Iodine Supplemented Diet Positively Affect Immune Response and Dairy Product Quality in Fresian Cow
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal and Study Design
2.2. Blood and Milk Sampling
2.3. Blood Analysis
2.4. Chemical Analysis of Milk
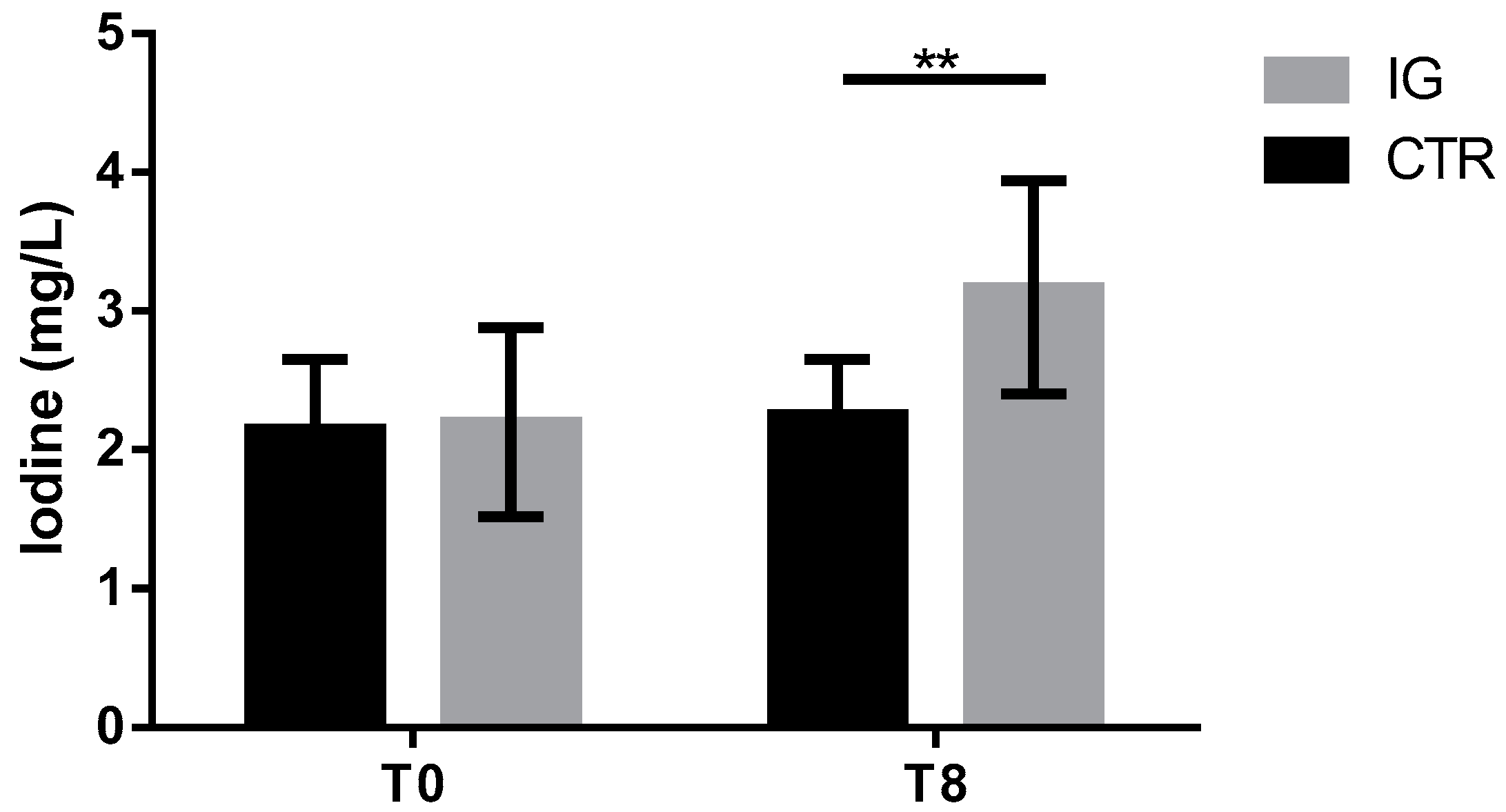
2.5. Iodine Determination in Milk
2.6. Library Preparation and RNA-Seq Analysis
2.7. Enriched Pathway Analysis
2.8. Ricotta Cheese-Making Procedure and Lipid Oxidation by Thiobarbituric Acid Reactive Substance Test
2.9. Statistics
3. Results
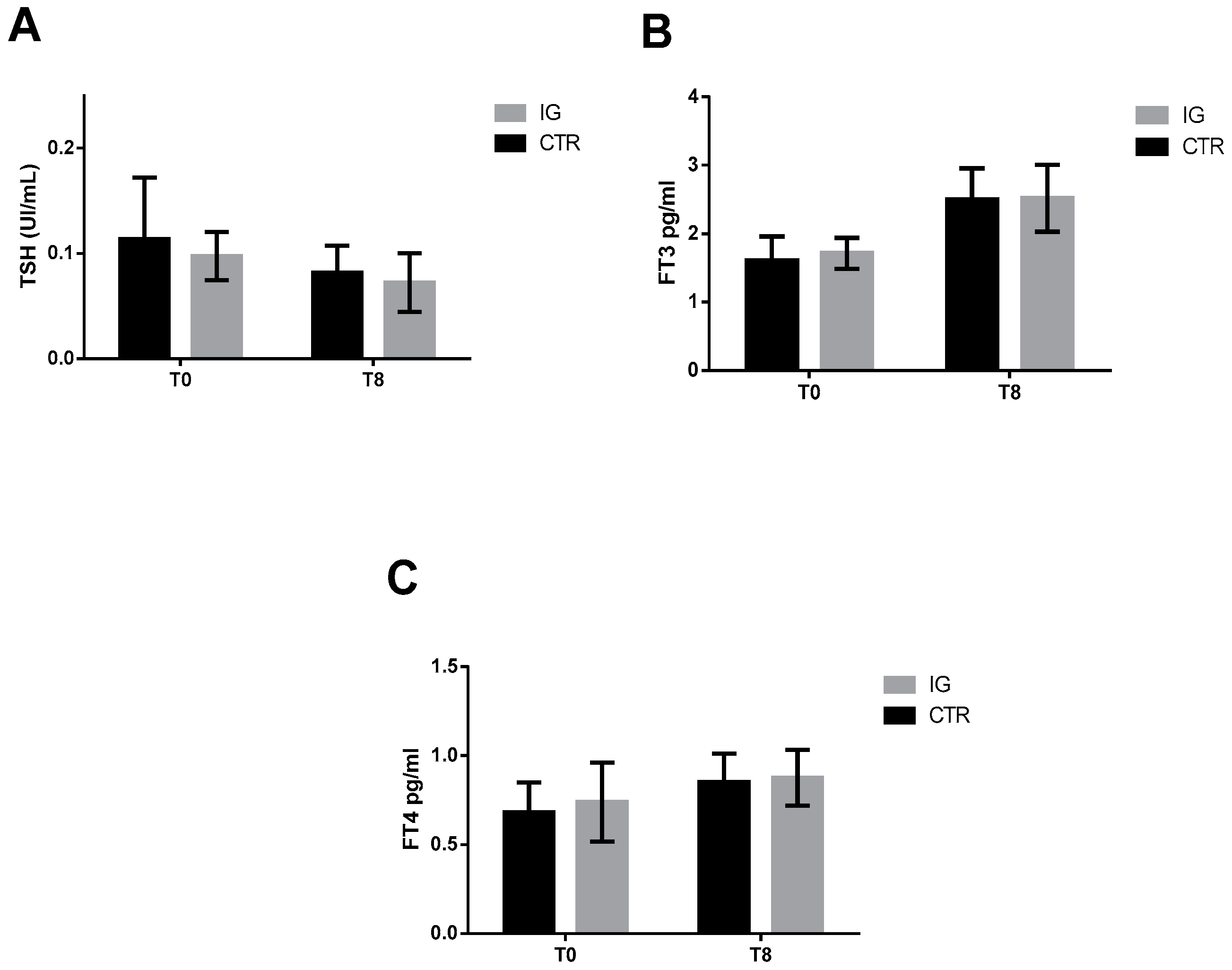
3.1. Serum Thyroid Hormone and Iodine Concentrations
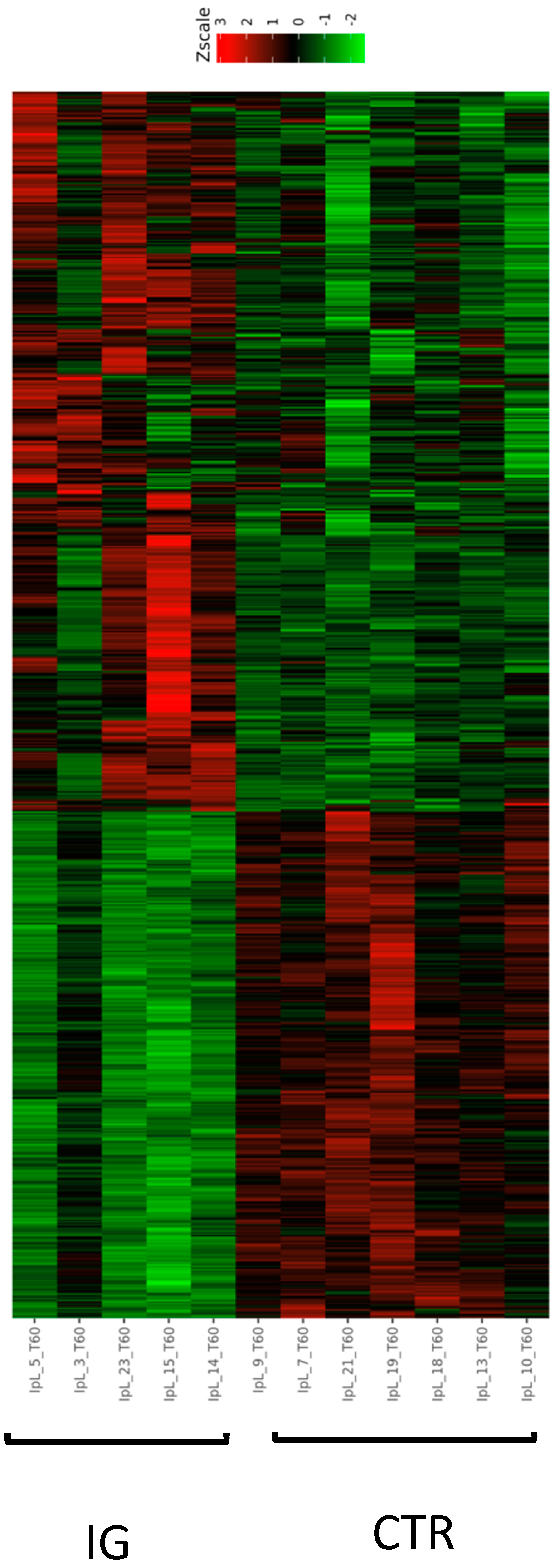
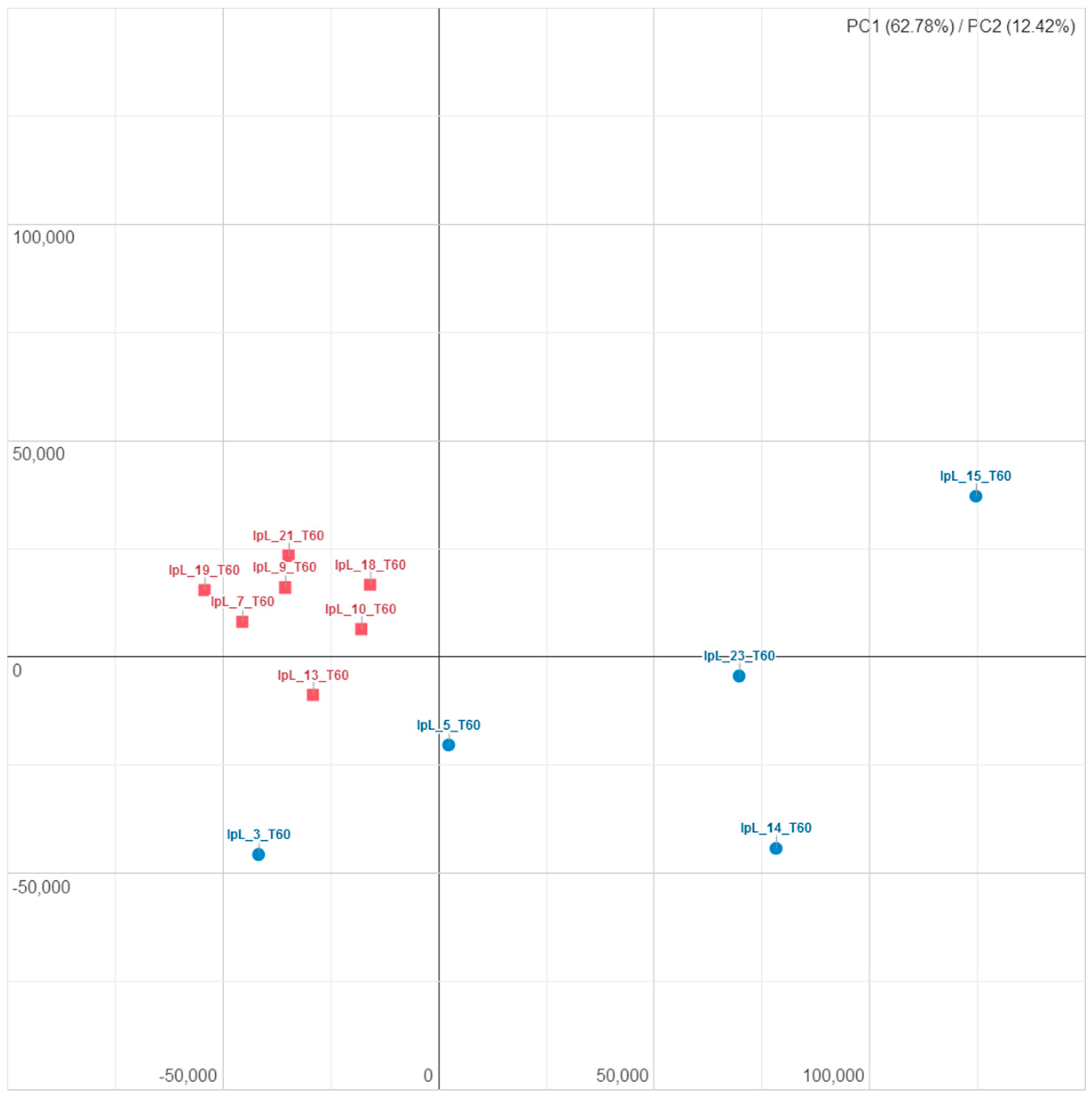
3.2. Influence of I-supplemented Diet on Blood Transcriptome
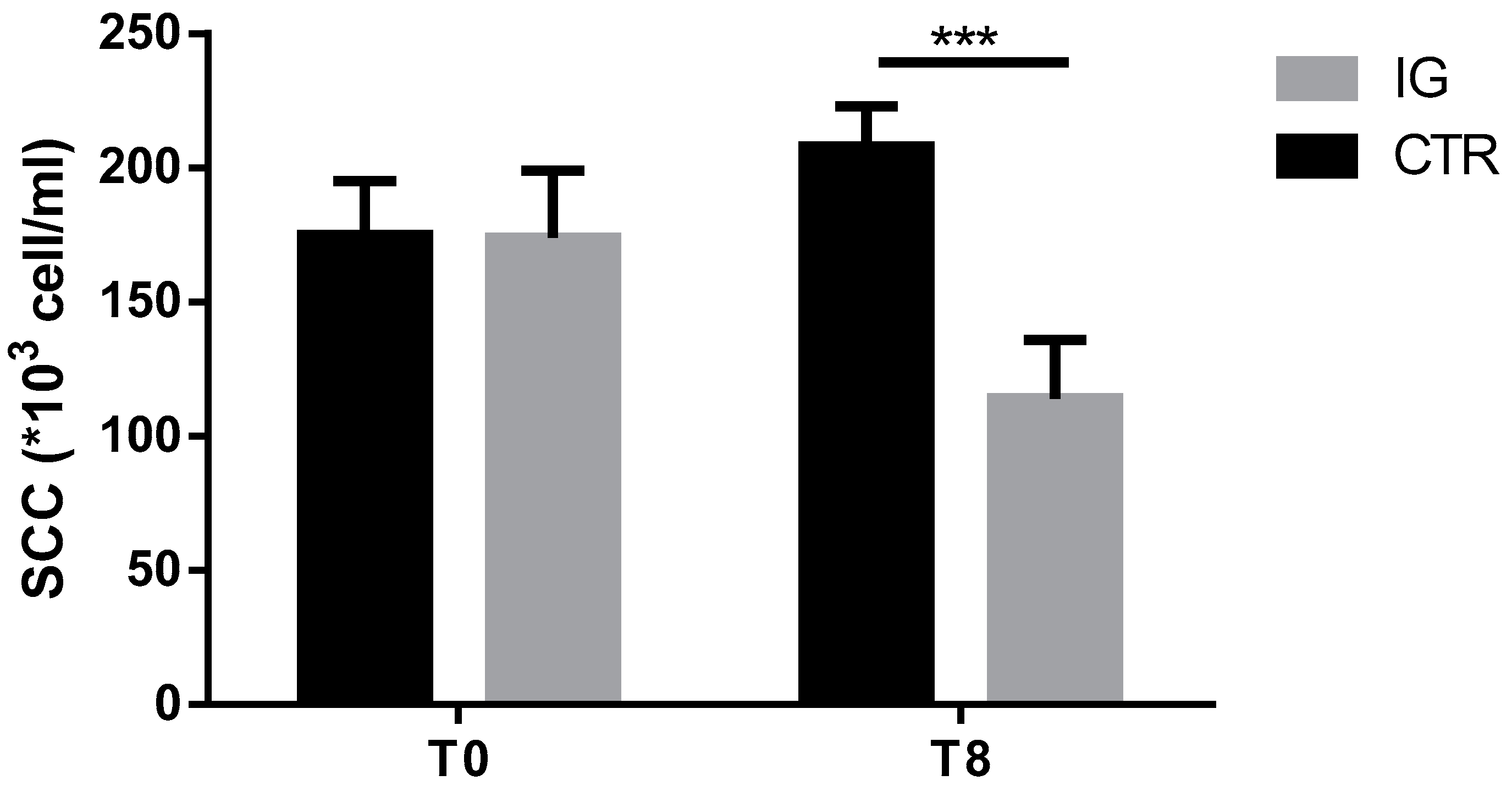
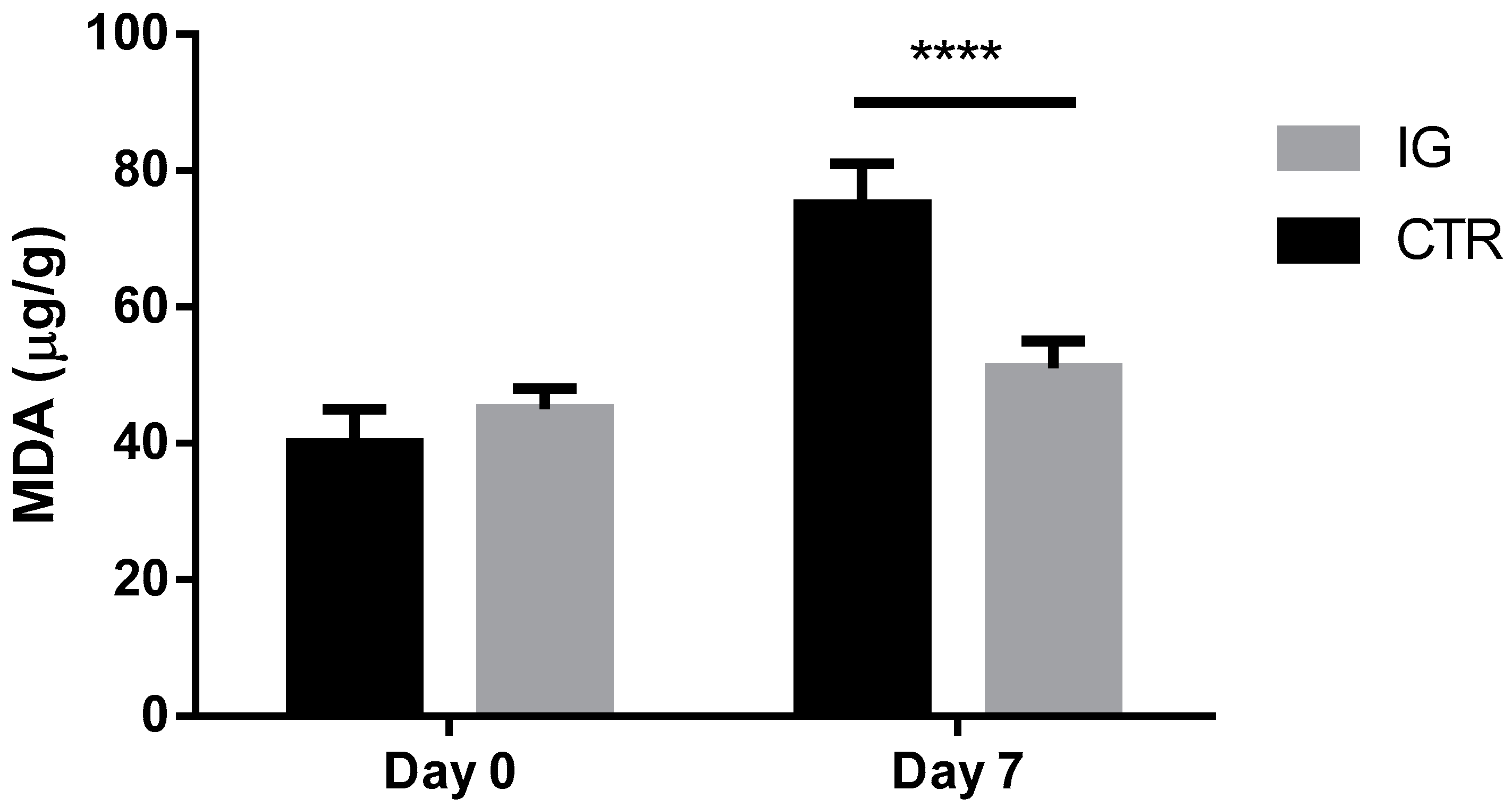
3.3. Effects of I-Supplementation on Quality of Dairy Products
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zimmermann, M.B. Iodine deficiency. Endocr. Rev. 2009, 30, 376–408. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsdottir, I.; Dahl, L. Iodine intake in human nutrition: A systematic literature review. Food Nutr. Res. 2012, 56. [Google Scholar] [CrossRef] [PubMed]
- Tetens, I. EFSA Panel on Dietetic Products Nutrition and Allergies. Scientific Opinion on Dietary Reference Values for iodine. EFSA J. 2014, 12, 3660. [Google Scholar]
