Genome-Wide Association Study on the Estimated Breeding Values for Udder and Longevity and the Candidate Genes in Holstein-Friesian Cows in Hungary
Simple Summary
Abstract
1. Introduction
1.1. Studies Related to Udder Health and Conformation
1.2. Studies Related to Longevity
2. Materials and Methods
3. Results
4. Discussion
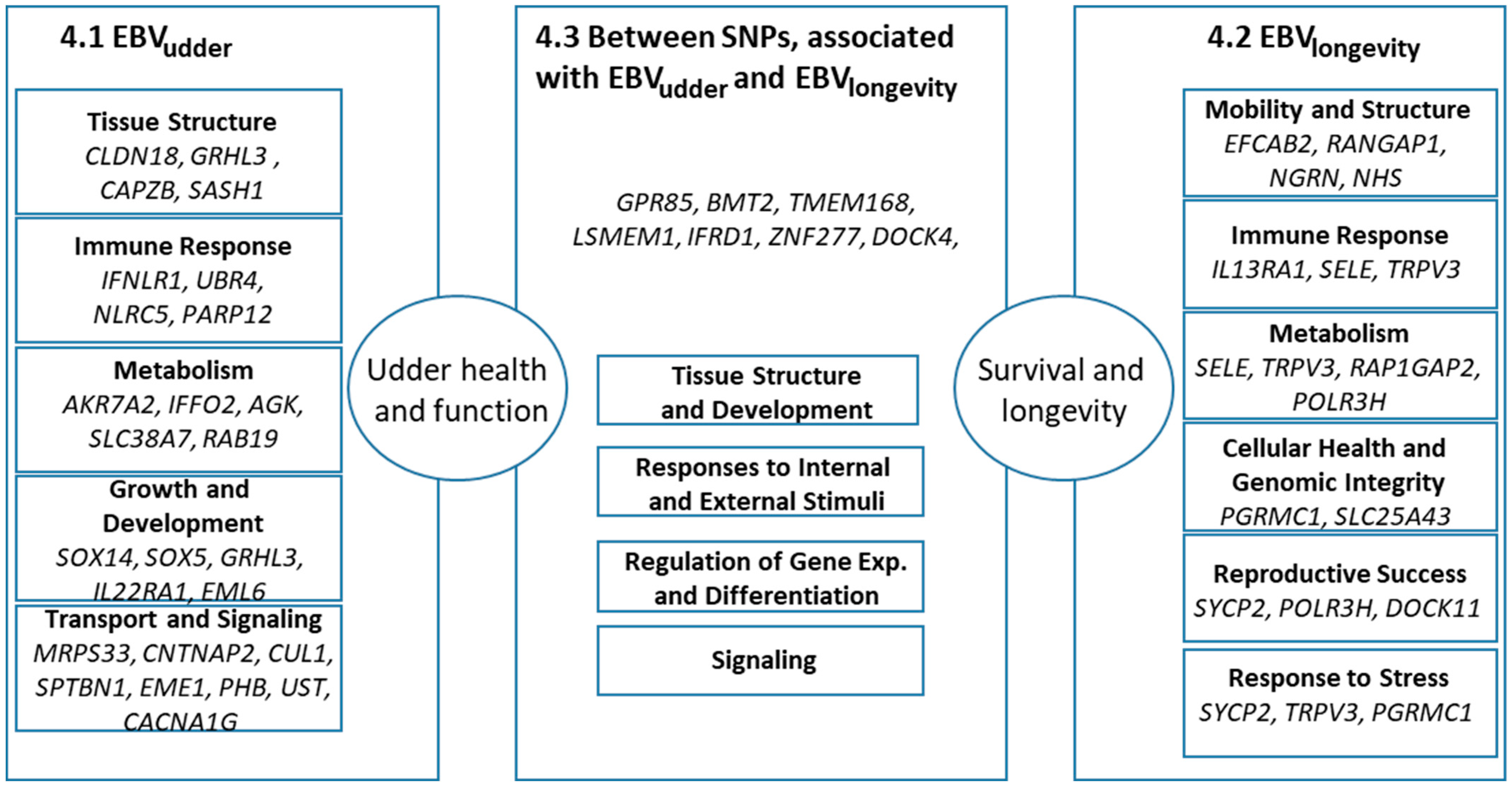
4.1. Genes Around SNPs Associated with EBVudder
4.1.1. Tissue Structure
4.1.2. Immune Response
4.1.3. Metabolism
4.1.4. Growth, Development, and Differentiation
4.1.5. Transport and Signaling
4.2. Genes Around SNPs Associated with EBVlongevity
4.2.1. Mobility and Structural Soundness
4.2.2. Immune Response
4.2.3. Metabolism
4.2.4. Cellular Health and Genomic Integrity
4.2.5. Reproductive Success
4.2.6. Responding to Environmental and Physiological Stress
4.3. Genes Between SNPs Associated with EBVudder and EBVlongevity
4.3.1. Tissue Structure and Development
4.3.2. Responses to Internal and External Stimuli
4.3.3. Regulation of Gene Expression and Differentiation
4.3.4. Signaling
4.4. Impressions Based on Recurring Functional Categories Across the Two EBVs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Elischer, M. History of Dairy Cow Breeds: Holstein; Michigan State University Extension: East Lansing, MI, USA, 2014. [Google Scholar]
- National Association of Hungarian Holstein Friesian Breeders. Available online: https://www.holstein.hu (accessed on 21 July 2025).
