Integrative Analysis of Methylation and Transcriptional Profiles to Reveal the Genetic Stability of Cashmere Traits in the Tβ4 Overexpression of Cashmere Goats
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Culture
2.3. Generation of Cloned Goats via Somatic Cell Nuclear Transfer (SCNT)
2.4. Measurement of Body-Related Indices
2.5. Blood Biochemistry
2.6. RNA Extraction and Quantitative Real Time-PCR (qRT-PCR)
2.7. Western Blot Analysis
2.8. Hematoxylin-Eosin (H&E) and Immunohistochemistry (IHC) Staining
2.9. DNA Extraction
2.10. Whole Genome Bisulphite Sequencing (WGBS) and Data Processing
2.11. Methylation Level Analysis
2.12. Differentially Methylated Regions (DMRs) Analysis
2.13. Transcriptome Analysis
2.14. Enrichment Analysis of Differentially Expressed Genes (DEGs) and Differentially Methylated Regions (DMRs)
2.15. Statistical Analyses
3. Results
3.1. Establishment of the Germline of Thymosin β4 Overexpression (Tβ4-OE) Cashmere Goats
3.2. Cashmere Yield and SHFs/PHFs Ratio Increased in F1-SCNT
3.3. Differential Analysis of Methylation and Expression
3.4. Roles of Methylation in Regulating Gene Expression
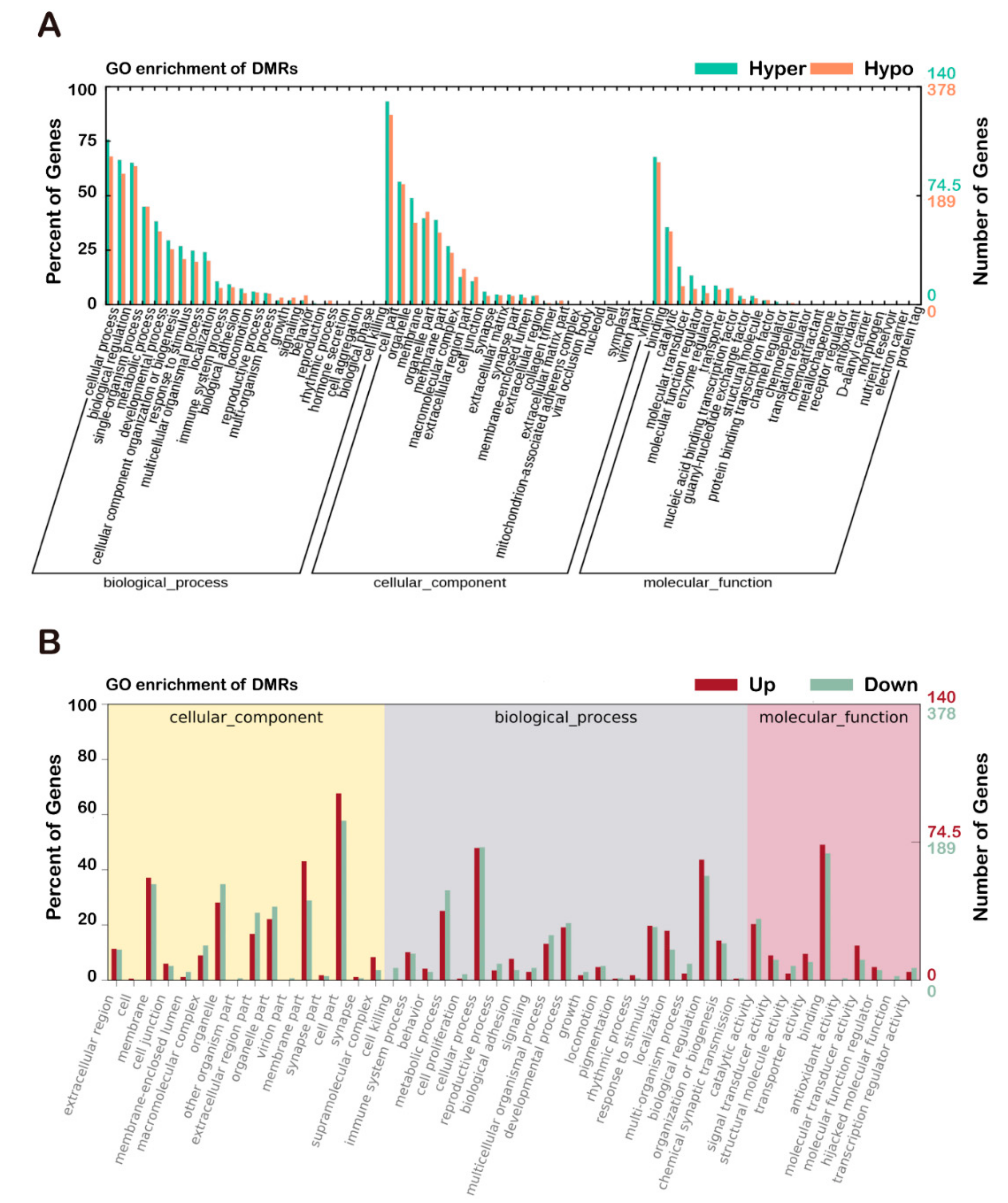
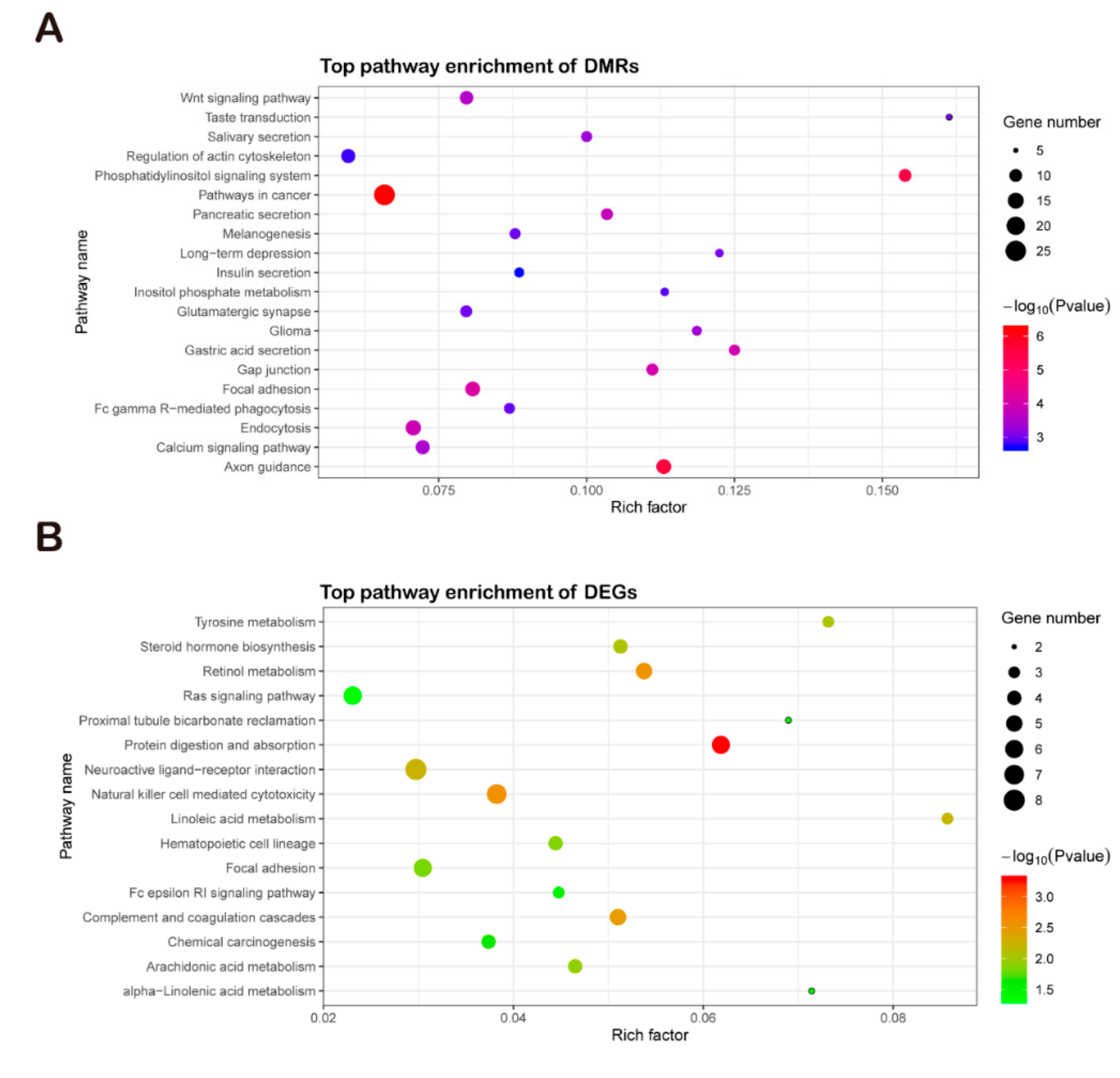
3.5. Comparison of Functional Analysis of DEGs and DMRs with GO and KEGG
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Reynolds, J.D. Animal breeding systems. Trends Ecol. Evol. 1996, 11, 68–72. [Google Scholar] [CrossRef]
