Comparative Profiling of the Fecal Bacteriome, Mycobiome, and Protist Community in Wild Versus Captive (Cervus canadensis)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Sample Collection
2.2. Microbial Sequencing and Data Quality Control
2.3. Data Analysis and Visualization
3. Results
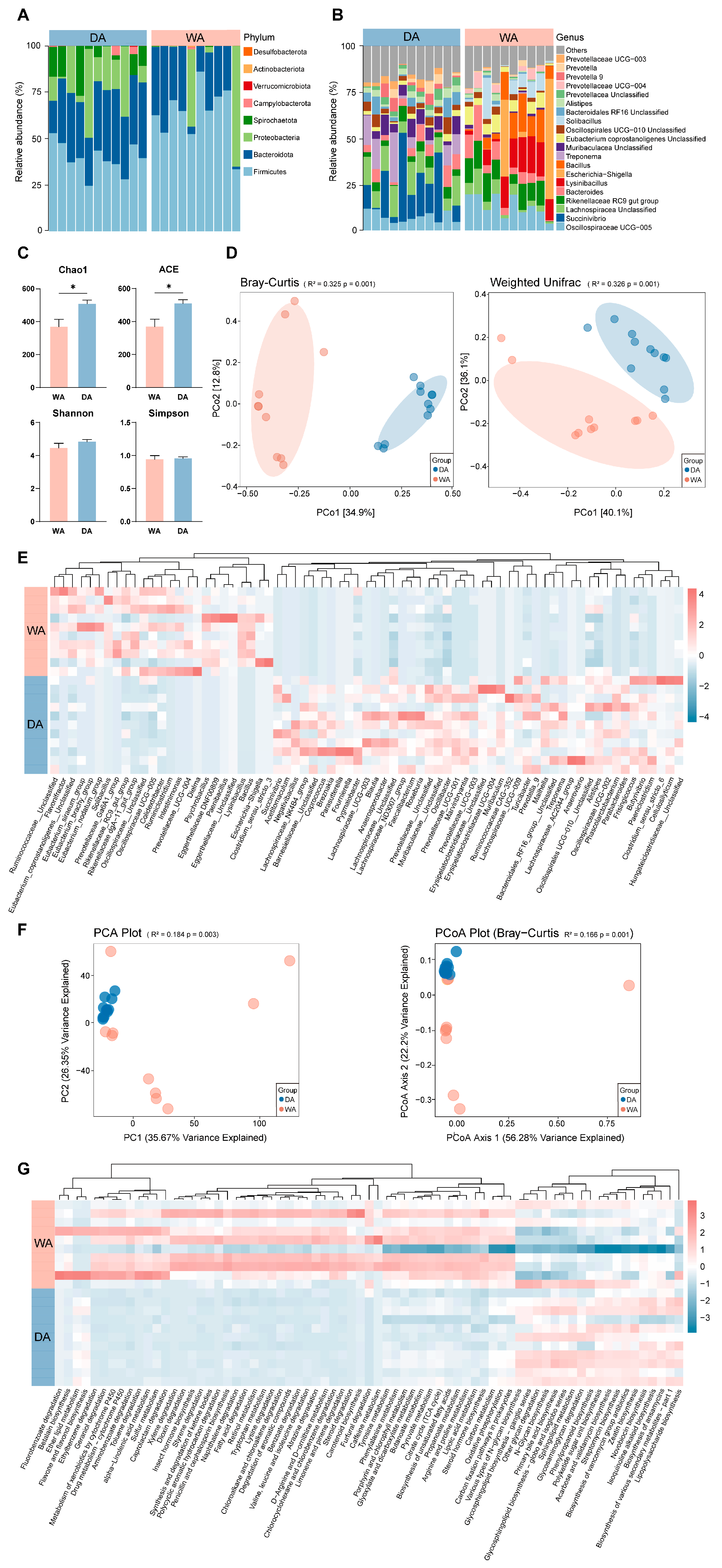
3.1. Compositional Characteristics and Predicted Metagenomic Functions in Bacteria
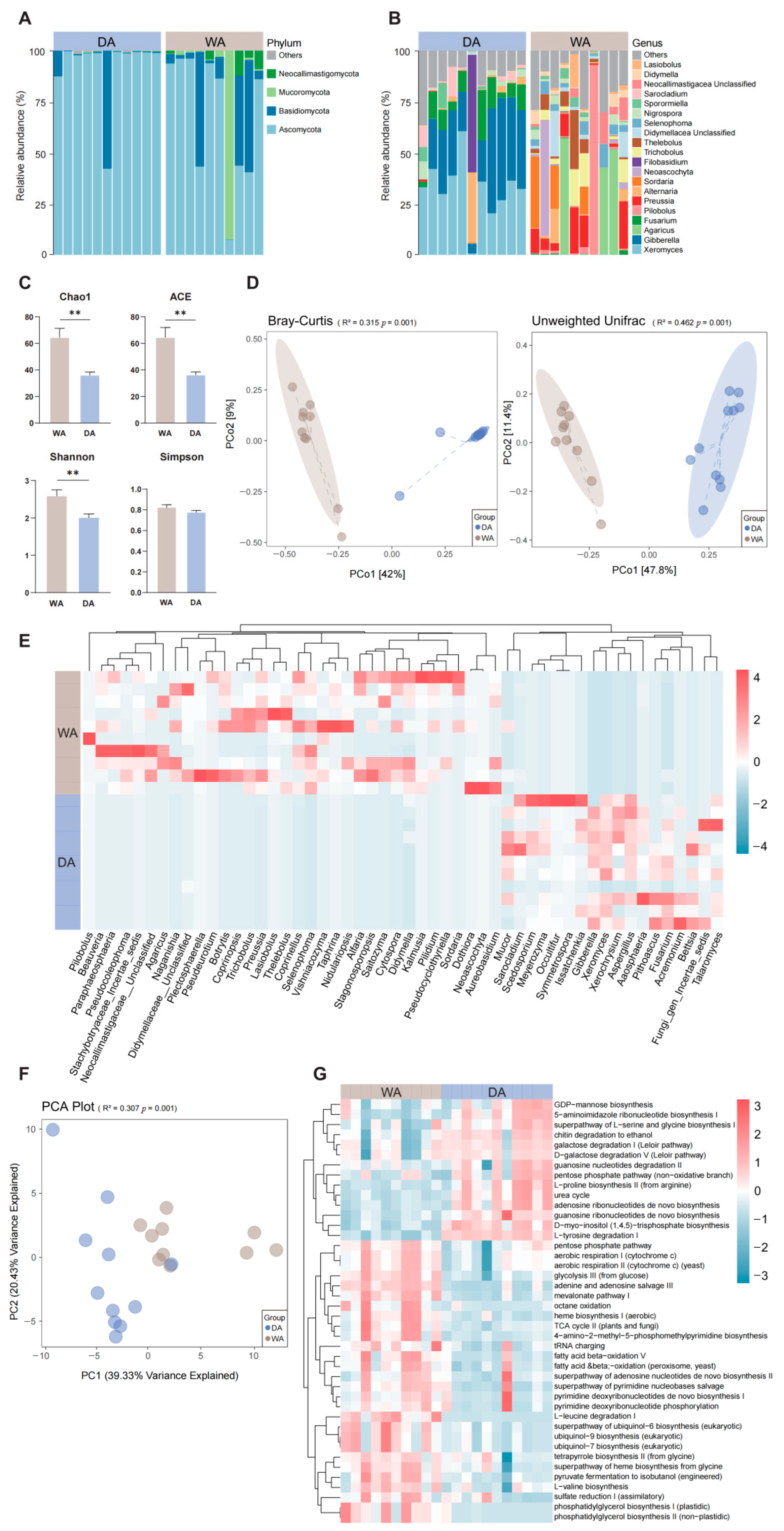
3.2. Compositional Characteristics and Predicted Metagenomic Functions in Fungi
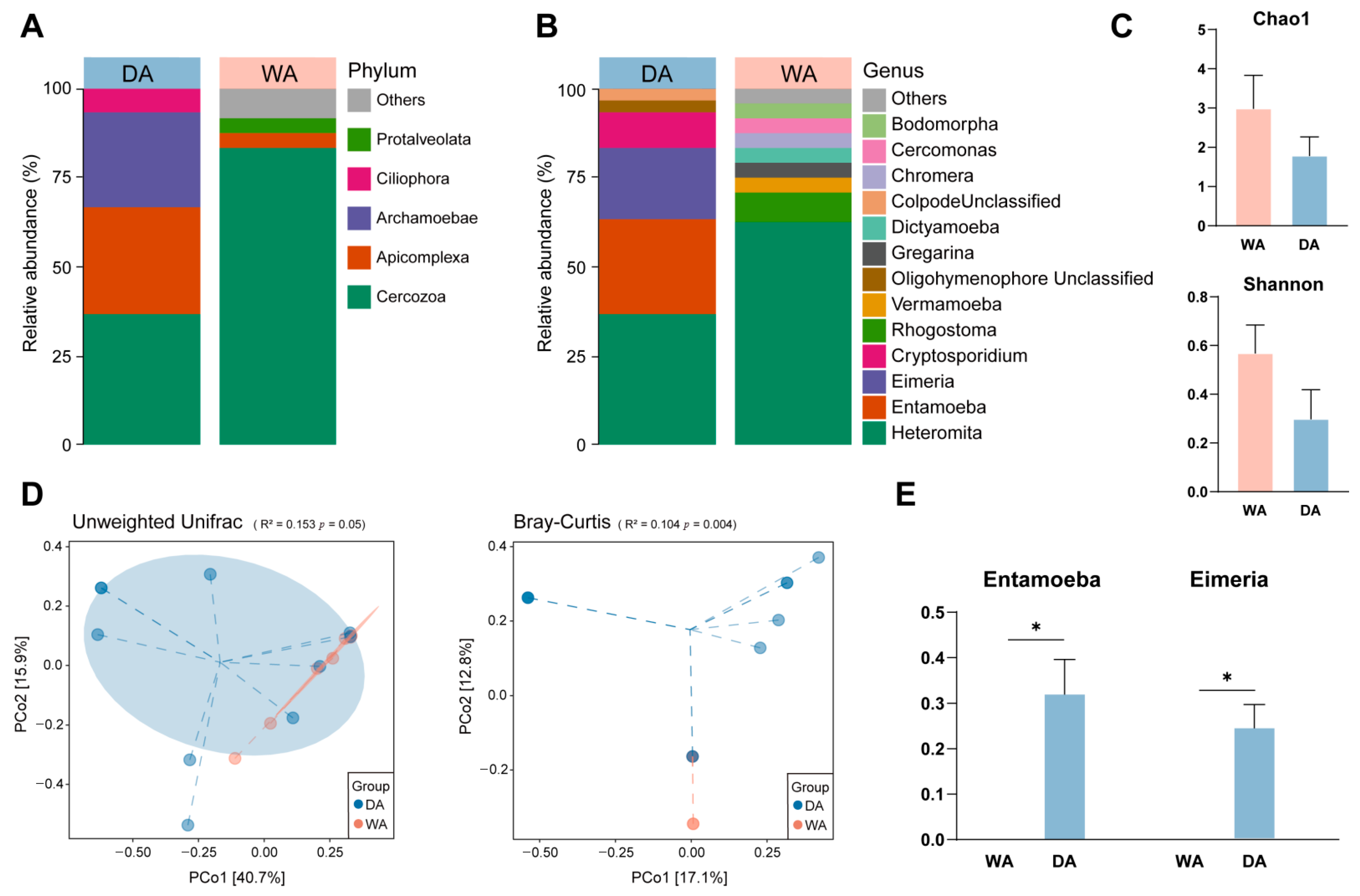
3.3. Exploration the Protozoa Composition in the Gut of Wapiti
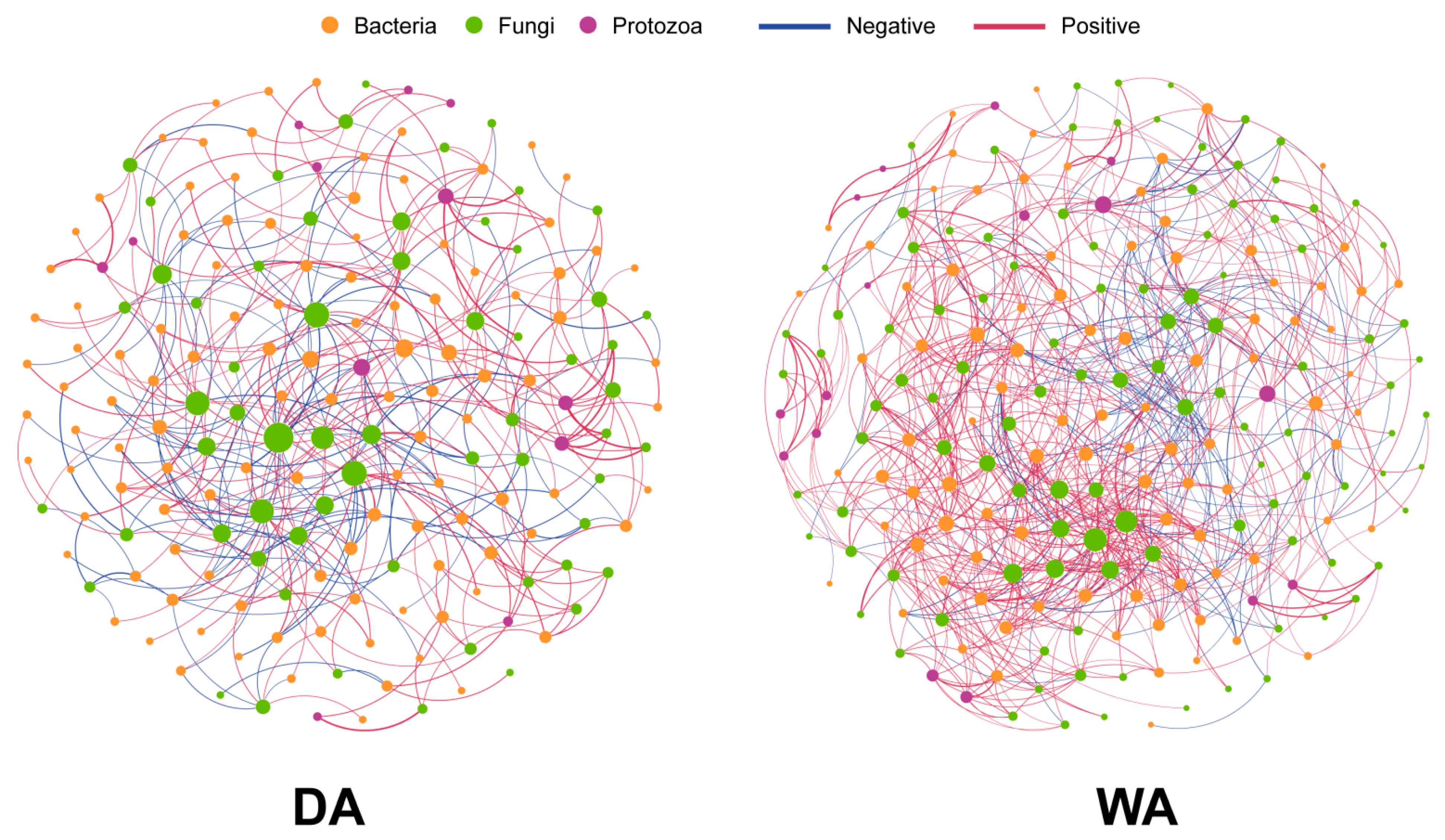
