Copy Number Variations of KLF6 Modulate Gene Transcription and Growth Traits in Chinese Datong Yak (Bos Grunniens)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Areas and Quantitative Traits
2.2. Sample Collection, Deoxyribonucleic Acid (DNA) and Ribonucleic Acid (RNA) Isolation and cDNA Synthesis
2.3. Primer Design and PCR Amplification
2.4. Analysis of Copy Number Variation and Expression of the KLF6 Gene
2.5. Statistical Analysis
3. Results
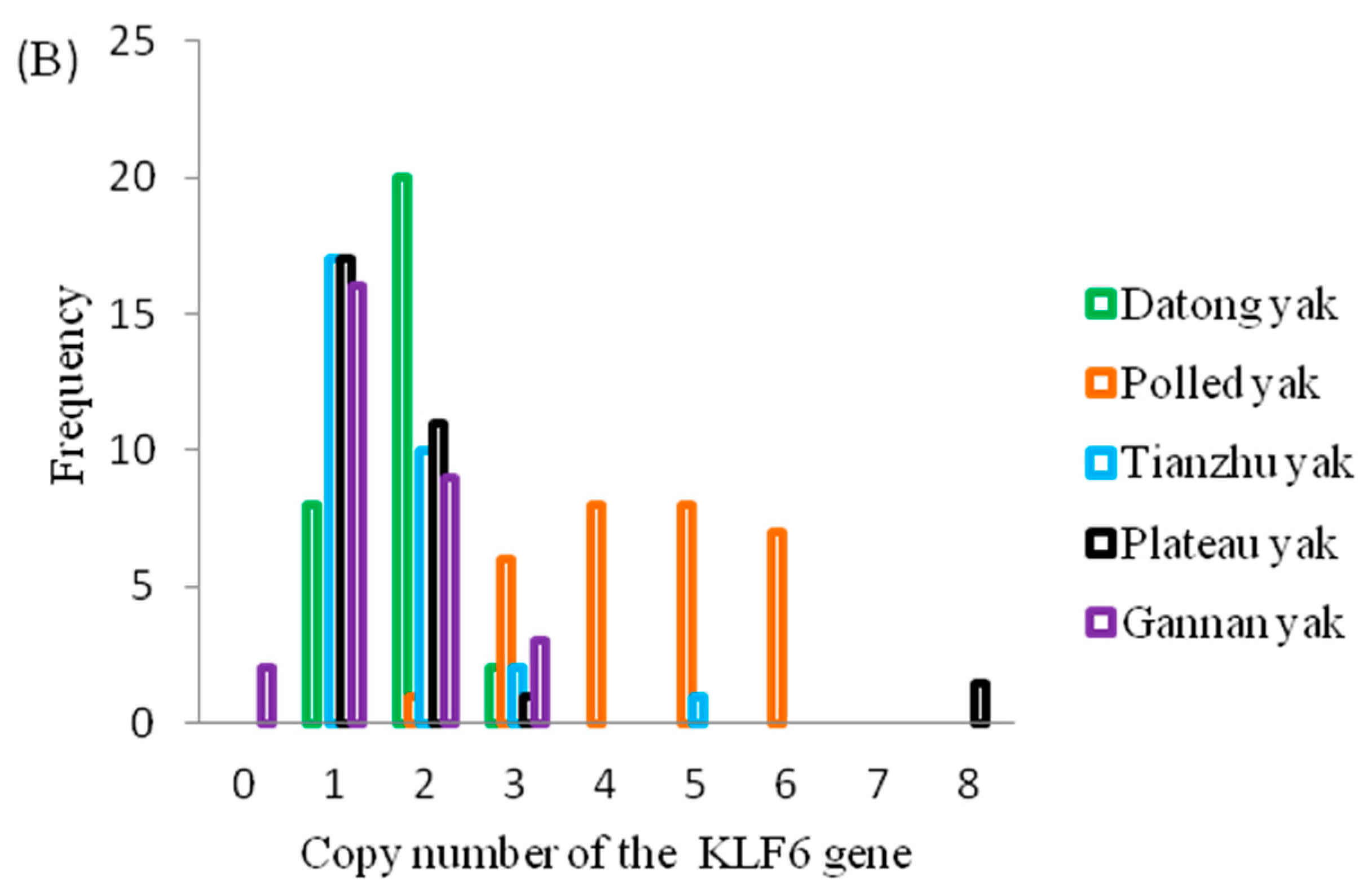
3.1. Association of Gene CNVs with Growth Traits in Yak
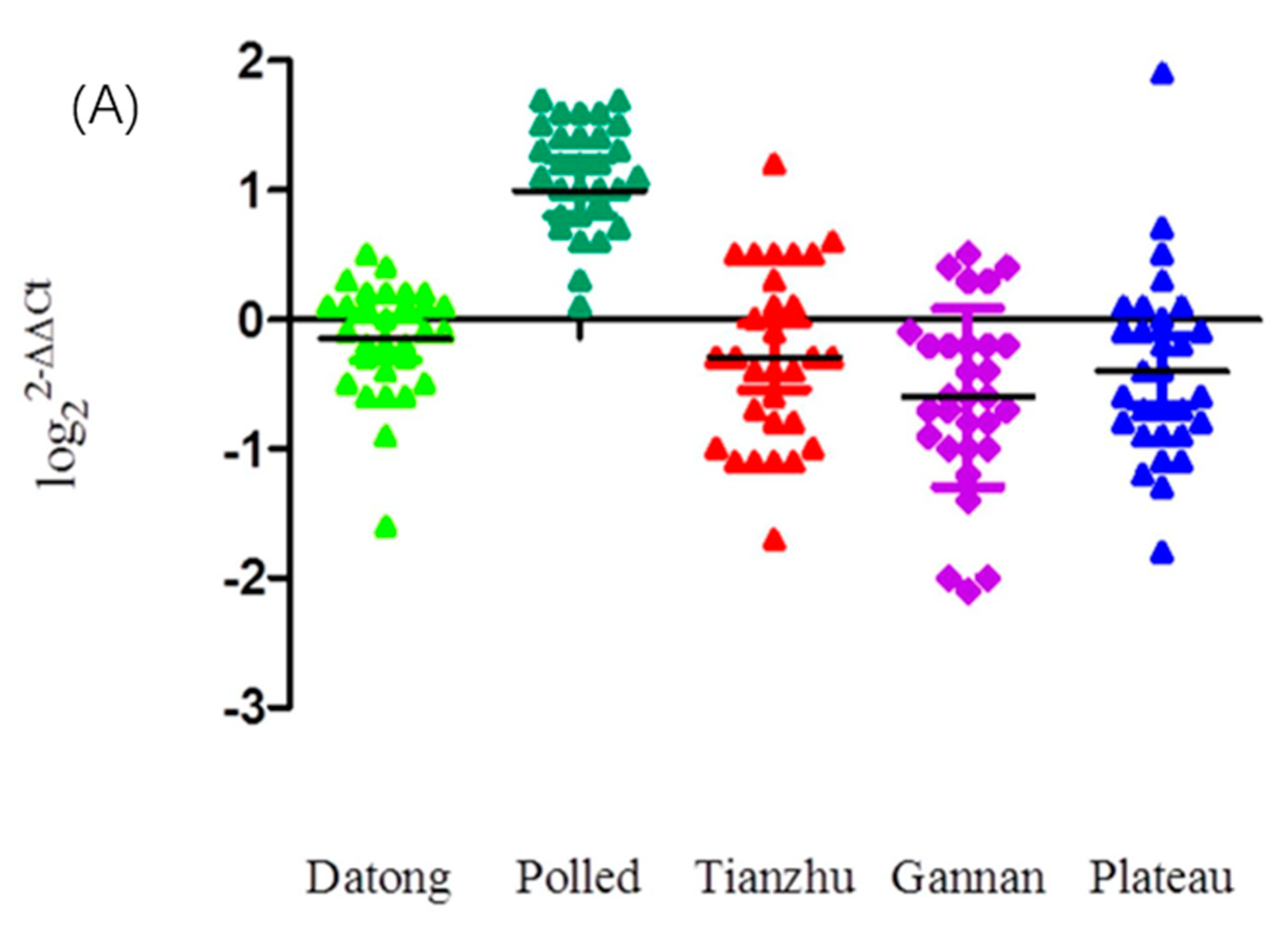
3.2. Population Genetic Analyses of CNVs
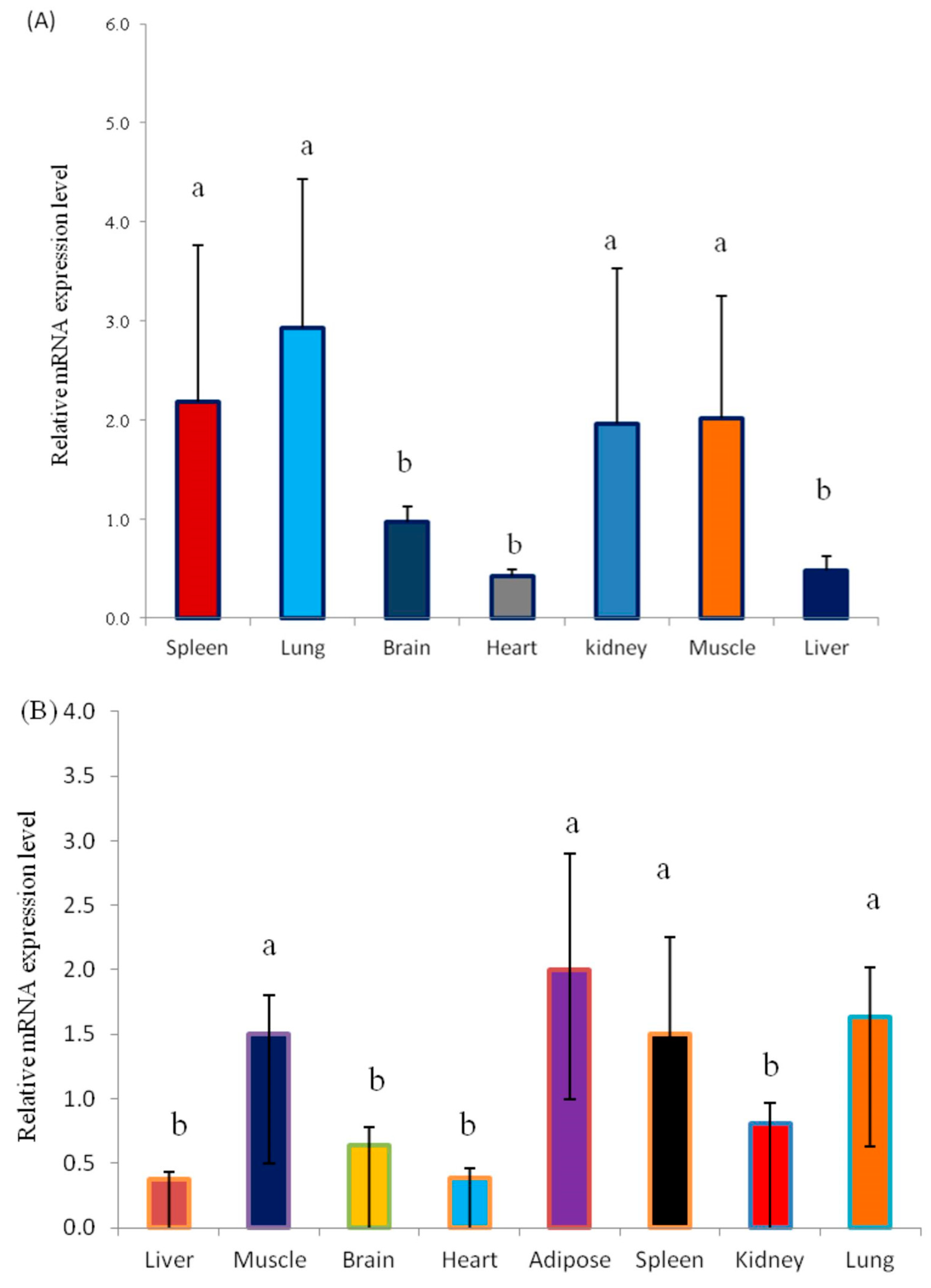
3.3. Analysis of the Expression Pattern of the KLF6 Gene in Different Tissues
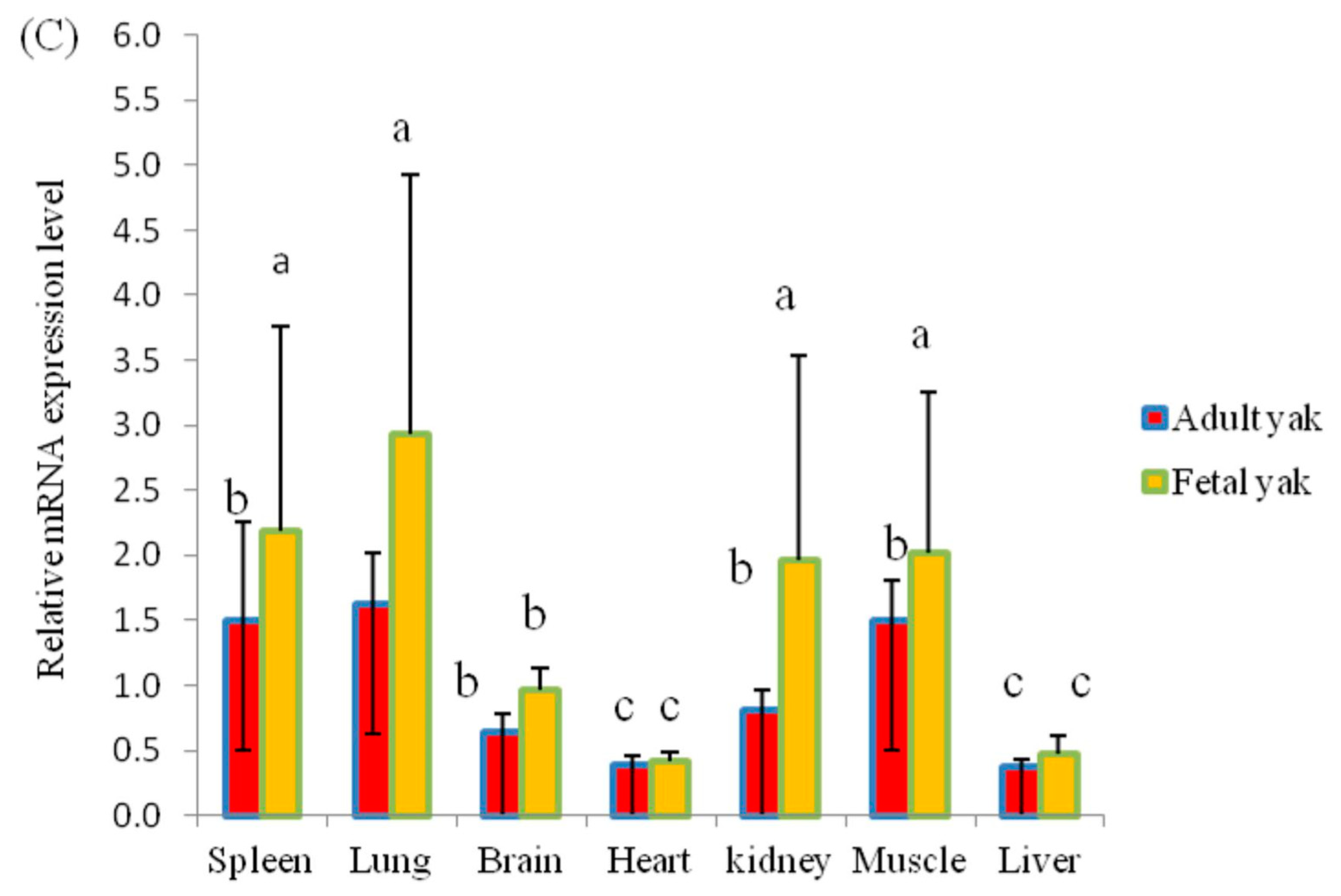
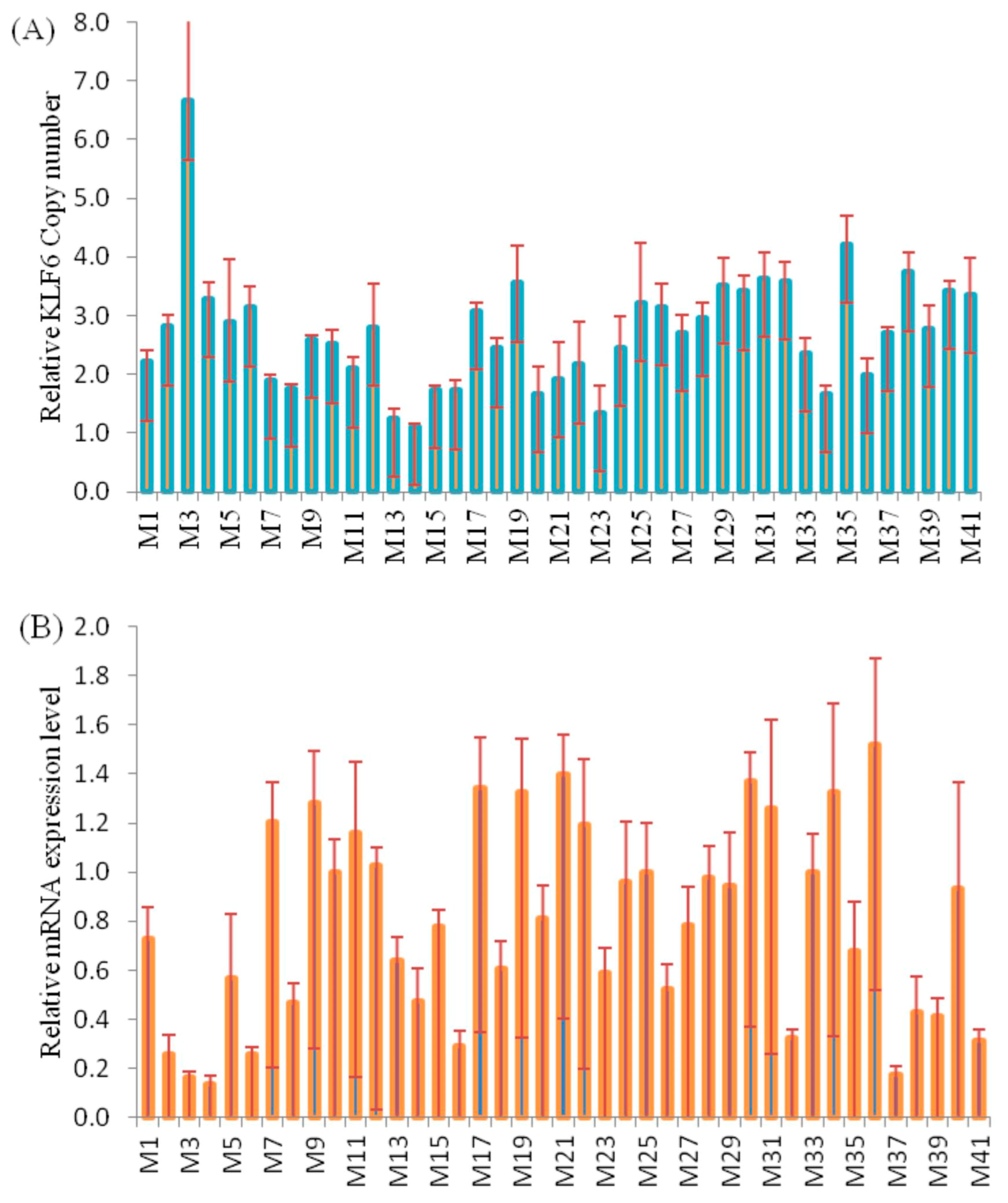
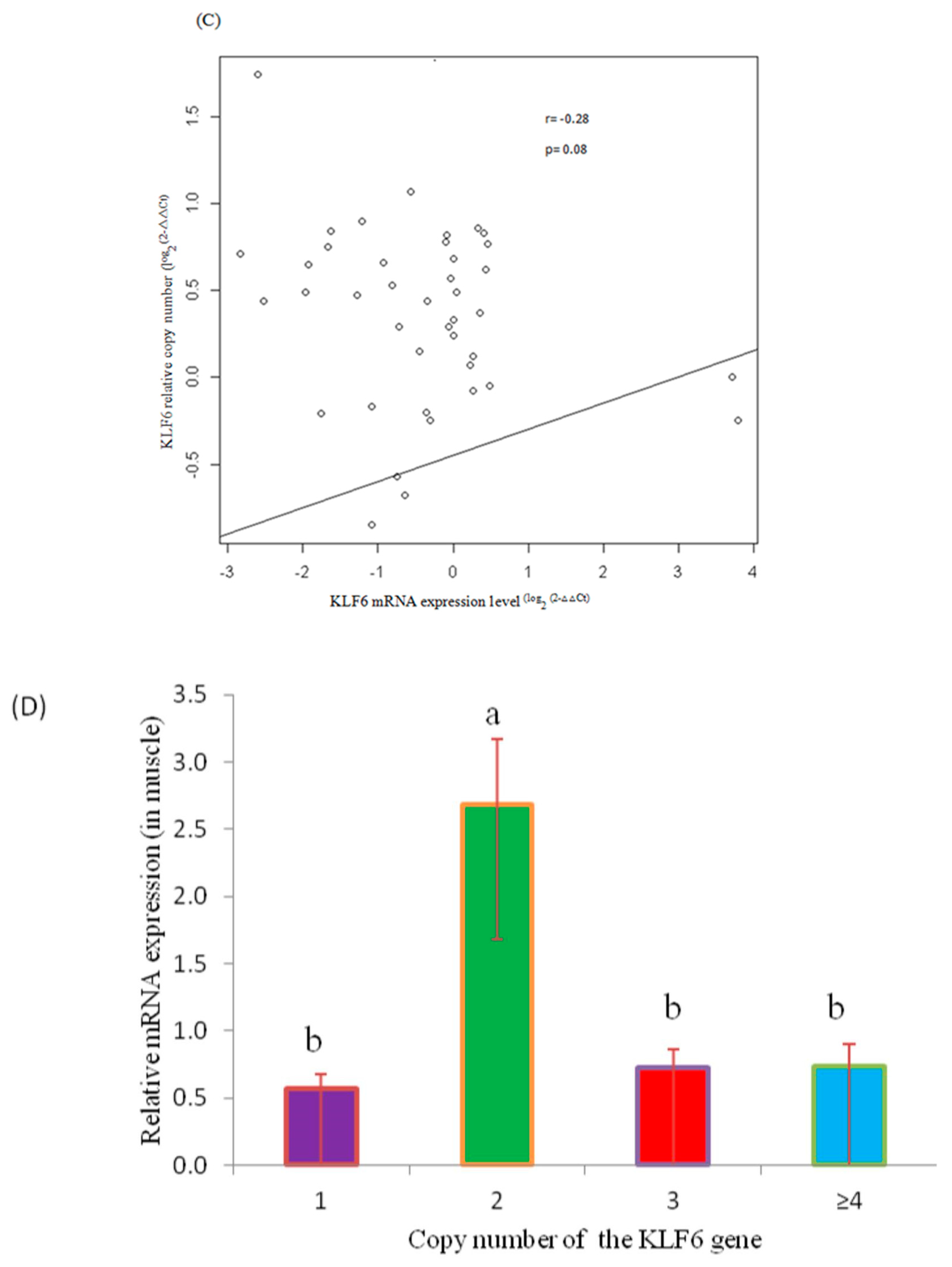
3.4. Correlation Analysis of CNVs and KLF6 mRNA Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Redon, R.; Ishikawa, S.; Fitch, K.R.; Feuk, L.; Perry, G.H.; Andrews, T.D.; Fiegler, H.; Shapero, M.H.; Carson, A.R.; Chen, W.; et al. Global variation in copy number in the human genome. Nature 2006, 444, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Durán Aguilar, M.; Román Ponce, S.I.; Ruiz López, F.J.; Gonzalez Padilla, E.; Vasquez Pelaez, C.G.; Bagnato, A.; Strillacci, M.G. Genome-wide association study for milk somatic cell score in Holstein cattle using copy number variation as markers. J. Anim. Breed. Genet. 2016, 134, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Ben Sassi, N.; González-Recio, Ó.; de Paz-del, R.R.; Rodríguez-Ramilo, S.T.; Fernández, A.I. Associated effects of copy number variants on economically important traits in Spanish Holstein dairy cattle. J. Dairy Sci. 2016, 99, 6371–6380. [Google Scholar] [CrossRef] [PubMed]
- Bickhart, D.M.; Hou, Y.; Schroeder, S.G.; Alkan, C.; Cardone, M.F.; Matukumalli, L.K.; Song, J.; Schnabel, R.D.; Ventura, M.; Taylor, J.F.; et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Res. 2012, 22, 778–790. [Google Scholar] [CrossRef] [PubMed]