- Zhang, J.; Cui, M.L.; Nie, Y.W.; Dai, B.; Li, F.R.; Liu, D.J.; Liang, H.; Cang, M. CRISPR/Cas9-mediated specific integration of fat-1 at the goat MSTN locus. FEBS J. 2018, 285, 2828–2839. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Li, H.; Xu, K.; Wu, T.; Wei, J.; Zhou, R.; Liu, Z.; Mu, Y.; Yang, S.; Ouyang, H.; et al. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci. Rep. 2015, 5, 14253. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Lin, J.; Huang, J.; Zhang, H.; Zhang, R.; Zhang, X.; Cao, C.; Hambly, C.; Qin, G.; Yao, J.; et al. Reconstitution of UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs decreases fat deposition and improves thermogenic capacity. Proc. Natl. Acad. Sci. USA 2017, 114, E9474–E9482. [Google Scholar] [CrossRef]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef]
- Han, K.; Liang, L.; Li, L.; Ouyang, Z.; Zhao, B.; Wang, Q.; Liu, Z.; Zhao, Y.; Ren, X.; Jiang, F.; et al. Generation of Hoxc13 knockout pigs recapitulates human ectodermal dysplasia-9. Hum. Mol. Genet. 2017, 26, 184–191. [Google Scholar] [CrossRef]
- Furner, I.J.; Sheikh, M.A.; Collett, C.E. Gene silencing and homology-dependent gene silencing in Arabidopsis: Genetic modifiers and DNA methylation. Genetics 1998, 149, 651–662. [Google Scholar]
- Sanz, J.J.P.J.; Moreno, J.; Merino, S.; Frias, O. Climate change and fitness components of a migratory bird breeding in the Mediterranean region. Glob. Chang. Biol. 2010, 9, 461–472. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. DNA methylation and genetic instability in colorectal cancer cells. Proc. Natl. Acad. Sci. USA 1997, 94, 2545–2550. [Google Scholar] [CrossRef]