- Ghirri, P.; Lunardi, S.; Boldrini, A. Iodine Supplementation in the Newborn. Nutrients 2014, 6, 382–390. [Google Scholar] [CrossRef] [PubMed]
- Flachowsky, G.; Franke, K.; Meyer, U.; Leiterer, M.; Schöne, F. Influencing factors on iodine content of cow milk. Eur. J. Nutr. 2014, 53, 351–365. [Google Scholar] [CrossRef] [PubMed]
- EU. Commission Implementing Regulation 2015/861 of 3 June 2015 concerning the authorisation of potassium iodide, calcium iodate anhydrous and coated granulated calcium iodate anhydrous as feed additives for all animal species. Off. J. Eur. Union. 2015, L137, 1–7. [Google Scholar]
- Weiss, W.P.; Wyatt, D.J.; Kleinschmit, D.H.; Socha, M.T. Effect of including canola meal and supplemental iodine in diets of dairy cows on short-term changes in iodine concentrations in milk. J. Dairy Sci. 2015, 98, 4841–4849. [Google Scholar] [CrossRef] [PubMed]
- Nudda, A.; Battacone, G.; Decandia, M.; Acciaro, M.; Aghini-Lombardi, F.; Frigeri, M.; Pulina, G. The effect of dietary iodine supplementation in dairy goats on milk production traits and milk iodine content. J. Dairy Sci. 2009, 92, 5133–5138. [Google Scholar] [CrossRef]
- Maas, J.; Davis, L.E.; Hempstead, C.; Berg, J.N.; Hoffman, K.A. Efficacy of ethylenediamine dihydriodide in the prevention of naturally occurring foot rot in cattle. Am. J. Vet. Res. 1984, 45, 2347–2350. [Google Scholar]
- Kulow, M.; Merkatoris, P.; Anklam, K.S.; Rieman, J.; Larson, C.; Branine, M.; Döpfer, D. Evaluation of the prevalence of digital dermatitis and the effects on performance in beef feedlot cattle under organic trace mineral supplementation. J. Anim. Sci. 2017, 95, 3435–3444. [Google Scholar] [CrossRef]