- Panigrahi, M.; Rajawat, D.; Nayak, S.S.; Jain, K.; Vaidhya, A.; Prakash, R.; Sharma, A.; Parida, S.; Bhushan, B.; Dutt, T. Genomic insights into key genes and QTLs involved in cattle reproduction. Gene 2024, 917, 148465. [Google Scholar] [CrossRef]
- Spellman, M.E.; Geary, C.M.; Somula, H.; Singh, A.; Wieland, M. The association between teat shape and clinical mastitis. J. Dairy Sci. 2025, 108, 773–780. [Google Scholar] [CrossRef]
- Azovtseva, A.I.; Pozovnikova, M.V.; Shcherbakov, Y.S.; Tulinova, O.V.; Romanova, E.A.; Ryabova, A.E. Genome-wide association study for conformation traits in Ayrshire cattle. Anim. Sci. J. 2024, 95, e13985. [Google Scholar] [CrossRef]
- Dominguez-Castano, P.; Fortes, M.; Tan, W.L.A.; Toro-Ospina, A.M.; Silva, J. Genome-wide association study for milk yield, frame, and udder-conformation traits of Gir dairy cattle. J. Dairy Sci. 2024, 107, 11127–11138. [Google Scholar] [CrossRef]
- Sinha, R.; Sinha, B.; Kumari, R.; Vineeth, M.R.; Sharma, N.; Verma, A.; Gupta, I.D. Association of udder type traits with single nucleotide polymorphisms in Sahiwal (Bos indicus) and Karan Fries (Bos taurus × Bos indicus) cattle. Anim. Biotechnol. 2023, 34, 2745–2756. [Google Scholar] [CrossRef]
- Nazar, M.; Lu, X.; Abdalla, I.M.; Ullah, N.; Fan, Y.; Chen, Z.; Arbab, A.A.I.; Mao, Y.; Yang, Z. Genome-Wide Association Study Candidate Genes on Mammary System-Related Teat-Shape Conformation Traits in Chinese Holstein Cattle. Genes 2021, 12, 2020. [Google Scholar] [CrossRef]
- Tribout, T.; Croiseau, P.; Lefebvre, R.; Barbat, A.; Boussaha, M.; Fritz, S.; Boichard, D.; Hoze, C.; Sanchez, M.P. Confirmed effects of candidate variants for milk production, udder health, and udder morphology in dairy cattle. Genet. Sel. Evol. 2020, 52, 55. [Google Scholar] [CrossRef]
- Marete, A.; Lund, M.S.; Boichard, D.; Ramayo-Caldas, Y. A system-based analysis of the genetic determinism of udder conformation and health phenotypes across three French dairy cattle breeds. PLoS ONE 2018, 13, e0199931. [Google Scholar] [CrossRef]
- Tolleson, M.W.; Gill, C.A.; Herring, A.D.; Riggs, P.K.; Sawyer, J.E.; Sanders, J.O.; Riley, D.G. Association of udder traits with single nucleotide polymorphisms in crossbred Bos indicus-Bos taurus cows. J. Anim. Sci. 2017, 95, 2399–2407. [Google Scholar] [CrossRef]
- Hu, H.H.; Li, F.; Mu, T.; Han, L.Y.; Feng, X.F.; Ma, Y.F.; Jiang, Y.; Xue, X.S.; Du, B.Q.; Li, R.R.; et al. Genetic analysis of longevity and their associations with fertility traits in Holstein cattle. Animal 2023, 17, 100851. [Google Scholar] [CrossRef]
- Kusaka, H.; Yamazaki, T.; Sakaguchi, M. Association of age at first calving with longevity, milk yield, and fertility up to the third lactation in a herd of Holstein dairy cows in Japan. J. Reprod. Dev. 2023, 69, 291–297. [Google Scholar] [CrossRef]
- Novak, K.; Valcikova, T.; Samake, K.; Bjelka, M. Association of Variants in Innate Immune Genes TLR4 and TLR5 with Reproductive and Milk Production Traits in Czech Simmental Cattle. Genes 2023, 15, 24. [Google Scholar] [CrossRef]
- Liu, D.; Chen, Z.; Zhao, W.; Guo, L.; Sun, H.; Zhu, K.; Liu, G.; Shen, X.; Zhao, X.; Wang, Q.; et al. Genome-wide selection signatures detection in Shanghai Holstein cattle population identified genes related to adaption, health and reproduction traits. BMC Genom. 2021, 22, 747. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, A.; Wang, Y.; Luo, H.; Yan, X.; Guo, X.; Li, X.; Liu, L.; Su, G. Genetic Parameters and Genome-Wide Association Studies of Eight Longevity Traits Representing Either Full or Partial Lifespan in Chinese Holsteins. Front. Genet. 2021, 12, 634986. [Google Scholar] [CrossRef]
- Steri, R.; Moioli, B.; Catillo, G.; Galli, A.; Buttazzoni, L. Genome-wide association study for longevity in the Holstein cattle population. Animal 2019, 13, 1350–1357. [Google Scholar] [CrossRef]
- Nayeri, S.; Sargolzaei, M.; Abo-Ismail, M.K.; Miller, S.; Schenkel, F.; Moore, S.S.; Stothard, P. Genome-wide association study for lactation persistency, female fertility, longevity, and lifetime profit index traits in Holstein dairy cattle. J. Dairy Sci. 2017, 100, 1246–1258. [Google Scholar] [CrossRef]