- Zingg, J.M.; Jones, P.A. Genetic and epigenetic aspects of DNA methylation on genome expression, evolution, mutation and carcinogenesis. Carcinogenesis 1997, 18, 869–882. [Google Scholar] [CrossRef]
- Taberlay, P.C.; Jones, P.A. DNA methylation and cancer. Fortschritte der Arzneimittelforschung. In Progress in Drug Research; Progrès des Recherches Pharmaceutiques; Springer Nature: Basel, Switzerland; Volume 67, pp. 1–23.
- Zeder, M.A.; Hesse, B. The initial domestication of goats (Capra hircus) in the Zagros mountains 10,000 years ago. Science 2000, 287, 2254–2257. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Wang, S.H.; Sun, B.; Zhang, Y.L.; Shen, W.; Khatib, H.; Wang, X. Melatonin promotes Cashmere goat (Capra hircus) secondary hair follicle growth: A view from integrated analysis of long non-coding and coding RNAs. Cell Cycle 2018, 17, 1255–1267. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.J.; Goldstein, A.L.; White, A. Effects of the thymus lymphocytopoietic factor. Ann. N. Y. Acad. Sci. 1966, 135, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.Y.; Hou, F.; Zhang, Z.P.; Nuo, M.T.; Liang, H.; Cang, M.; Wang, Z.G.; Wang, X.; Xu, T.; Yan, L.Y.; et al. Role of thymosin beta 4 in hair growth. Mol. Genet. Genomics 2016, 291, 1639–1646. [Google Scholar] [CrossRef]
- Gao, X.; Liang, H.; Hou, F.; Zhang, Z.; Nuo, M.; Guo, X.; Liu, D. Thymosin beta-4 induces mouse hair growth. PLoS ONE 2015, 10, e0130040. [Google Scholar] [CrossRef]
- Deborah, P.; Mychi, N.; Brooke, S.; Sharleen, S.S.; Villa, A.M.; Adam, O.; Kleinman, H.K.; Michael, E. Thymosin beta4 increases hair growth by activation of hair follicle stem cells. FASEB J. 2004, 18, 385–387. [Google Scholar]