- Antaya, N.T.; Soder, K.J.; Kraft, J.; Whitehouse, N.L.; Guindon, N.E.; Erickson, P.S.; Conroy, A.B.; Brito, A.F. Incremental amounts of Ascophyllum nodosum meal do not improve animal performance but do increase milk iodine output in early lactation dairy cows fed high-forage diets. J. Dairy Sci. 2015, 98, 1991–2004. [Google Scholar] [CrossRef] [PubMed]
- Derscheid, R.J.; van Geelen, A.; Berkebile, A.R.; Gallup, J.M.; Hostetter, S.J.; Banfi, B.; McCray, P.B., Jr.; Ackermann, M.R. Increased Concentration of Iodide in Airway Secretions Is Associated with Reduced Respiratory Syncytial Virus Disease Severity. Am. J. Respir. Cell Mol. Biol. 2014, 50, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Iannaccone, M.; Elgendy, R.; Ianni, A.; Martino, C.; Palazzo, F.; Giantin, M.; Grotta, L.; Dacasto, M.; Martino, G. Whole-transcriptome profiling of sheep fed with a high iodine-supplemented diet. Animal 2019. [Google Scholar] [CrossRef] [PubMed]
- Iannaccone, M.; Ianni, A.; Ramazzotti, S.; Grotta, L.; Marone, E.; Cichelli, A.; Martino, G. Whole Blood Transcriptome Analysis Reveals Positive Effects of Dried Olive Pomace-Supplemented Diet on Inflammation and Cholesterol in Laying Hens. Animals 2019, 9, 427. [Google Scholar] [CrossRef] [PubMed]