- Keel, B.N.; Lindholm-Perry, A.K.; Snelling, W.M. Evolutionary and functional features of copy number variation in the cattle genome. Front. Genet. 2016, 7, 207. [Google Scholar] [CrossRef] [PubMed]
- Prinse, R.T.M.M.; Strillacci, M.G.; Schiavini, F.; Santus, E.; Rossoni, A.; Maurer, V.; Bieber, A.; Gredler, B.; Dolezal, M.; Bagnato, A. A genome-wide scan of copy number variants using high-density SNPs in Brown Swiss dairy cattle. Livest. Sci. 2016, 191, 153–160. [Google Scholar] [CrossRef]
- Wu, Y.; Fan, H.; Jing, S.; Xia, J.; Chen, Y.; Zhang, L.; Gao, X.; Li, J.; Gao, H.; Ren, H.A. Genome-wide scan for copy number variations using high-density single nucleotide polymorphism array in Simmental cattle. Anim. Genet. 2015, 46, 289–298. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Hou, Y.; Bickhart, D.M.; Zhou, Y.; Hay, H.E.; Song, J.; Sonstegard, T.S.; Van Tassell, C.P.; Liu, G.E. Population-genetic properties of differentiated copy number variations in cattle. Sci. Rep. 2016, 6, 23161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, K.; Wang, L.; Yang, Y.; Ni, Z.; Xie, X.; Shao, X.; Han, J.; Wan, D.; Qiu, Q. Genome-wide patterns of copy number variation in the Chinese yak genome. BMC Genom. 2016, 17, 379. [Google Scholar] [CrossRef] [PubMed]
- Prinsen, R.T.M.M.; Rossoni, A.; Gredler, B.; Bieber, A.; Bagnato, A.; Strillacci, M.G. A genome-wide association study between CNVs and quantitative traits in Brown Swiss cattle. Livest. Sci. 2017, 202, 7–12. [Google Scholar] [CrossRef]
- Scherer, S.W.; Lee, C.; Birney, E.; Altshuler, D.M.; Eichler, E.E.; Carter, N.P.; Hurles, M.E.; Feuk, L. Challenges and standards in integrating surveys of structural variation. Nat. Genet. 2007, 39, S7–S15. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Liu, G.E.; Bickhart, D.M.; Cardone, M.F.; Wang, K.; Kim, E.S.; Matukumalli, L.K.; Ventura, M.; Song, J.; VanRaden, P.M.; et al. Genomic characteristics of cattle copy number variations. BMC Genom. 