- Shi, B.; Ding, Q.; He, X.; Zhu, H.; Niu, Y.; Cai, B.; Cai, J.; Lei, A.; Kang, D.; Yan, H. Tβ4-overexpression based on the piggyBac transposon system in cashmere goats alters hair fiber characteristics. Transgenic Res. 2016, 26, 1–9. [Google Scholar] [CrossRef]
- Hao, F.; Yan, W.; Li, X.; Wang, H.; Wang, Y.; Hu, X.; Liu, X.; Liang, H.; Liu, D. Generation of cashmere goats carrying anEDARGene mutant using CRISPR-Cas9-mediated genome editing. Int. J. Biol. Sci. 2018, 14, 427–436. [Google Scholar] [CrossRef]
- He, N.; Dong, Z.; Tai, D.; Liang, H.; Guo, X.; Cang, M.; Liu, D. The role of Sox9 in maintaining the characteristics and pluripotency of Arbas Cashmere goat hair follicle stem cells. Cytotechnology 2018, 70, 1155–1165. [Google Scholar] [CrossRef]
- Nie, Y.; Liu, D. N-Glycosylation is required for FDNC5 stabilization and irisin secretion. Biochem. J. 2017, 474, 3167–3177. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, T.W.; Gown, A.M.; Yaziji, H.; Barnes, M.J.; Schnitt, S.J. Comparison of fluorescence in situ hybridization and immunohistochemistry for the evaluation of HER-2/neu in breast cancer. J. Clin. Oncol. 1999, 17, 1974–1982. [Google Scholar] [CrossRef] [PubMed]
- Nosrati, A.; Naghshvar, F.; Khanari, S. Cancer stem cell markers CD44, CD133 in primary gastric adenocarcinoma. Int. J. Mol. Cell. Med. 2014, 3, 279–286. [Google Scholar] [PubMed]
- Lu, J.; Johnston, A.; Berichon, P.; Ru, K.; Korbie, D.; Trau, M. PrimerSuite: A high-throughput web-based primer design program for multiplex Bisulfite PCR. Sci. Rep. 2017, 7, 41328. [Google Scholar] [CrossRef] [PubMed]