- Lopez, C.C.; Serio, A.; Rossi, C.; Mazzarrino, G.; Marchetti, S.; Castellani, F.; Grotta, L.; Fiorentino, F.P.; Paparella, A.; Martino, G. Effect of diet supplementation with Ascophyllum nodosum on cow milk composition and microbiota. J. Dairy Sci. 2016, 99, 6285–6297. [Google Scholar] [CrossRef] [PubMed]
- EU. Directive 2010/63/EU of the European Parliament and of the Council of 22 September 2010 on the protection of animals used for scientific purposes. Off. J. Eur. Union. 2010, L276, 33–79. [Google Scholar]
- EC. No 1459/2005 of 8 September 2005 amending the conditions for authorisation of a number of feed additives belonging to the group of trace elements. Off. J. Eur. Union. 2005, L233, 8–10. [Google Scholar]
- Fecher, P.A.; Goldmann, I.; Nagengast, A. Determination of iodine in food samples by inductively coupled plasma mass spectrometry after alkaline extraction. J. Anal. At. Spectrom. 1998, 13, 977–982. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ianni, A.; Innosa, D.; Martino, C.; Grotta, L.; Bennato, F.; Martino, G. Zinc supplementation of Friesian cows: Effect on chemical-nutritional composition and aromatic profile of dairy products. J. Dairy Sci. 2019, 102, 2918–2927. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, M.B. The Effects of Iodine Deficiency in Pregnancy and Infancy. Paediatr. Perinat. Epidemiol. 2012, 26, 108–117. [Google Scholar] [CrossRef] [PubMed]