- Hamidi Hay, E.; Roberts, A. Genomic prediction and genome-wide association analysis of female longevity in a composite beef cattle breed. J. Anim. Sci. 2017, 95, 1467–1471. [Google Scholar] [CrossRef]
- Zhang, Q.; Guldbrandtsen, B.; Thomasen, J.R.; Lund, M.S.; Sahana, G. Genome-wide association study for longevity with whole-genome sequencing in 3 cattle breeds. J. Dairy Sci. 2016, 99, 7289–7298. [Google Scholar] [CrossRef]
- Fontanesi, L.; Calo, D.G.; Galimberti, G.; Negrini, R.; Marino, R.; Nardone, A.; Ajmone-Marsan, P.; Russo, V. A candidate gene association study for nine economically important traits in Italian Holstein cattle. Anim. Genet. 2014, 45, 576–580. [Google Scholar] [CrossRef]
- Szyda, J.; Morek-Kopec, M.; Komisarek, J.; Zarnecki, A. Evaluating markers in selected genes for association with functional longevity of dairy cattle. BMC Genet. 2011, 12, 30. [Google Scholar] [CrossRef]
- Garcia, M.D.; Michal, J.J.; Gaskins, C.T.; Reeves, J.J.; Ott, T.L.; Liu, Y.; Jiang, Z. Significant association of the calpastatin gene with fertility and longevity in dairy cattle. Anim. Genet. 2006, 37, 304–305. [Google Scholar] [CrossRef]
- Appendix 1 of Section 5 of the ICAR Guidelines—The Standard Trait Definition for Dairy Cattle. Available online: https://www.icar.org/Guidelines/05-Conformation-recording-Appendix-1.pdf (accessed on 19 October 2024).
- Bognar, L.; Korosi, Z.J.; Bene, S.A.; Szabo, F.; Anton, I.; Zsolnai, A. Simultaneous Effects of Single-Nucleotide Polymorphisms on the Estimated Breeding Value of Milk, Fat, and Protein Yield of Holstein Friesian Cows in Hungary. Animals 2024, 14, 3518. [Google Scholar] [CrossRef]
- Vilhjalmsson, B.J. Mixmogam [Internet]. Available online: https://github.com/bvilhjal/mixmogam (accessed on 21 July 2025).
- Excoffier, L.; Slatkin, M. Maximum-likelihood estimation of molecular haplotype frequencies in a diploid population. Mol. Biol. Evol. 1995, 12, 921–927. [Google Scholar] [CrossRef]
- Braz, C.U.; Taylor, J.F.; Bresolin, T.; Espigolan, R.; Feitosa, F.L.B.; Carvalheiro, R.; Baldi, F.; de Albuquerque, L.G.; de Oliveira, H.N. Sliding window haplotype approaches overcome single SNP analysis limitations in identifying genes for meat tenderness in Nelore cattle. BMC Genet. 2019, 20, 8. [Google Scholar] [CrossRef]
- Bordbar, F.; Jensen, J.; Wadood, A.A.; Yao, Z. Linkage Disequilibrium Decay in Selected Cattle Breeds. Animals 2024, 14, 3317. [Google Scholar] [CrossRef]
- El Hou, A.; Rocha, D.; Venot, E.; Blanquet, V.; Philippe, R. Long-range linkage disequilibrium in French beef cattle breeds. Genet. Sel. Evol. 2021, 53, 63. [Google Scholar] [CrossRef]
- LaFemina, M.J.; Sutherland, K.M.; Bentley, T.; Gonzales, L.W.; Allen, L.; Chapin, C.J.; Rokkam, D.; Sweerus, K.A.; Dobbs, L.G.; Ballard, P.L.; et al. Claudin-18 deficiency results in alveolar barrier dysfunction and impaired alveologenesis in mice. Am. J. Respir. Cell Mol. Biol. 2014, 51, 550–558. [Google Scholar] [CrossRef]
- Boglev, Y.; Wilanowski, T.; Caddy, J.; Parekh, V.; Auden, A.; Darido, C.; Hislop, N.R.; Cangkrama, M.; Ting, S.B.; Jane, S.M. The unique and cooperative roles of the Grainy head-like transcription factors in epidermal development reflect unexpected target gene specificity. Dev. Biol. 2011, 349, 512–522. [Google Scholar] [CrossRef]
- Mukherjee, K.; Ishii, K.; Pillalamarri, V.; Kammin, T.; Atkin, J.F.; Hickey, S.E.; Xi, Q.J.; Zepeda, C.J.; Gusella, J.F.; Talkowski, M.E.; et al. Actin capping protein CAPZB regulates cell morphology, differentiation, and neural crest migration in craniofacial morphogenesis. Hum. Mol. Genet. 2016, 25, 1255–1270. [Google Scholar] [CrossRef]
- Clements, C.M.; Vogeli, B.; Shellman, Y.G.; Henen, M.A. SAM1 domain of SASH1 harbors distinctive structural heterogeneity. J. Struct. Biol. 2022, 214, 107914. [Google Scholar] [CrossRef]