- Kemal, A.; Thomas, H.; Silvia, G.; Jan, V.; Achim, T. Genome-wide quantitative analysis of DNA methylation from bisulfite sequencing data. Bioinformatics 2014, 30, 1933–1934. [Google Scholar]
- Siren, J.; Välimäki, N.; Mäkinen, V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans. Comput. Biol. Bioinform. 2014, 11, 375–388. [Google Scholar] [CrossRef] [PubMed]
- Likun, W.; Zhixing, F.; Xi, W.; Xiaowo, W.; Xuegong, Z. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar]
- Zhang, Y.; Qiujin, X.U.; Beidou amp, X.I.; Zhang, L. Major problems and control measures of water ecological environment in Inner Mongolia-Xinjiang Plateau. J. Lake Sci. 2011, 23, 828–836. [Google Scholar]
- Zhang, J.; Wu, J.; Zeng, W.; Yao, K.; Zu, H.; Zhao, Y. Function of thymosin beta-4 in Ethanol-induced microglial activation. Cell. Physiol. Biochem. 2016, 38, 2230–2238. [Google Scholar] [CrossRef]
- Djekidel, M.N.; Inoue, A.; Matoba, S.; Suzuki, T.; Zhang, C.; Lu, F.; Jiang, L.; Zhang, Y. Reprogramming of chromatin accessibility in somatic cell nuclear transfer is DNA replication independent. Cell Rep. 2018, 23, 1939–1947. [Google Scholar] [CrossRef]
- Chung, Y.G.; Matoba, S.; Liu, Y.; Eum, J.H.; Lu, F.; Jiang, W.; Lee, J.E.; Sepilian, V.; Cha, K.Y.; Lee, D.R.; et al. Histone demethylase expression enhances human somatic cell nuclear transfer efficiency and promotes derivation of pluripotent stem cells. Cell Stem Cell 2015, 17, 758–766. [Google Scholar] [CrossRef] [PubMed]
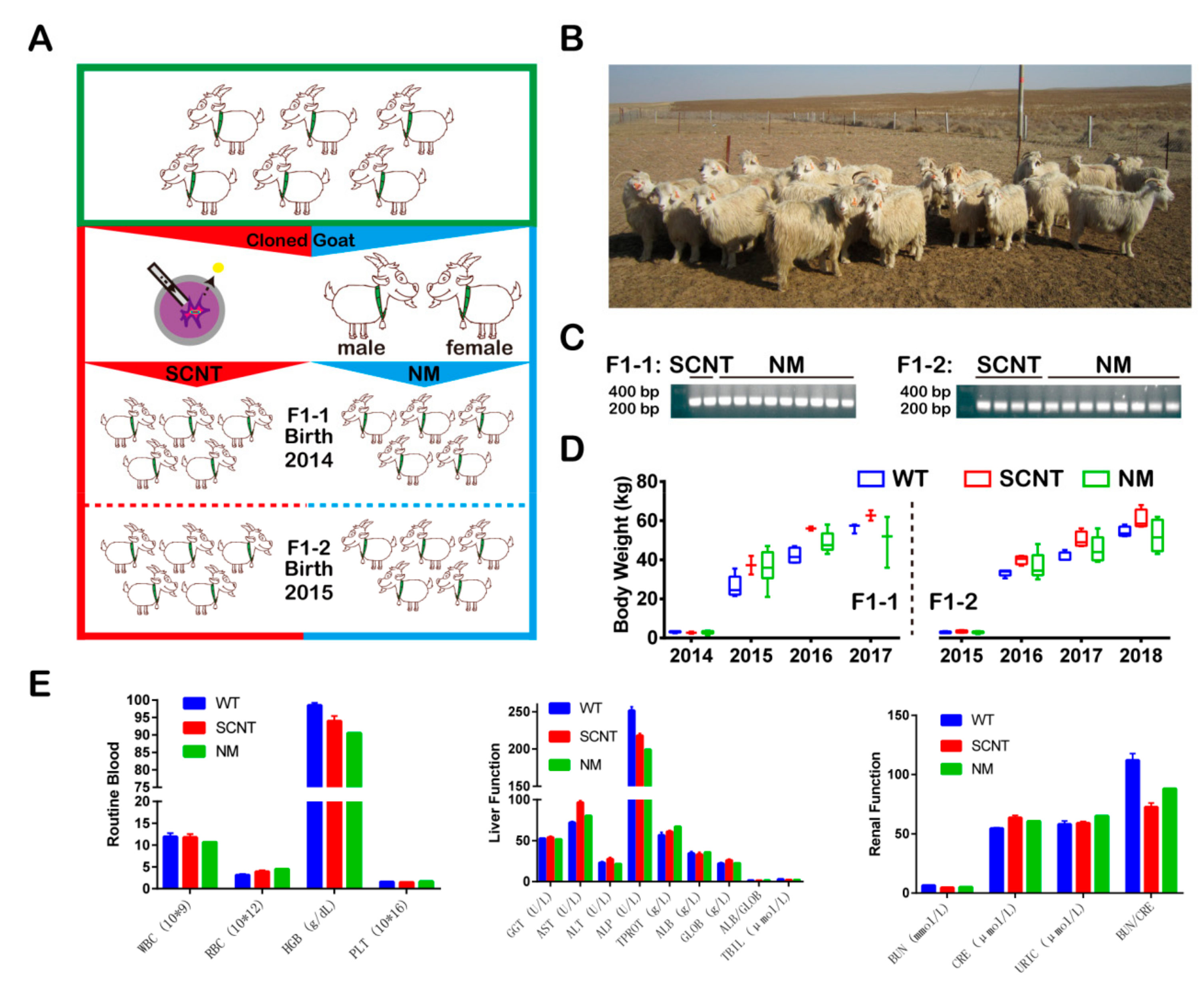