2011, 12, 127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Gu, W.; Hurles, M.E.; Lupski, J.R. Copy number variation in human health, disease, and evolution. Annu. Rev. Genom. Hum. Genet. 2009, 10, 4514–4581. [Google Scholar] [CrossRef] [PubMed]
- Reymond, A.; Henrichsen, C.N.; Harewood, L.; Merla, G. Side effects of genome structural changes. Curr. Opin. Genet. Dev. 2007, 17, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Weischenfeldt, J.; Symmons, O.; Spitz, F.; Korbel, J.O. Phenotypic impact of genomic structural variation: Insights from and for human disease. Nat. Rev. Genet. 2013, 14, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Bagnato, A.; Strillacci, M.G.; Pellegrino, L.; Schiavini, F.; Frigo, E.; Rossoni, A.; Fontanesi, L.; Maltecca, C.; Prinsen, R.T.M.M.; Doleza, M.A. Identification and validation of copy number variants in Italian Brown Swiss dairy cattle using Illumina bovine SNP50 Beadchip. Ital. J. Anim. Sci. 2015, 14, 3900. [Google Scholar] [CrossRef]
- Da Silva, J.M.; Giachetto, P.F.; da Silva, L.O.; Cintra, L.C.; Paiva, S.R.; Yamagishi, M.E.B.; Caetano, A.R. Genome-wide copy number variation (CNV) detection in Nelore cattle reveals highly frequent variants in genome regions harboring QTLs affecting production traits. BMC Genom. 2016, 17, 454. [Google Scholar] [CrossRef] [PubMed]
- De Almeida Santana, M.H.; Oliveira Junior, G.A.; Mello Cesar, A.S.; Freua, M.C.; Gomes, R.C.; Silva, S.L.; Leme, P.R.; Fukumasu, H.; Carvalho, M.E. Copy number variations and genome-wide associations reveal putative genes and metabolic pathways involved with the feed conversion ratio in beef cattle. J. Appl. Genet. 2016, 57, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Cole, J.B.; Bickhart, D.M.; Hou, Y.; Song, J.; van Raden, P.M.; Sonstegard, T.S.; van Tassell, C.P.; Liu, G.E. Genome wide CNV analysis reveals additional variants associated with milk production traits in Holsteins. BMC Genom. 