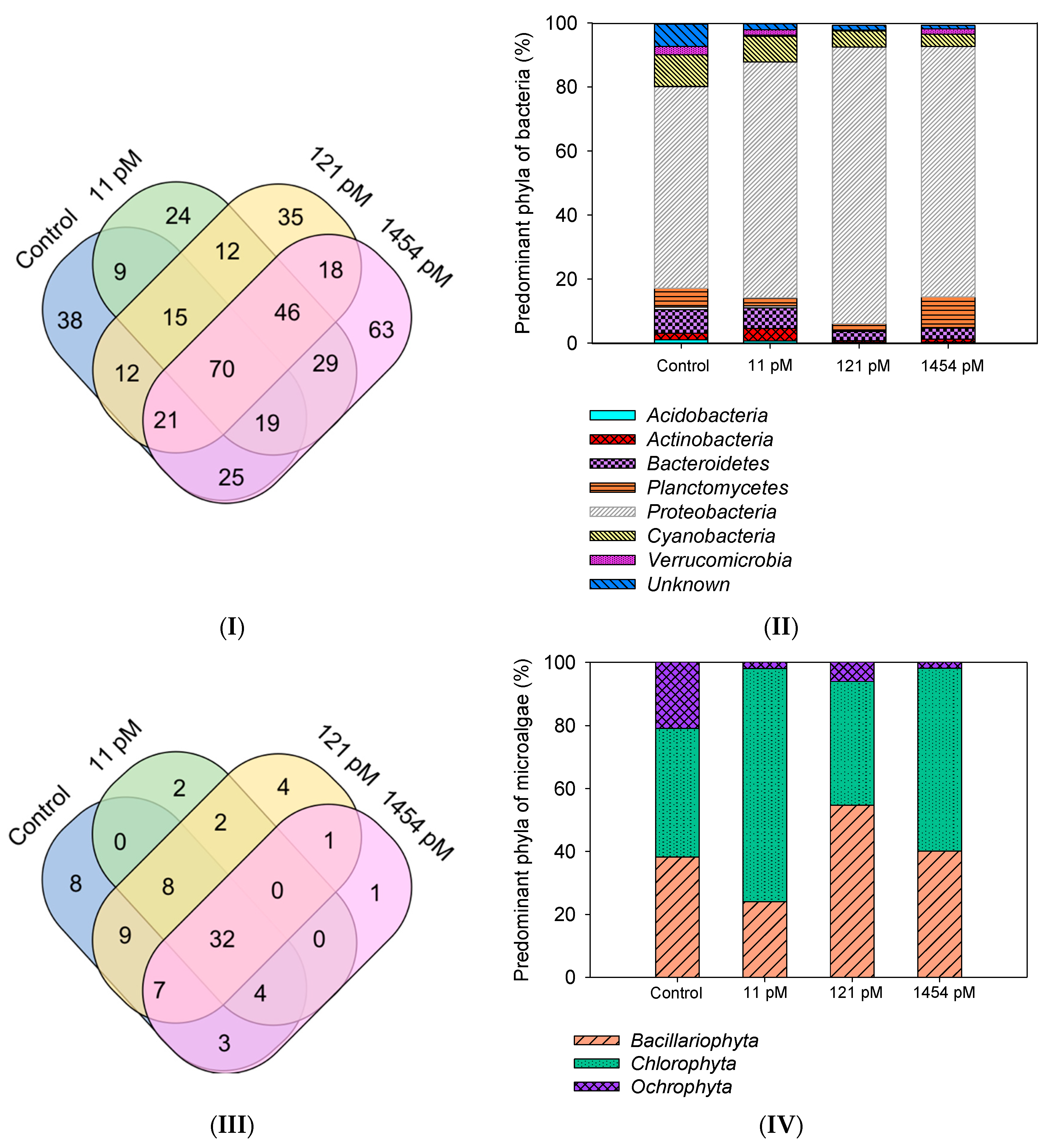
| Taxa | Parameters | Control | 11 pM | 121 pM | 1454 pM |
|---|---|---|---|---|---|
| Bacteria | Total sequences | 1269 | 21,441 | 25,731 | 45,251 |
| OTUs richness | 209 | 224 | 229 | 291 | |
| Rarefied richness | 209 ± 1 | 191 ± 4 | 180 ± 5 | 210 ± 6 | |
| Simpson’s index | 0.97 | 0.96 | 0.94 | 0.97 | |
| Species turnover 1 | - | 0.65 | 0.63 | 0.63 | |
| Algae | Total sequences | 12023 | 2006 | 5624 | 1520 |
| OTUs richness | 71 | 48 | 63 | 48 | |
| Rarefied richness | 56 ± 2 | 46 ± 1 | 50 ± 5 | 48 ± 1 | |
| Simpson’s index | 0.87 | 0.77 | 0.92 | 0.85 | |
| Species turnover 1 | - | 0.41 | 0.28 | 0.37 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dranguet, P.; Freiburghaus, A.; Slaveykova, V.I.; Le Faucheur, S. Long-Term Effects of Mercury on Biofilms Grown in Contaminated Microcosms: A Pilot Study. Environments 2019, 6, 28. https://doi.org/10.3390/environments6030028
Dranguet P, Freiburghaus A, Slaveykova VI, Le Faucheur S. Long-Term Effects of Mercury on Biofilms Grown in Contaminated Microcosms: A Pilot Study. Environments. 2019; 6(3):28. https://doi.org/10.3390/environments6030028
Chicago/Turabian StyleDranguet, Perrine, Aline Freiburghaus, Vera I. Slaveykova, and Séverine Le Faucheur. 2019. "Long-Term Effects of Mercury on Biofilms Grown in Contaminated Microcosms: A Pilot Study" Environments 6, no. 3: 28. https://doi.org/10.3390/environments6030028
APA StyleDranguet, P., Freiburghaus, A., Slaveykova, V. I., & Le Faucheur, S. (2019). Long-Term Effects of Mercury on Biofilms Grown in Contaminated Microcosms: A Pilot Study. Environments, 6(3), 28. https://doi.org/10.3390/environments6030028