- De Vito, P.; Incerpi, S.; Pedersen, J.Z.; Luly, P.; Davis, F.B.; Davis, P.J. Thyroid Hormones as Modulators of Immune Activities at the Cellular Level. Thyroid 2011, 21, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Singh, N.K.; Bhadwal, M.S. Relationship of Somatic Cell Count and Mastitis: An Overview. Asian-Australas. J. Anim. Sci. 2011, 24, 429–438. [Google Scholar] [CrossRef]
- Iannaccone, M.; Elgendy, R.; Giantin, M.; Martino, C.; Giansante, D.; Ianni, A.; Dacasto, M.; Martino, G. RNA Sequencing-Based Whole-Transcriptome Analysis of Friesian Cattle Fed with Grape Pomace-Supplemented Diet. Animals 2018, 8, 188. [Google Scholar] [CrossRef]
- Elgendy, R.; Giantin, M.; Castellani, F.; Grotta, L.; Palazzo, F.; Dacasto, M.; Martino, G. Transcriptomic signature of high dietary organic selenium supplementation in sheep: A nutrigenomic insight using a custom microarray platform and gene set enrichment analysis. J. Anim. Sci. 2016, 94, 3169–3184. [Google Scholar] [CrossRef]
- Elgendy, R.; Palazzo, F.; Castellani, F.; Giantin, M.; Grotta, L.; Cerretani, L.; Dacasto, M.; Martino, G. Transcriptome profiling and functional analysis of sheep fed with high zinc-supplemented diet: A nutrigenomic approach. Anim. Feed Sci. Tech. 2017, 234, 195–2042. [Google Scholar] [CrossRef]
- Bilal, M.Y.; Dambaeva, S.; Kwak-Kim, J.; Gilman-Sachs, A.; Beaman, K. A Role for Iodide and Thyroglobulin in Modulating the Function of Human Immune Cells. Front. Immunol. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Weetman, A.P.; Mcgregor, A.M.; Campbell, H.; Lazarus, J.H.; Ibbertson, H.K.; Hall, R. Iodide enhances IgG synthesis by human peripheral blood lymphocytes in vitro. Eur. J. Endocrinol. 1983, 103, 210–215. [Google Scholar] [CrossRef]