2014, 15, 683. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, L.Z.; Shi, T.; Zhou, Y.; Cai, H.F.; Lan, X.; Zhang, C.; Lei, C.; Chen, H. Copy number variations of MICAL-L2 shaping gene expression contribute to different phenotypes of cattle. Mamm. Genome 2013, 24, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Xu, Y.; Yang, M.; Huang, Y.; Lan, X.; Lei, C.; Qi, X.; Yang, X.; Chen, H. Copy number variations at LEPR gene locus associated with gene expression and phenotypic traits in Chinese cattle. Anim. Sci. 2016, 87, 336–343. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shi, T.; Cai, H.; Zhou, Y.; Lan, X.; Zhang, C.; Lei, C.; Qi, X.; Chen, H. Associations of MYH3 gene copy number variations with transcriptional expression and growth traits in Chinese cattle. Gene 2014, 535, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Utsunomiya, Y.T.; Xu, L.; abdel Hay, E.H.; Bickhart, D.M.; Alexandre, P.A.; Rosen, B.D.; Schroeder, S.G.; Carvalheiro, R.; Neves, H.H.R.; et al. Genome-wide CNV analysis reveals variants associated with growth traits in Bos indicus. BMC Genom. 2016, 17, 419. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Lia, B.; Huanga, Y.; Yang, M.; Lana, X.; Lei, C.; Qub, W.; Bai, Y.; Chena, H. Copy number variation of bovine MAPK10 modulates the transcriptional activity and affects growth traits. Livest. Sci. 2016, 194, 44–50. [Google Scholar] [CrossRef]
- Zhang, L.; Jia, S.; Yang, M.; Xu, Y.; Li, C.; Sun, J.; Huang, Y.; Lan, X.; Lei, C.; Zhou, Y.; et al. Detection of copy number variations and their effects in Chinese bulls. BMC Genom. 2014, 15, 480. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Lv, J.; Zhang, L.; Li, M.; Zhou, Y.; Lan, X.; Lei, C.; Chen, H. Association study and expression analysis of CYP4A11 gene copy number variation in Chinese cattle. Sci. Rep. 2017, 7, 46599. [Google Scholar] [CrossRef] [PubMed]