- Philip, D.T.; Goins, N.M.; Catanzaro, N.J.; Misumi, I.; Whitmire, J.K.; Atkins, H.M.; Lazear, H.M. Interferon lambda restricts herpes simplex virus skin disease by suppressing neutrophil-mediated pathology. mBio 2024, 15, e0262323. [Google Scholar] [CrossRef]
- Sallam, A.M.; Abou-Souliman, I.; Reyer, H.; Wimmers, K.; Rabee, A.E. New insights into the genetic predisposition of brucellosis and its effect on the gut and vaginal microbiota in goats. Sci. Rep. 2023, 13, 20086. [Google Scholar] [CrossRef]
- Guo, B.; Zhu, H.; Xiao, C.; Zhang, J.; Liu, X.; Fang, Y.; Wei, B.; Zhang, J.; Cao, Y.; Zhan, L. NLRC5 exerts anti-endometriosis effects through inhibiting ERβ-mediated inflammatory response. BMC Med. 2024, 22, 351. [Google Scholar] [CrossRef]
- Jiang, B.; Li, L.; Wu, Y.; Wang, X.; Gao, N.; Xu, Z.; Guo, C.; He, S.; Zhang, G.; Chen, Y.; et al. Unveiling Shared Immune Responses in Porcine Alveolar Macrophages during ASFV and PRRSV Infection Using Single-Cell RNA-seq. Microorganisms 2024, 12, 563. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Khan, M.Z.; Kou, X.; Chen, Y.; Liang, H.; Ullah, Q.; Khan, N.; Khan, A.; Chai, W.; Wang, C. Enhancing Metabolism and Milk Production Performance in Periparturient Dairy Cattle Through Rumen-Protected Methionine and Choline Supplementation. Metabolites 2023, 13, 1080. [Google Scholar] [CrossRef]
- Sakatani, M.; Bonilla, L.; Dobbs, K.B.; Block, J.; Ozawa, M.; Shanker, S.; Yao, J.; Hansen, P.J. Changes in the transcriptome of morula-stage bovine embryos caused by heat shock: Relationship to developmental acquisition of thermotolerance. Reprod. Biol. Endocrinol. 2013, 11, 3. [Google Scholar] [CrossRef]
- Penagaricano, F.; Valente, B.D.; Steibel, J.P.; Bates, R.O.; Ernst, C.W.; Khatib, H.; Rosa, G.J. Exploring causal networks underlying fat deposition and muscularity in pigs through the integration of phenotypic, genotypic and transcriptomic data. BMC Syst. Biol. 2015, 9, 58. [Google Scholar] [CrossRef]
- Santana, M.H.; Gomes, R.C.; Utsunomiya, Y.T.; Neves, H.H.; Novais, F.J.; Bonin, M.N.; Fukumasu, H.; Garcia, J.F.; Alexandre, P.A.; Oliveira Junior, G.A.; et al. Genome-wide association with residual body weight gain in Bos indicus cattle. Genet. Mol. Res. 2015, 14, 5229–5233. [Google Scholar] [CrossRef]
- Zhou, P.; Chi, H. AGK Unleashes CD8(+) T Cell Glycolysis to Combat Tumor Growth. Cell Metab. 2019, 30, 233–234. [Google Scholar] [CrossRef]
- Jiang, Q.; Sherlock, D.N.; Guyader, J.; Loor, J.J. Abundance of Amino Acid Transporters and mTOR Pathway Components in the Gastrointestinal Tract of Lactating Holstein Cows. Animals 2023, 13, 1189. [Google Scholar] [CrossRef]
- Dias, M.S.; Pedrosa, V.B.; Rocha da Cruz, V.A.; Silva, M.R.; Batista Pinto, L.F. Genome-wide association and functional annotation analysis for the calving interval in Nellore cattle. Theriogenology 2024, 218, 214–222. [Google Scholar] [CrossRef]
- Huang, Q.X.; Yang, J.; Hu, M.; Lu, W.; Zhong, K.; Wang, Y.; Yang, G.; Loor, J.J.; Han, L. Milk fat globule membrane proteins are involved in controlling the size of milk fat globules during conjugated linoleic acid-induced milk fat depression. J. Dairy Sci. 2022, 105, 9179–9190. [Google Scholar] [CrossRef]
- Wilmore, H.P.; Smith, M.J.; Wilcox, S.A.; Bell, K.M.; Sinclair, A.H. SOX14 is a candidate gene for limb defects associated with BPES and Mobius syndrome. Hum. Genet. 2000, 106, 269–276. [Google Scholar] [CrossRef]
- Zhang, Z.; Chu, M.; Bao, Q.; Bao, P.; Guo, X.; Liang, C.; Yan, P. Two Different Copy Number Variations of the SOX5 and SOX8 Genes in Yak and Their Association with Growth Traits. Animals 2022, 12, 1587. [Google Scholar] [CrossRef]
- Goldie, S.J.; Arhatari, B.D.; Anderson, P.; Auden, A.; Partridge, D.D.; Jane, S.M.; Dworkin, S. Mice lacking the conserved transcription factor Grainyhead-like 3 (Grhl3) display increased apposition of the frontal and parietal bones during embryonic development. BMC Dev. Biol. 2016, 16, 37. [Google Scholar] [CrossRef]
- Rios, A.C.H.; Nasner, S.L.C.; Londono-Gil, M.; Gonzalez-Herrera, L.G.; Lopez-Herrera, A.; Florez, J.C.R. Genome-wide association study for reproduction traits in Colombian Creole Blanco Orejinegro cattle. Trop. Anim. Health Prod. 2023, 55, 429. [Google Scholar] [CrossRef]