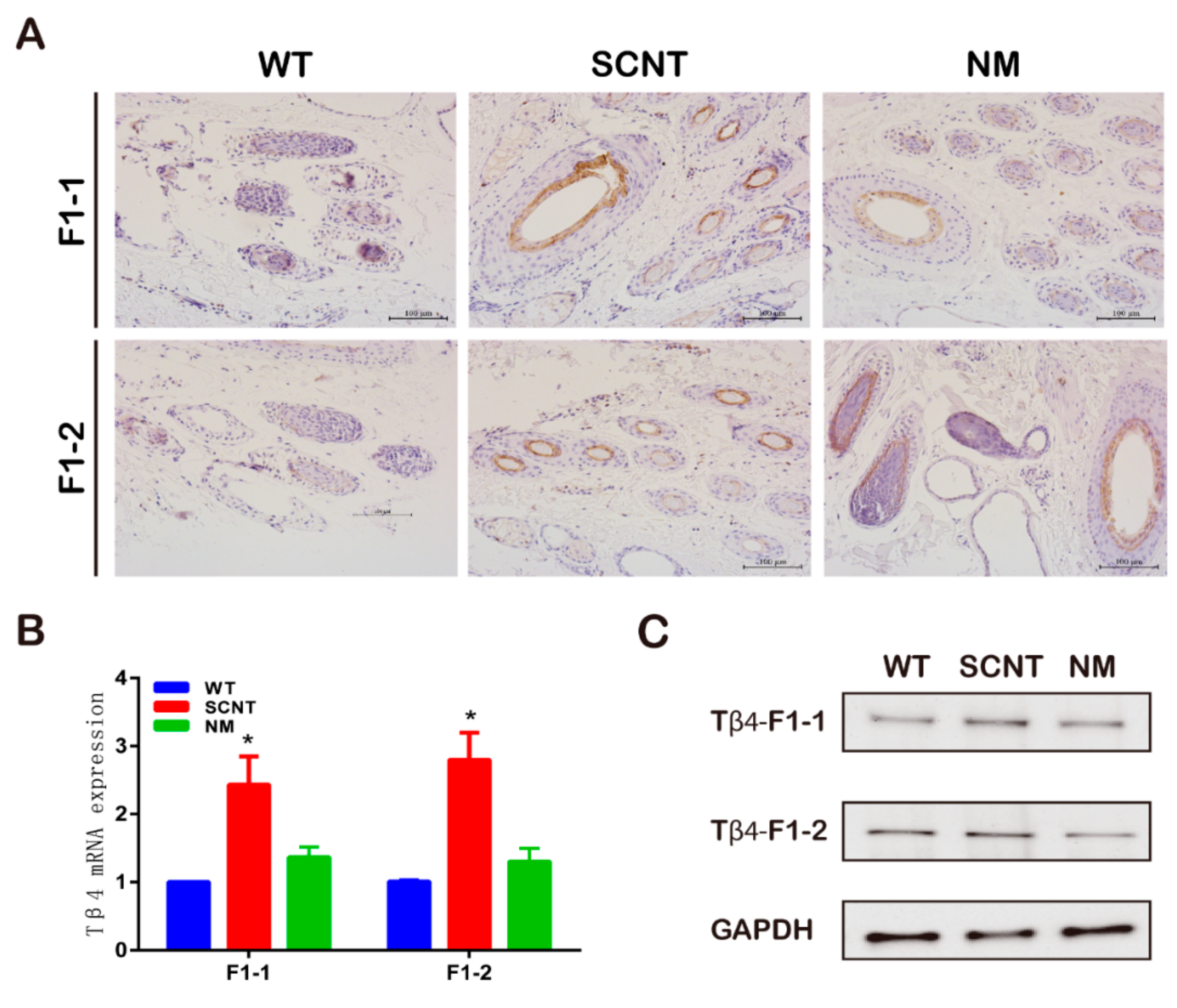
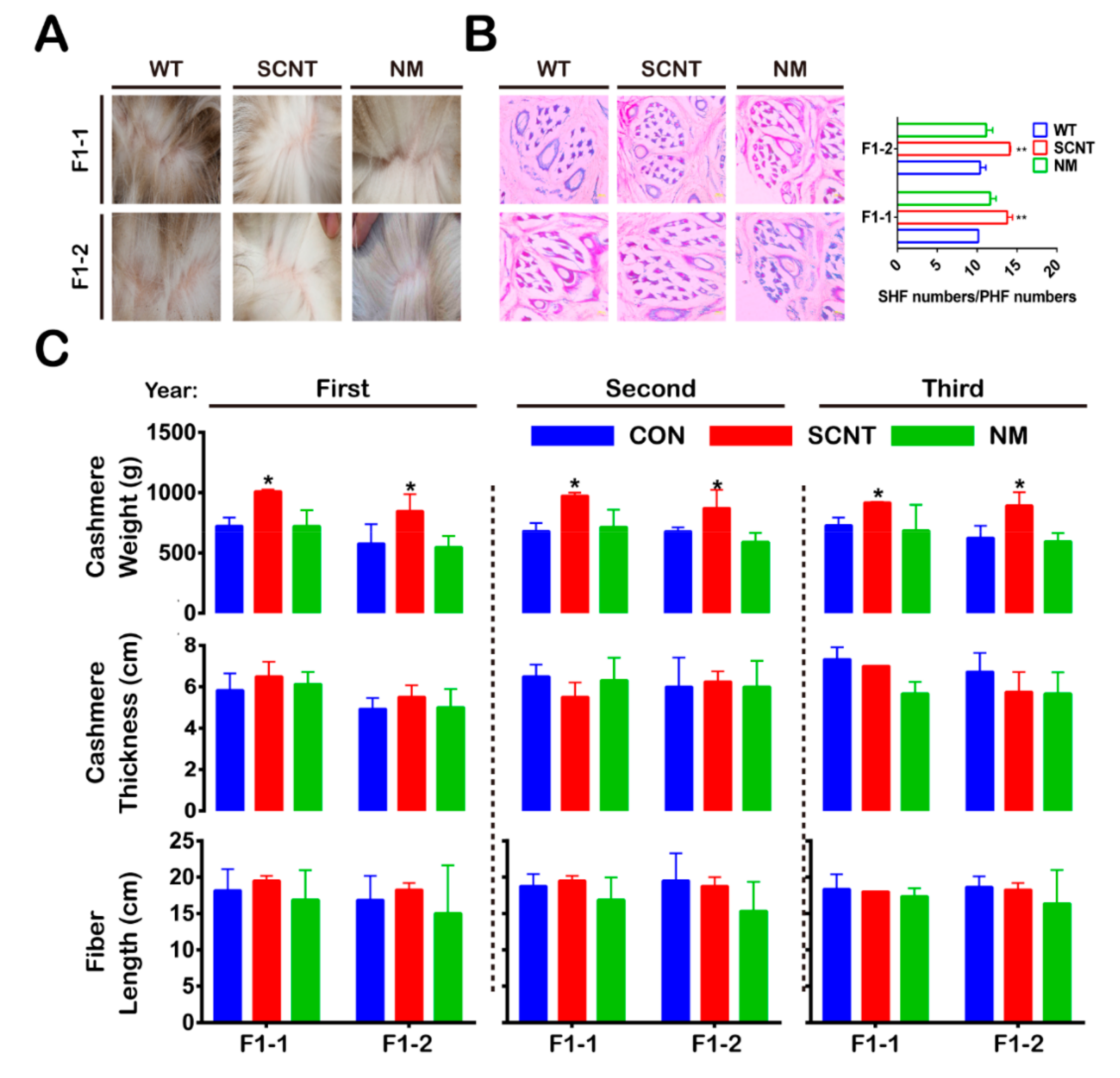

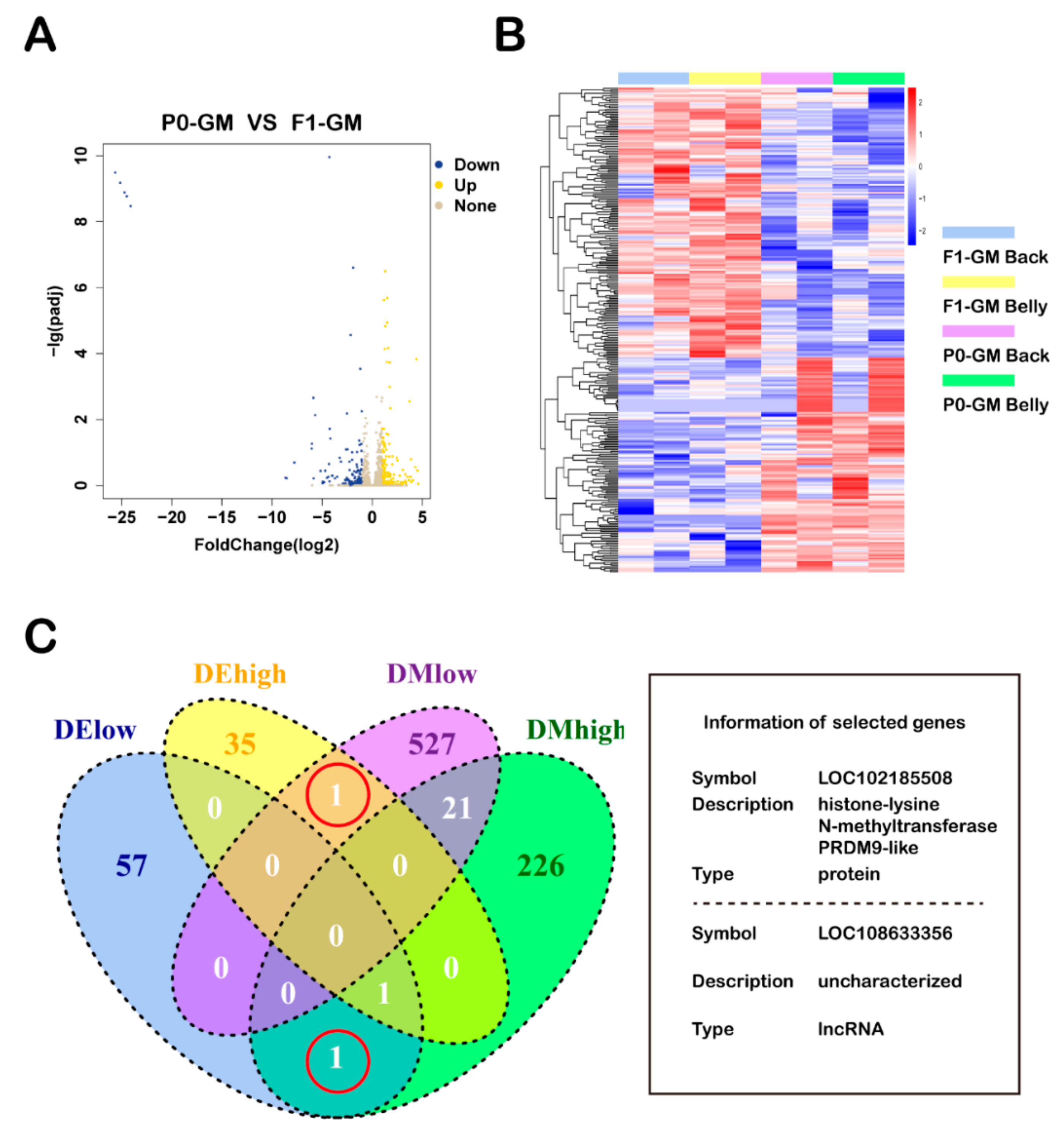
| Birth Year | Type | Group | Number | Number of Experiments-1 12 | Number of Experiments-2 13 | Number of Experiments-3 14 |
|---|---|---|---|---|---|---|
| 2010 | P0 1 | P0-GM 4 | 2 | NA 15 | NA 15 | 2 |
| 2014 | F1-1 2 | F1-1-NM 5 | 9 | 9 | 1 | NA 15 |
| F1-1-SCNT 6 | 2 | 2 | 1 | NA 15 | ||
| F1-1-WT 7 | 7 | 7 | 1 | NA 15 | ||
| 2015 | F1-2 3 | F1-2-NM 8/F1-GM 11 | 8 | 8 | 1 | 2 |
| F1-2-SCNT 9 | 4 | 4 | 1 | NA 15 | ||
| F1-2-WT 10 | 10 | 10 | 1 | NA 15 | ||
| Overall | 42 | 40 | 7 | 4 | ||
| Group | Mature 2 | Clone 3 | Fusion 4 | Cleavage 5 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Failure (n) | Success (n) | Ratio (%) | Failure (n) | Success (n) | Ratio (%) | Failure (n) | Success (n) | Ratio (%) | Failure (n) | 2C 6 (n) | 4C 7 (n) | ≥8C 8 (n) | Success (n) | Ratio (%) | |
| 2014 SCNT 1 | 86 | 150 | 63.56 | 19 | 131 | 87.33 | 33 | 98 | 74.81 | 44 | 0 | 47 | 7 | 54 | 55.10 |
| 65 | 118 | 64.48 | 13 | 105 | 88.98 | 26 | 79 | 75.24 | 18 | 0 | 42 | 19 | 61 | 77.22 | |
| 50 | 85 | 62.96 | 17 | 68 | 80.00 | 18 | 50 | 73.53 | 29 | 0 | 12 | 9 | 21 | 42.00 | |
| 41 | 82 | 66.67 | 4 | 78 | 95.12 | 19 | 59 | 75.64 | 25 | 0 | 14 | 20 | 34 | 57.63 | |
| 40 | 56 | 58.33 | 2 | 54 | 96.43 | 15 | 39 | 72.22 | 19 | 0 | 6 | 14 | 20 | 51.28 | |
| 30 | 35 | 53.85 | 2 | 33 | 94.29 | 8 | 25 | 75.76 | 13 | 0 | 4 | 8 | 12 | 48.00 | |
| 50 | 68 | 57.63 | 15 | 53 | 77.94 | 20 | 33 | 62.26 | 23 | 0 | 9 | 1 | 10 | 30.30 | |
| 32 | 117 | 78.52 | 17 | 100 | 85.47 | 30 | 70 | 70.00 | 42 | 0 | 20 | 8 | 28 | 40.00 | |