- Klebanoff, S.J.; Kettle, A.J.; Rosen, H.; Winterbourn, C.C.; Nauseef, W.M. Myeloperoxidase: A front-line defender against phagocytosed microorganisms. J. Leukoc. Biol. 2013, 93, 185–198. [Google Scholar] [CrossRef]
- Soboll, S. Thyroid hormone action on mitochondrial energy transfer. Biochim. Biophis. Acta Bioenerg. 1993, 1144, 1–16. [Google Scholar] [CrossRef]
- Venturi, S. Evolutionary Significance of Iodine. Curr. Chem. Biol. 2011, 5, 155–162. [Google Scholar] [CrossRef]
- Grace, N.D.; Waghorn, G.C. Impact of iodine supplementation of dairy cows on milk production and iodine concentrations in milk. N. Z. Vet. J. 2005, 53, 10–13. [Google Scholar] [CrossRef] [PubMed]
| Ingredients of TMR | |
| Corn silage, % | 23.7 |
| First cut, alfalfa hay, % | 5.3 |
| Corn meal, % | 3.4 |
| Soybean, meal, % | 3.2 |
| Fine bran, % | 3.0 |
| Barley, meal, % | 1.9 |
| CaCO3, % | 0.2 |
| Vitamins and minerals, % | 0.4 |
| Kg of dry matter/head per day | 22.41 |
| Chemical Composition of TMR | |
| Dry Matter, % | 56.76 |
| Crude protein 1, % | 15.34 |
| Ether extract 1, % | 2.97 |
| Ash 1, % | 5.31 |
| Neutral detergent fiber 1, % | 32.51 |
| Acid detergent fiber 1, % | 20.03 |
| Starch 1, % | 27.02 |
| Iodine (mg/head/day) 1 | 20 (+65) * |
| Pathway | FDR | Genes |
|---|---|---|