- Yin, H.; Hou, X.; Zhang, T.; Shi, L.; Su, Y.Q. Participation of EML6 in the regulation of oocyte meiotic progression in mice. J. Biomed. Res. 2019, 34, 44–53. [Google Scholar] [CrossRef]
- Boutinaud, M.; Herve, L.; Quesnel, H.; Lollivier, V.; Finot, L.; Dessauge, F.; Chanat, E.; Lacasse, P.; Charton, C.; Guinard-Flament, J. Review: The cellular mechanisms underlying mammary tissue plasticity during lactation in ruminants. Animal 2019, 13, s52–s64. [Google Scholar] [CrossRef]
- Cheong, A.; Lingutla, R.; Mager, J. Expression analysis of mammalian mitochondrial ribosomal protein genes. Gene Expr. Patterns 2020, 38, 119147. [Google Scholar] [CrossRef]
- Robinson, B.G.; Oster, B.A.; Robertson, K.; Kaltschmidt, J.A. Loss of ASD-related molecule Cntnap2 affects colonic motility in mice. Front. Neurosci. 2023, 17, 1287057. [Google Scholar] [CrossRef]
- Goldenberg, S.J.; Cascio, T.C.; Shumway, S.D.; Garbutt, K.C.; Liu, J.; Xiong, Y.; Zheng, N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell 2004, 119, 517–528. [Google Scholar] [CrossRef]
- Yang, P.; Yang, Y.; Sun, P.; Tian, Y.; Gao, F.; Wang, C.; Zong, T.; Li, M.; Zhang, Y.; Yu, T.; et al. βII spectrin (SPTBN1): Biological function and clinical potential in cancer and other diseases. Int. J. Biol. Sci. 2021, 17, 32–49. [Google Scholar] [CrossRef]
- Zhou, X.; DeLucia, M.; Ahn, J. SLX4-SLX1 Protein-independent Down-regulation of MUS81-EME1 Protein by HIV-1 Viral Protein R (Vpr). J. Biol. Chem. 2016, 291, 16936–16947. [Google Scholar] [CrossRef] [PubMed]
- Signorile, A.; Sgaramella, G.; Bellomo, F.; De Rasmo, D. Prohibitins: A Critical Role in Mitochondrial Functions and Implication in Diseases. Cells 2019, 8, 71. [Google Scholar] [CrossRef]
- Chen, J.; Sun, T.; Lin, B.; Wu, B.; Wu, J. The Essential Role of Proteoglycans and Glycosaminoglycans in Odontogenesis. J. Dent. Res. 2024, 103, 345–358. [Google Scholar] [CrossRef]
- Han, J.; Li, D.; Qu, C.; Wang, D.; Wang, L.; Guo, X.; Lammi, M.J. Altered expression of chondroitin sulfate structure modifying sulfotransferases in the articular cartilage from adult osteoarthritis and Kashin-Beck disease. Osteoarthr. Cartil. 2017, 25, 1372–1375. [Google Scholar] [CrossRef]
- Kehribar, L.; Betul Celik, Z.; Yalcin Kehribar, D.; Eseoglu, I.; Gunaydin, C.; Aydin, M. The relationship of promoter methylation of calcium voltage-gated channel alpha 1 and interleukin-16 to primary osteoarthritis. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 4436–4441. [Google Scholar] [CrossRef]
- Hu, H.; Mu, T.; Ma, Y.; Wang, X.; Ma, Y. Analysis of Longevity Traits in Holstein Cattle: A Review. Front. Genet. 2021, 12, 695543. [Google Scholar] [CrossRef]
- Sitz, T.; DelCurto-Wyffels, H.; Van Emon, M.; Wyffels, S.; Retallick, K.; Tarpoff, E.; Kangas, K.; DelCurto, T. Importance of Foot and Leg Structure for Beef Cattle in Forage-Based Production Systems. Animals 2023, 13, 495. [Google Scholar] [CrossRef] [PubMed]
- Kachi, E.; Kazama, K.; Murakami, M.; Onda, K. Opsin 3, encoding a non-visual photoreceptor, is a pseudogene in cattle. Res. Vet. Sci. 2025, 186, 105586. [Google Scholar] [CrossRef]
- Huang, M.; Chen, B.; Chen, X.; Liu, T.; Liang, S.; Hu, H.; Bai, X.; Gong, Y. RanGAP1 maintains chromosome stability in limb bud mesenchymal cells during bone development. Cell. Signal. 2024, 120, 111222. [Google Scholar] [CrossRef]
- Jacinto, J.G.P.; Letko, A.; Hafliger, I.M.; Akyurek, E.E.; Sacchetto, R.; Gentile, A.; Drogemuller, C. Whole genome sequencing reveals candidate causal genetic variants for spastic syndrome in Holstein cattle. Sci. Rep. 2024, 14, 31188. [Google Scholar] [CrossRef]
- Garvey, M. Lameness in Dairy Cow Herds: Disease Aetiology, Prevention and Management. Dairy 2022, 3, 199–210. [Google Scholar] [CrossRef]
- Kilili, H.; Padilla-Morales, B.; Castillo-Morales, A.; Monzon-Sandoval, J.; Diaz-Barba, K.; Cornejo-Paramo, P.; Vincze, O.; Giraudeau, M.; Bush, S.J.; Li, Z.; et al. Maximum lifespan and brain size in mammals are associated with gene family size expansion related to immune system functions. Sci. Rep. 2025, 15, 15087. [Google Scholar] [CrossRef]