- Stankiewicz, P.; Lupski, J.R. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010, 61, 437–455. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Chung, E.K.; Wu, Y.L.; Savelli, S.L.; Nagaraja, H.N.; Zhou, B.; Hebert, M.; Jones, K.N.; Shu, Y.; Kitzmiller, K.; et al. Gene copy-number variation and associated polymorphisms of complement component C4 in human systemic lupus erythematosus (SLE): Low copy number is a risk factor for and high copy number is a protective factor against SLE susceptibility in European Americans. Am. J. Hum. Genet. 2007, 80, 1037–1054. [Google Scholar] [PubMed]
- Wang, K.; Hu, Q.; Ma, H.; Wang, L.; Yang, Y.; Luo, W.; Qiu, Q. Genome-wide variation within and between wild and domestic yak. Mol. Ecol. Resour. 2014, 14, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Schaller, G.B.; Liu, W. Distribution, status, and conservation of wild yak Bos grunniens. Biol. Conserv. 1996, 7, 61–68. [Google Scholar] [CrossRef]
- Wiener, G.; Jianlin, H.; Ruijun, L. The Yak, 2nd ed.; FAO Regional Office for Asia and the Pacific Food and Agriculture Organization of the United Nations: Bangkok, Thailand, 2003; p. 99. [Google Scholar]
- Guo, X.; Long, R.; Kreuzer, M.; Ding, L.; Shang, Z.; Zhang, Y.; Yang, Y.; Cui, G. Importance of functional ingredients in yak milk-derived food on the health of Tibetan nomads living under high-altitude stress: A review. Crit. Rev. Food Sci. Nutr. 2014, 54, 292–302. [Google Scholar] [CrossRef] [PubMed]
- Han Jianlin, R.C.; Hanotte, O.; McVeigh, C.; Rege, J.E.O. (Eds.) Yak production in central Asian highlands. In Proceedings of the Third International Congress on Yak, Lhasa, China, 4–9 September 2000; ILRI (International Livestock Research Institute): Nairobi, Kenya, 2000; pp. 40–49. [Google Scholar]
- Matsumoto, N.; Kubo, A.; Liu, H.; Akita, K.; Laub, F.; Ramirez, F.; Keller, G.; Friedman, S.L. Developmental regulation of yolk Sac hematopoiesis by Krüppel-like factor 6. Blood 2006, 107, 1357–1365. [Google Scholar] [CrossRef] [PubMed]
- Bieker, J.J. Kruppel-like factors: Three fingers in many pies. J. Biol. Chem. 2001, 276, 34355–34358. [Google Scholar] [CrossRef] [PubMed]