| regulation of transcription, DNA-templated | 1.30 × 10−5 | ZNF93, SBNO1, ZBTB6, SNAPC3, ZFX, ZNF12, LOC530973, ZNF655, LOC104968476, ZNF175, MAP3K7, ZNF184, JADE1, ZNF182, ZNF148, RBAK, ZNF248, ZNF286A, DNTTIP2, MYNN, ZNF605, ZNF572, ZNF436 |
| cellular response to DNA damage stimulus | 7.45 × 10−4 | SHPRH, HELB, ZMAT3, FMR1, RNF168, SPRTN, USP16, NEK4 |
| transcription, DNA-templated | 0.0057 | CCNT2, ZKSCAN8, ZBTB6, SNAPC3, ZNF131, ZFX, SCAI, ZNF518A, MAP3K7, ZNF184, JADE1, ZNF148, PSIP1, DNTTIP2, USP16, MYNN, ZNF572 |
| RNA processing | 0.0153 | DHX29, U2SURP, YTHDC2, DHX36 |
| negative regulation of transforming growth factor beta receptor signaling pathway | 0.0186 | ZNF451, LEMD3, SMURF2, SIRT1 |
| cell division | 0.0473 | CDC40, USP37, USP16, CENPJ, SPICE1, SMC3 |
| Pathway | FDR | Genes |
|---|---|---|
| Fc gamma R-mediated phagocytosis | 2.63 × 10−6 | AKT1, AKT2, ARF6, ARPC1B, CFL1, CRKL, GSN, LIMK1, RAC2, SCIN, VASP |
| Non-alcoholic fatty liver disease (NAFLD) | 1.33 × 10−5 | AKT1, AKT2, COX8A, GSK3A, MAP3K11, NDUFA7, NDUFB7, NDUFS6, NDUFS7, NDUFS8, SDHA, TGFB1, UQCR11 |
| Oxidative phosphorylation | 0.0005 | ATP5D, ATP5G2, COX8A, NDUFA7, NDUFB7, NDUFS6, NDUFS7, NDUFS8, SDHA, UQCR11 |
| Rap1 signaling pathway | 0.0008 | ACTB, ACTG1, AKT1, AKT2, CRKL, MAP2K3, RAC2, RAPGEF1, RASSF5, SIPA1, TLN1, VASP |
| Bacterial invasion of epithelial cells | 0.0016 | ACTB, ACTG1, ARPC1B, CRKL, PXN, RHOG, SEPT9 |
| Proteasome | 0.0078 | PSMB10, PSMB4, PSMB8, PSMB9, PSMD4 |
| Carbon metabolism | 0.0079 | ENO1, GPI, HK1, IDH2, PFKL, SDHA, TPI1 |