- Yao, M.; Gao, W.; Tao, H.; Yang, J.; Liu, G.; Huang, T. Regulation signature of miR-143 and miR-26 in porcine Salmonella infection identified by binding site enrichment analysis. Mol. Genet. Genom. 2016, 291, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.M.; He, Y.; Hwang, S.; Bertola, A.; Mackowiak, B.; Ahmed, Y.A.; Seo, W.; Ma, J.; Wang, X.; Park, S.H.; et al. E-Selectin-Dependent Inflammation and Lipolysis in Adipose Tissue Exacerbate Steatosis-to-NASH Progression via S100A8/9. Cell. Mol. Gastroenterol. Hepatol. 2022, 13, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Saif, L.J. Bovine Immunology: Implications for Dairy Cattle. Front. Immunol. 2021, 12, 643206. [Google Scholar] [CrossRef]
- Hu, Y.; Zou, W.; Zhang, L.; Zhang, S.; Hu, L.; Song, Z.; Kong, S.; Gao, Y.; Zhang, J.; Yang, Y.; et al. TRPV3 facilitates lipolysis and attenuates diet-induced obesity via activation of the NRF2/FSP1 signaling axis. Free Radic. Biol. Med. 2024, 221, 155–168. [Google Scholar] [CrossRef]
- Justice, A.E.; Chittoor, G.; Gondalia, R.; Melton, P.E.; Lim, E.; Grove, M.L.; Whitsel, E.A.; Liu, C.T.; Cupples, L.A.; Fernandez-Rhodes, L.; et al. Methylome-wide association study of central adiposity implicates genes involved in immune and endocrine systems. Epigenomics 2020, 12, 1483–1499. [Google Scholar] [CrossRef]
- Palombo, V.; Milanesi, M.; Sgorlon, S.; Capomaccio, S.; Mele, M.; Nicolazzi, E.; Ajmone-Marsan, P.; Pilla, F.; Stefanon, B.; D’Andrea, M. Genome-wide association study of milk fatty acid composition in Italian Simmental and Italian Holstein cows using single nucleotide polymorphism arrays. J. Dairy Sci. 2018, 101, 11004–11019. [Google Scholar] [CrossRef] [PubMed]
- Rauw, W.M.; Baumgard, L.H.; Dekkers, J.C.M. Review: Feed efficiency and metabolic flexibility in livestock. Animal 2025, 19, 101376. [Google Scholar] [CrossRef]
- Liu, H.; Franken, A.; Bielfeld, A.P.; Fehm, T.; Niederacher, D.; Cheng, Z.; Neubauer, H.; Stamm, N. Progesterone-induced progesterone receptor membrane component 1 rise-to-decline changes are essential for decidualization. Reprod. Biol. Endocrinol. 2024, 22, 20. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, Y.; Tian, Y.; Geng, M.; Liu, Y.; Zhang, W.; Shuai, L. Genome-wide screening in the haploid system reveals Slc25a43 as a target gene of oxidative toxicity. Cell Death Dis. 2022, 13, 284. [Google Scholar] [CrossRef]
- Fall, N.; Forslund, K.; Emanuelson, U. Reproductive performance, general health, and longevity of dairy cows at a Swedish research farm with both organic and conventional production. Livest. Sci. 2008, 118, 11–19. [Google Scholar] [CrossRef]
- Li, Y.; Yang, Y.; Yu, B.; Gao, R.; Wang, X. Transcriptome and Metabolome Analyses Reveal High-Altitude Adaptation Mechanism of Epididymis Sperm Maturation in Tibetan Sheep. Animals 2024, 14, 3117. [Google Scholar] [CrossRef]
- Franca, M.M.; Han, X.; Funari, M.F.A.; Lerario, A.M.; Nishi, M.Y.; Fontenele, E.G.P.; Domenice, S.; Jorge, A.A.L.; Garcia-Galiano, D.; Elias, C.F.; et al. Exome Sequencing Reveals the POLR3H Gene as a Novel Cause of Primary Ovarian Insufficiency. J. Clin. Endocrinol. Metab. 2019, 104, 2827–2841. [Google Scholar] [CrossRef]
- Saowaphak, P.; Duangjinda, M.; Plaengkaeo, S.; Suwannasing, R.; Boonkum, W. Genetic correlation and genome-wide association study (GWAS) of the length of productive life, days open, and 305-days milk yield in crossbred Holstein dairy cattle. Genet. Mol. Res. 2017, 16, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Dallago, G.M.; Wade, K.M.; Cue, R.I.; McClure, J.T.; Lacroix, R.; Pellerin, D.; Vasseur, E. Keeping Dairy Cows for Longer: A Critical Literature Review on Dairy Cow Longevity in High Milk-Producing Countries. Animals 2021, 11, 808. [Google Scholar] [CrossRef]