- Pearson, R.; Fleetwood, J.; Eaton, S.; Crossley, M.; Bao, S. Krüppel-like transcription factors: A functional family. Int. J. Biochem. Cell Biol. 1996, 40, 1996–2001. [Google Scholar] [CrossRef] [PubMed]
- Gehrau, R.C.; D’Asolfo, D.S.; Adreoli, V.; Bocco, J.L.; Koritschnoer, N.P. Differential expression of klf6 tumor suppressor gene upon cell-damaging treatments in cancer cells. Mutat. Res. 2010, 707, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Andreoli, V.; Gehrau, R.C.; Bocco, J.L. Biology of Kruppel-like factor 6 transcription regulator in cell life and death. IUBMB Life 2010, 62, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Dayton, W.R.; White, M. Cellular and molecular regulation of muscle growth and development in meat animals. Anim. Sci. 2008, 86, E217–E225. [Google Scholar] [CrossRef] [PubMed]
- Serão, N.V.L.; González-Peña, D.; Beever, J.E.; Bollero, G.A.; Southey, B.R.; Faulkner, D.B.; Rodriguez-Zas, S.L. Bivariate genome-wide association analysis of the growth and intake components of feed efficiency. PLoS ONE 2013, 8, e78530. [Google Scholar] [CrossRef] [PubMed]
- Botella, L.M.; Sanz-Rodriguez, F.; Komi, Y.; Fernandez, A.; Varela, E.; Garrido-Martin, E.M.; Narla, G.; Friedman, S.L.; Kojima, S. TGF-β regulates the expression of transcription factor KLF6 and its splice variants and promotes cooperative transactivation of common target genes through Smad3-Sp1 -KLF6 interaction. Biochem. J. 2009, 419, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, R.P.; Bailey, D.R.C.; Shannon, N.H. Linear body measurements of cattle before and after 20 years of selection for postweaning gain when fed two different diets. Anim. Sci. 1993, 71, 1712–1720. [Google Scholar] [CrossRef]
- U.S. National Library of Medicine. Available online: https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=blastHome (accessed on 20 August 2018).
- dbVar. Available online: https://www.ncbi.nlm.nih.gov/dbvar (accessed on 20 August 2018).