| Focal adhesion | 0.0079 | ACTB, ACTG1, AKT1, AKT2, CRKL, PXN, RAC2, RAPGEF1, TLN1, VASP |
| Regulation of actin cytoskeleton | 0.0079 | ACTB, ACTG1, ARPC1B, CFL1, CRKL, GSN, LIMK1, PXN, RAC2, SCIN |
| B cell receptor signaling pathway | 0.0024 | AKT1, AKT2, CD81, NFKBIB, RAC2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iannaccone, M.; Ianni, A.; Elgendy, R.; Martino, C.; Giantin, M.; Cerretani, L.; Dacasto, M.; Martino, G. Iodine Supplemented Diet Positively Affect Immune Response and Dairy Product Quality in Fresian Cow. Animals 2019, 9, 866. https://doi.org/10.3390/ani9110866
Iannaccone M, Ianni A, Elgendy R, Martino C, Giantin M, Cerretani L, Dacasto M, Martino G. Iodine Supplemented Diet Positively Affect Immune Response and Dairy Product Quality in Fresian Cow. Animals. 2019; 9(11):866. https://doi.org/10.3390/ani9110866
Chicago/Turabian StyleIannaccone, Marco, Andrea Ianni, Ramy Elgendy, Camillo Martino, Mery Giantin, Lorenzo Cerretani, Mauro Dacasto, and Giuseppe Martino. 2019. "Iodine Supplemented Diet Positively Affect Immune Response and Dairy Product Quality in Fresian Cow" Animals 9, no. 11: 866. https://doi.org/10.3390/ani9110866
APA StyleIannaccone, M., Ianni, A., Elgendy, R., Martino, C., Giantin, M., Cerretani, L., Dacasto, M., & Martino, G. (2019). Iodine Supplemented Diet Positively Affect Immune Response and Dairy Product Quality in Fresian Cow. Animals, 9(11), 866. https://doi.org/10.3390/ani9110866