- Song, Y.; Zhao, X.; Aihemaiti, A.; Haire, A.; Gao, Y.; Niu, C.; Yang, P.; Liu, G.; Jia, G.; Wusiman, A. The Mechanism of Heat Stress Resistance During Spermatogenesis in Turpan Black Sheep. Front. Vet. Sci. 2022, 9, 846981. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Yoshimoto, R.U.; Matsui, T.; Amagai, M.; Kido, M.A.; Tominaga, M. Involvement of skin TRPV3 in temperature detection regulated by TMEM79 in mice. Nat. Commun. 2023, 14, 4104. [Google Scholar] [CrossRef]
- Paital, B.; Panda, S.K.; Hati, A.K.; Mohanty, B.; Mohapatra, M.K.; Kanungo, S.; Chainy, G.B. Longevity of animals under reactive oxygen species stress and disease susceptibility due to global warming. World J. Biol. Chem. 2016, 7, 110–127. [Google Scholar] [CrossRef]
- Ballan, M.; Schiavo, G.; Bovo, S.; Schiavitto, M.; Negrini, R.; Frabetti, A.; Fornasini, D.; Fontanesi, L. Comparative analysis of genomic inbreeding parameters and runs of homozygosity islands in several fancy and meat rabbit breeds. Anim. Genet. 2022, 53, 849–862. [Google Scholar] [CrossRef] [PubMed]
- Fashemi, B.E.; Rougeau, A.K.; Salazar, A.M.; Bark, S.J.; Chappidi, R.; Brown, J.W.; Cho, C.J.; Mills, J.C.; Mysorekar, I.U. IFRD1 is required for maintenance of bladder epithelial homeostasis. iScience 2024, 27, 111282. [Google Scholar] [CrossRef]
- Kim, K.H.; Kim, A.; Park, C.I. Data on molecular characterisation and expression analysis of the interferon-related developmental regulator 2 (IFRD2) gene from red sea bream, Pagrus major. Data Brief 2019, 25, 104142. [Google Scholar] [CrossRef] [PubMed]
- Onishi, Y.; Park, G.; Iezaki, T.; Horie, T.; Kanayama, T.; Fukasawa, K.; Ozaki, K.; Hinoi, E. The transcriptional modulator Ifrd1 is a negative regulator of BMP-2-dependent osteoblastogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 329–334. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, Q.; Shi, J.; Fu, L.; Cheng, S. Screening of Genes Related to Growth, Development and Meat Quality of Sahan Crossbred F1 Sheep Based on RNA-Seq Technology. Front. Vet. Sci. 2022, 9, 831519. [Google Scholar] [CrossRef]
- Timberlake, A.T.; Kiziltug, E.; Jin, S.C.; Nelson-Williams, C.; Loring, E.; Yale Center for Genome, A.; Allocco, A.; Marlier, A.; Banka, S.; Stuart, H.; et al. De novo mutations in the BMP signaling pathway in lambdoid craniosynostosis. Hum. Genet. 2023, 142, 21–32. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, Y.; Zhao, Y.; Guo, D.; Chen, L.; Shi, L.; Xu, G. DOCK4 regulates ghrelin production in gastric X/A-like cells. J. Endocrinol. Investig. 2022, 45, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Motsinger-Reif, A.A.; Reif, D.M.; Akhtari, F.S.; House, J.S.; Campbell, C.R.; Messier, K.P.; Fargo, D.C.; Bowen, T.A.; Nadadur, S.S.; Schmitt, C.P.; et al. Gene-environment interactions within a precision environmental health framework. Cell Genom. 2024, 4, 100591. [Google Scholar] [CrossRef]
- Gu, X.; Orozco, J.M.; Saxton, R.A.; Condon, K.J.; Liu, G.Y.; Krawczyk, P.A.; Scaria, S.M.; Harper, J.W.; Gygi, S.P.; Sabatini, D.M. SAMTOR is an S-adenosylmethionine sensor for the mTORC1 pathway. Science 2017, 358, 813–818. [Google Scholar] [CrossRef]
- Peiris, M.; Aktar, R.; Raynel, S.; Hao, Z.; Mumphrey, M.B.; Berthoud, H.R.; Blackshaw, L.A. Effects of Obesity and Gastric Bypass Surgery on Nutrient Sensors, Endocrine Cells, and Mucosal Innervation of the Mouse Colon. Nutrients 2018, 10, 1529. [Google Scholar] [CrossRef] [PubMed]
- Fu, K.; Miyamoto, Y.; Sumi, K.; Saika, E.; Muramatsu, S.I.; Uno, K.; Nitta, A. Overexpression of transmembrane protein 168 in the mouse nucleus accumbens induces anxiety and sensorimotor gating deficit. PLoS ONE 2017, 12, e0189006. [Google Scholar] [CrossRef]
- Teteau, O.; Vitorino Carvalho, A.; Papillier, P.; Mandon-Pepin, B.; Jouneau, L.; Jarrier-Gaillard, P.; Desmarchais, A.; Lebachelier de la Riviere, M.E.; Vignault, C.; Maillard, V.; et al. Bisphenol A and bisphenol S both disrupt ovine granulosa cell steroidogenesis but through different molecular pathways. J. Ovarian Res. 2023, 16, 30. [Google Scholar] [CrossRef] [PubMed]