- Butte, A.J.; Dzau, V.J.; Glueck, S.B. Further defining housekeeping, or “maintenance,” genes focus on “A compendium of gene expression in normal human tissues”. Physiol. Genom. 2001, 7, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.H.; Chung, Y.J.; Jin, E.H.; Shim, S.C.; Kim, J.Y.; Kim, Y.S.; Hu, H.J.; Shin, S.H.; Pae, H.O.; Zouali, M.; et al. The potential role of VPREB1 gene copy number variation in susceptibility to rheumatoid arthritis. Mol. Immunol. 2011, 48, 1338–1343. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.S.; Cheong, H.S.; Kim, L.H.; NamGung, S.; Park, T.J.; Chun, J.Y.; Kim, J.Y.; Pasaje, C.F.; Lee, J.S.; Shin, H.D. Identification of copy number variations and common deletion polymorphisms in cattle. BMC Genom. 2010, 11, 232. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1107. [Google Scholar] [CrossRef] [PubMed]
- Knezevic, S.Z.; Streibig, J.C.; Ritz, C. Utilizing R software package for dose-response studies: The concept and data analysis. Weed Technol. 2007, 21, 840–848. [Google Scholar] [CrossRef]
- Stranger, B.E.; Forrest, M.S.; Dunning, M.; Ingle, C.E.; Beazley, C.; Thorne, N.; Redon, R.; Bird, C.P.; de Grassi, A.; Lee, C.; et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science 2007, 315, 848–853. [Google Scholar] [CrossRef] [PubMed]
- Quanwei, Z.; Youji, M.; Yong, Z.; Xingxu, Z. Cross-species Analysis of Copy Number Variations in domestic yak genome based study on Heterologous Hybridization using Bovine HD Genotyping bead Chip Array. In Proceedings of the Fifth International Conference on Yak, Lanzhou, China, 28–30 August 2014; pp. 131–138. [Google Scholar]
- Bickhart, D.M.; Liu, G.E. The challenges and importance of structural variation detection in livestock. Front. Genet. 2014, 5, 37. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.E.; Hou, Y.L.; Zhu, B.; Cardone, M.F.; Jiang, L.; Cellamare, A.; Mitra, A.; Alexander, L.J.; Coutinho, L.L.; Dell’Aquila, M.E.; et al. Analysis of copy number variations among diverse cattle breeds. Genome Res. 2010, 20. [Google Scholar] [CrossRef] [PubMed]
- Gout, J.-F.; Kahn, D.; Duret, L.; Paramecium Post-Genomics Consortium. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010, 6, e1000944. [Google Scholar] [CrossRef]
- Dionyssiou, M.G.; Salma, J.; Bevzyuk, M.; Wales, S.; Zakharyan, L.; McDermott, J.C. Krüppel-like factor 6 (KLF6) promotes cell proliferation in skeletal myoblasts in response to TGFβ/Smad3 signaling. Skeletal Muscle 2013, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Antúnez-Ortiz, D.L.; Flores-Alfaro Burguete-García, A.I.; Bonnefond, A.; Peralta-Romero, J.; Froguel, P.; Espinoza-Rojo, M.; Cruz, M. Copy number variations in candidate genes and intergenic regions affect body mass index and abdominal obesity in Mexican children. Biomed. Res. Int. 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Bickhart, D.M.; Chung, H.; Hutchison, J.L.; Norman, H.D.; Connor, E.E.; Liu, G.E. Analysis of copy number variations in Holstein cows identify potential mechanisms contributing to differences in residual feed intake. Funct. Integr. Genom. 2012, 12, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Lehnert, S.A.; Reverter, A.; Byrne, K.A.; Wang, Y.; Nattrass, G.S.; Hudson, N.J.; Greenwood, P.L. Gene expression studies of developing bovine longissimus muscle from two different beef cattle breeds. BMC Dev. Biol. 2007, 7, 95. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Liu, J.; Liang, C.N.; Guo, X.; Bao, P.J.; Chu, M.; Ding, X.Z.; Wang, H.B.; Zhu, X.S.; Yan, P. Associations of single nucleotide polymorphisms in candidate genes with the polled trait in Datong domestic yaks. Anim. Genet. 2013, 45, 138–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.M.; Zheng, L.; He, H.; Song, C.C.; Zhang, Z.J.; Cao, X.K.; Lei, C.Z.; Lan, X.Y.; Qi, X.L.; Chen, H.; et al. Associations of GBP2 gene copy number variations with growth traits and transcriptional expression in Chinese cattle. Gene 2018, 647, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, M.; da Silva, V.H.; Megens, H.J.; Visker, M.H.P.W.; Ajmone-Marsan, P.; Bâlteanu, V.A.; Dunner, S.; Garcia, J.F.; Ginja, C.; Kantanen, J.; et al. Distribution and functionality of copy number variation across European cattle populations. Front. Genet. 2017, 8, 108. [Google Scholar] [CrossRef] [PubMed]
- Koritschoner, N.P.; Bocco, J.L.; Panzetta-Dutari, G.M.; Dumur, C.I.; Flury, A.; Patrito, L.C. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J. Biol. Chem. 1997, 272, 9573–9580. [Google Scholar] [CrossRef] [PubMed]
- Slavin, D.; Sapin, V.; López-Diaz, F.; Jacquemin, P.; Koritschoner, N.; Dastugue, B.; Davidson, I.; Chatton, B.; Bocco, J.L. The Krüppel-like core promoter binding protein gene is primarily expressed in the placenta during mouse development. Biol. Reprod. 1999, 6, 1586–1591. [Google Scholar] [CrossRef]