| 46 | 70 | 60.34 | 27 | 43 | 61.43 | 6 | 37 | 86.05 | 23 | 0 | 8 | 6 | 14 | 37.84 | |
| 28 | 42 | 60.00 | 1 | 41 | 97.62 | 18 | 23 | 56.10 | 15 | 0 | 8 | 0 | 8 | 34.78 | |
| 16 | 49 | 75.38 | 7 | 42 | 85.71 | 13 | 29 | 69.05 | 14 | 0 | 13 | 2 | 15 | 51.72 | |
| 53 | 39 | 42.39 | 7 | 32 | 82.05 | 14 | 18 | 56.25 | 5 | 0 | 9 | 4 | 13 | 72.22 | |
| Overall | 537 | 911 | 62.91 | 131 | 780 | 85.62 | 220 | 560 | 71.79 | 270 | 0 | 192 | 98 | 290 | 51.79 |
| 2015 SCNT 1 | 27 | 79 | 74.53 | 0 | 79 | 100.00 | 27 | 52 | 65.82 | 40 | 12 | 0 | 0 | 12 | 23.08 |
| 54 | 130 | 70.65 | 0 | 130 | 100.00 | 55 | 75 | 57.69 | 66 | 9 | 0 | 0 | 9 | 12.00 | |
| 46 | 119 | 72.12 | 0 | 119 | 100.00 | 42 | 77 | 64.71 | 35 | 42 | 0 | 0 | 42 | 54.55 | |
| 51 | 93 | 64.58 | 0 | 93 | 100.00 | 40 | 53 | 56.99 | 43 | 4 | 0 | 0 | 4 | 8.51 | |
| 36 | 107 | 74.83 | 0 | 107 | 100.00 | 49 | 58 | 54.21 | 41 | 17 | 0 | 0 | 17 | 29.31 | |
| 28 | 135 | 82.82 | 0 | 135 | 100.00 | 67 | 68 | 50.37 | 30 | 32 | 0 | 0 | 32 | 51.61 | |
| 33 | 118 | 78.15 | 0 | 118 | 100.00 | 45 | 73 | 61.86 | 47 | 23 | 0 | 0 | 23 | 32.86 | |
| 36 | 84 | 70.00 | 0 | 84 | 100.00 | 21 | 63 | 75.00 | 37 | 23 | 0 | 0 | 23 | 38.33 | |
| Overall | 311 | 865 | 73.55 | 0 | 865 | 100.00 | 346 | 519 | 60.00 | 339 | 162 | 0 | 0 | 162 | 32.34 |
| Birth Year | Group | Oocytes(n) | SCNT 1 Embryos (n) | Embryos Transferred (n) | Surrogates (n) | Pregnancies (n) | Live Births (n) | Surviving Offspring (n) | Sex (♀/♂) |
|---|---|---|---|---|---|---|---|---|---|
| 2014 | F1-1-NM 2 | NA 8 | NA 8 | NA 8 | NA 8 | 7 | 9 | 9 | 1/8 |