- Iezaki, T.; Fukasawa, K.; Park, G.; Horie, T.; Kanayama, T.; Ozaki, K.; Onishi, Y.; Takahata, Y.; Nakamura, Y.; Takarada, T.; et al. Transcriptional Modulator Ifrd1 Regulates Osteoclast Differentiation Through Enhancing the NF-κB/NFATc1 Pathway. Mol. Cell. Biol. 2016, 36, 2451–2463. [Google Scholar] [CrossRef]
- Roth, A.; Gill, R.; Certa, U. Temporal and spatial gene expression patterns after experimental stroke in a rat model and characterization of PC4, a potential regulator of transcription. Mol. Cell. Neurosci. 2003, 22, 353–364. [Google Scholar] [CrossRef]
- Franceschetti, T.; Kessler, C.B.; Lee, S.K.; Delany, A.M. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J. Biol. Chem. 2013, 288, 33347–33360. [Google Scholar] [CrossRef]
- Gosztyla, M.L.; Zhan, L.; Olson, S.; Wei, X.; Naritomi, J.; Nguyen, G.; Street, L.; Goda, G.A.; Cavazos, F.F.; Schmok, J.C.; et al. Integrated multi-omics analysis of zinc-finger proteins uncovers roles in RNA regulation. Mol. Cell 2024, 84, 3826–3842.e8. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, T.; Lu, X.; Lan, X.; Chen, Z.; Lu, S. G protein-coupled receptors (GPCRs): Advances in structures, mechanisms, and drug discovery. Signal Transduct. Target. Ther. 2024, 9, 88. [Google Scholar] [CrossRef]
- Huang, L.; Chambliss, K.L.; Gao, X.; Yuhanna, I.S.; Behling-Kelly, E.; Bergaya, S.; Ahmed, M.; Michaely, P.; Luby-Phelps, K.; Darehshouri, A.; et al. SR-B1 drives endothelial cell LDL transcytosis via DOCK4 to promote atherosclerosis. Nature 2019, 569, 565–569. [Google Scholar] [CrossRef] [PubMed]


| EBVudder | EBVlongevity |
|---|---|
| 34 | 28 |
| EBV Category | Tissue Structure | Immune Response | Metabolism | Growth, Development, and Differentiation | Tranport and Signaling | Cellular Health and Genomic Integrity | Reproductive Success | Responding to Environmental and Physiological Stress | Mobility and Structural Soundness | Responses to Internal and External Stimuli | Regulation of Gene Expression and Differentiation | Signaling |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EBVudder | CLDN18, GRHL3, CAPZB, SASH1, IFRD1, EFCAB2, DOCK4 | IFNLR1, UBR4, NLRC5 | AKR7A2, IFFO2, AGK, SLC38A7, RAB19 | SOX14, SOX5, GRHL3, IL22RA1, EML6 | MRPS33, CNTNAP2, CUL1, SPTBN1, EME1, PHB, ABCC6, UST, CACNA1G | BMT2, GPR85, TMEM168, LSMEM1 | IFRD1, GPR85, ZNF277, LSMEM1 | GPR85, BMT2, DOCK4, TMEM168 | ||||
| EBVlongevity | IFRD1, EFCAB2, DOCK4 | IL13RA1, SELE | SELE, RAP1GAP2, POLR3H | PGRMC1, SLC25A43 | SYCP2, POLR3H, DOCK11 | SYCP2, TRPV3, PGRMC1, | EFCAB2, RANGAP1, NGRN, NHS, | BMT2, GPR85, TMEM168, LSMEM1 | IFRD1, GPR85, ZNF277, LSMEM1 | GPR85, BMT2, DOCK4, TMEM168 | ||
| Between BTB-01738708 (EBVlongevity) and ARS-BFGL-NGS-111478 (EBVudder) | IFRD1, EFCAB2, DOCK4 | BMT2, GPR85, TMEM168, LSMEM1 | IFRD1, GPR85, ZNF277, LSMEM1 | GPR85, BMT2, DOCK4, TMEM168 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zsolnai, A.; Bognár, L.; Bene, S.A.; Rózsa, L.; Póti, P.; Szabó, F.; Anton, I. Genome-Wide Association Study on the Estimated Breeding Values for Udder and Longevity and the Candidate Genes in Holstein-Friesian Cows in Hungary. Animals 2026, 16, 73. https://doi.org/10.3390/ani16010073
Zsolnai A, Bognár L, Bene SA, Rózsa L, Póti P, Szabó F, Anton I. Genome-Wide Association Study on the Estimated Breeding Values for Udder and Longevity and the Candidate Genes in Holstein-Friesian Cows in Hungary. Animals. 2026; 16(1):73. https://doi.org/10.3390/ani16010073
Chicago/Turabian StyleZsolnai, Attila, László Bognár, Szabolcs Albin Bene, Laszló Rózsa, Péter Póti, Ferenc Szabó, and István Anton. 2026. "Genome-Wide Association Study on the Estimated Breeding Values for Udder and Longevity and the Candidate Genes in Holstein-Friesian Cows in Hungary" Animals 16, no. 1: 73. https://doi.org/10.3390/ani16010073
APA StyleZsolnai, A., Bognár, L., Bene, S. A., Rózsa, L., Póti, P., Szabó, F., & Anton, I. (2026). Genome-Wide Association Study on the Estimated Breeding Values for Udder and Longevity and the Candidate Genes in Holstein-Friesian Cows in Hungary. Animals, 16(1), 73. https://doi.org/10.3390/ani16010073