- Atkins, G.B.; Jain, M.K. Role of Kruppel-like transcription factors in endothelial. Biol. Circ. Res. 2007, 100, 1686–1695. [Google Scholar] [CrossRef] [PubMed]
- Laub, F.; Aldabe, R.; Friedrich, V., Jr.; Ohnishi, S.; Yoshida, T.; Ramirez, F. Developmental expression of mouse Kruppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 2001, 233, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Narla, G.; DiFeo, A.; Yao, S.; Banno, A.; Hod, E.; Reeves, H.L.; Qiao, R.F.; Camacho-Vanegas, O.; Levine, A.; Kirschenbaum, A.; et al. Targeted inhibition of the KLF6 splice variant, KLF6 SV1, suppresses prostate cancer cell growth and spread. Cancer Res. 2005, 65, 5761–5768. [Google Scholar] [CrossRef] [PubMed]
- Merla, G.; Howald, C.; Henrichsen, C.N.; Lyle, R.; Wyss, C.; Zabot, M.T.; Antonarakis, S.E.; Reymond, A. Submicroscopic deletion in patients with Williams-Beuren syndrome influences expression levels of the nonhemizygous flanking genes. Am. J. Hum. Genet. 2006, 79, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Guryev, V.; Saar, K.; Adamovic, T.; Verheul, M.; van Heesch, S.A.; Cook, S.; Pravenec, M.; Aitman, T.; Jacob, H.; Shull, J.D.; et al. Distribution and functional impact of DNA copy number variation in the rat. Nat. Genet. 2008, 40, 538–545. [Google Scholar] [CrossRef] [PubMed]
- Somerville, M.J.; Mervis, C.B.; Young, E.J.; Seo, E.-J.; Campo, M.; Bamforth, S.; Peregrine, E.; Loo, E.; Lilley, M.; Pérez-Jurado, L.A.; et al. Severe expressive-language delay related to duplication of the Williams-Beuren locus. N. Engl. J. Med. 2005, 353, 1694–1701. [Google Scholar] [CrossRef] [PubMed]
| Level | Accession No. | Gene Name | Primer Sequence (5′–>3′) | Product Length (bp) | Annealing Temperature (°C) |
|---|---|---|---|---|---|
| DNA | XM_005901807.2 | KLF6 | F: ATGCTCATGGGAAGGGTGTG | 82 | 57.45 |
| R: CTTGGCACCAGTGTGCTTTC | 57.45 | ||||
| NM_001034034.2 | BTF3 | F: AACCAGGAGAAACTCGCCAA | 166 | 55.40 | |
| R: TTCGGTGAAATGCCCTCTCG | 57.45 | ||||
| mRNA | NM_001035271.3 | KLF6 | F: GTGACAGGTGTTTCTCCAGG | 91 | 57.45 |
| R: TTTTAGCCCGCAGGAGTTGT | 55.40 | ||||
| NM_001034034.2 | GAPDH | F: AATGAAAGGGCCATCACCATC | 204 | 55.85 | |
| R: GTGGTTCACGCCCATCACA | 60.00 |
| Age | Growth Trait | CNV Type (Mean ± STD) | p-Value | ||
|---|---|---|---|---|---|
| Loss (n = 70) | Normal (n = 71) | Gain (n = 81) | |||
| 6 months (n = 222) | Body height (cm) | 93.9 ± 2.4 a | 90.9 ± 3.6 b,c | 91.2 ± 4.3 b | 0.0001 |
| Body length (cm) | 92.3 ± 1.8 a | 89.2 ± 3.8 b | 89.5 ± 4.3 b | 0.0001 | |
| Chest girth (cm) | 122.2 ± 4.8 a | 119.7 ± 6.8 b | 118.7 ± 6.0 b | 0.002 | |
| Cannon width (cm) | 12.0 ± 0.24 | 12.1 ± 0.4 | 12.1 ± 0.32 | 0.100 | |
| Body weight (kg) | 93.9 ± 9.2 a | 89.2 ± 13 b | 87.6 ± 10.7 b | 0.002 | |
| 3 years (n = 72) | Loss (n = 25) | Normal (n = 31) | Gain (n = 16) | ||
| Body height (cm) | 104.0 ± 3.2 | 104.4 ± 3.2 | 104.4 ± 3.5 | 0.89 | |
| Body length (cm) | 114.0 ± 4.9 | 110.4 ± 19.3 | 114.1 ± 4.9 | 0.53 | |
| Chest girth (cm) | 147.2 ± 6.9 | 148.8 ± 6.4 | 147.2 ± 6.3 | 0.59 | |
| Cannon width (cm) | 16.4 ± 0.51 | 16.5 ± 0.51 | 16.5 ± 0.52 | 0.92 | |
| Body weight (kg) | 176.0 ± 20.6 | 174.6 ± 36.4 | 175.9 ± 17.7 | 0.98 | |
| 5 years (n = 93) | Loss (n = 4) | Normal (n = 4) | Gain (n = 85) | ||
| Body height (cm) | 111.8 ± 2.1 | 112.0 ± 0.82 | 109.3 ± 3.1 | 0.065 | |
| Body length (cm) | 133.0 ± 10 a | 124.9 ± 1.2 b | 123.4 ± 3.5 b | 0.0001 | |
| Chest girth (cm) | 166.8 ± 15.5 a | 157.0 ± 10 a,b | 155.3 ± 5.3 b | 0.002 | |
| Cannon width (cm) | 16.4 ± 2.2 | 16.5 ± 0.5 | 16.5 ± 0.9 | 0.43 | |
| Body weight (kg) | 257.4 ± 83.1 a | 220.2 ± 29.7 b | 211.3 ± 20.7 b | 0.003 | |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Goshu, H.A.; Wu, X.; Chu, M.; Bao, P.; Ding, X.; Yan, P. Copy Number Variations of KLF6 Modulate Gene Transcription and Growth Traits in Chinese Datong Yak (Bos Grunniens). Animals 2018, 8, 145. https://doi.org/10.3390/ani8090145
Goshu HA, Wu X, Chu M, Bao P, Ding X, Yan P. Copy Number Variations of KLF6 Modulate Gene Transcription and Growth Traits in Chinese Datong Yak (Bos Grunniens). Animals. 2018; 8(9):145. https://doi.org/10.3390/ani8090145
Chicago/Turabian StyleGoshu, Habtamu Abera, Xiaoyun Wu, Min Chu, Pengjia Bao, Xuezhi Ding, and Ping Yan. 2018. "Copy Number Variations of KLF6 Modulate Gene Transcription and Growth Traits in Chinese Datong Yak (Bos Grunniens)" Animals 8, no. 9: 145. https://doi.org/10.3390/ani8090145
APA StyleGoshu, H. A., Wu, X., Chu, M., Bao, P., Ding, X., & Yan, P. (2018). Copy Number Variations of KLF6 Modulate Gene Transcription and Growth Traits in Chinese Datong Yak (Bos Grunniens). Animals, 8(9), 145. https://doi.org/10.3390/ani8090145