| F1-1-SCNT 3 | 1448 | 290 | 177 | 46 | 2 | 2 | 2 | 0/2 | |
| F1-1-1-WT 4 | NA 8 | NA 8 | NA 8 | NA 8 | 4 | 7 | 7 | 1/6 | |
| 2015 | F1-2-NM 5 | NA 8 | NA 8 | NA 8 | NA 8 | 5 | 8 | 8 | 5/3 |
| F1-2-SCNT 6 | 1176 | 501 | 367 | 99 | 4 | 4 | 4 | 2/2 | |
| F1-2-WT 7 | NA 8 | NA 8 | NA 8 | NA 8 | 6 | 10 | 10 | 3/7 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dai, B.; Zhang, M.; Yuan, J.-L.; Ren, L.-Q.; Han, X.-Y.; Liu, D.-J. Integrative Analysis of Methylation and Transcriptional Profiles to Reveal the Genetic Stability of Cashmere Traits in the Tβ4 Overexpression of Cashmere Goats. Animals 2019, 9, 1002. https://doi.org/10.3390/ani9121002
Dai B, Zhang M, Yuan J-L, Ren L-Q, Han X-Y, Liu D-J. Integrative Analysis of Methylation and Transcriptional Profiles to Reveal the Genetic Stability of Cashmere Traits in the Tβ4 Overexpression of Cashmere Goats. Animals. 2019; 9(12):1002. https://doi.org/10.3390/ani9121002
Chicago/Turabian StyleDai, Bai, Meng Zhang, Jian-Long Yuan, Li-Qing Ren, Xiao-Yu Han, and Dong-Jun Liu. 2019. "Integrative Analysis of Methylation and Transcriptional Profiles to Reveal the Genetic Stability of Cashmere Traits in the Tβ4 Overexpression of Cashmere Goats" Animals 9, no. 12: 1002. https://doi.org/10.3390/ani9121002
APA StyleDai, B., Zhang, M., Yuan, J.-L., Ren, L.-Q., Han, X.-Y., & Liu, D.-J. (2019). Integrative Analysis of Methylation and Transcriptional Profiles to Reveal the Genetic Stability of Cashmere Traits in the Tβ4 Overexpression of Cashmere Goats. Animals, 9(12), 1002. https://doi.org/10.3390/ani9121002