3.4. Interaction Analysis of Bacterial, Fungal, and Protozoan Microbial Networks Between Wild and Captive Wapiti
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schwab, C.; Cristescu, B.; Boyce, M.S.; Stenhouse, G.B.; Gänzle, M. Bacterial Populations and Metabolites in the Feces of Free Roaming and Captive Grizzly Bears. Can. J. Microbiol. 2009, 55, 1335–1346. [Google Scholar] [CrossRef]
- Bae, D.-Y.; Moon, S.-H.; Lee, T.G.; Ko, Y.-S.; Cho, Y.-C.; Kang, H.; Park, C.-S.; Kang, J.-S.; Oh, Y.; Cho, H.-S. Consequences of Domestication on Gut Microbiome: A Comparative Analysis between Wild Boars and Domestic Pigs. Animals 2025, 15, 747. [Google Scholar] [CrossRef]
- Shah, T.; Guo, X.; Song, Y.; Fang, Y.; Ding, L. Comparative Analysis of Gut Bacterial Diversity in Wild and Domestic Yaks on the Qinghai-Tibetan Plateau. Animals 2024, 14, 2380. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, H.; Liu, Z.; McMahon, C. Deer Antler—A Novel Model for Studying Organ Regeneration in Mammals. Int. J. Biochem. Cell Biol. 2014, 56, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Zhao, Q.; Li, S.; Zhu, Y.; Si, H.; Feng, J.; Li, Z. Comparison of Fecal Microbiota and Metabolites between Captive and Grazing Male Reindeer. Animals 2024, 14, 3606. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Yang, H.; Han, S.; Feng, L.; Wang, T.; Ge, J. Comparison of the Gut Microbiota Composition between Wild and Captive Sika Deer (Cervus nippon hortulorum) from Feces by High-Throughput Sequencing. AMB Express 2017, 7, 212. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, Y.; Guo, J.; Zhong, L.; Zhang, M. Alterations in Fecal Microbiota Linked to Environment and Sex in Red Deer (Cervus elaphus). Animals 2023, 13, 929. [Google Scholar] [CrossRef]
- Li, B.; Gao, H.; Song, P.; Liang, C.; Jiang, F.; Xu, B.; Liu, D.; Zhang, T. Captivity Shifts Gut Microbiota Communities in White-Lipped Deer (Cervus albirostris). Animals 2022, 12, 431. [Google Scholar] [CrossRef]
- Huang, J.; Sheng, Y.; Xue, P.; Yu, D.; Guan, P.; Ren, J.; Qian, W. Patterns of Spatial Variation in Rumen Microbiology, Histomorphology, and Fermentation Parameters in Tarim Wapiti (Cervus elaphus yarkandensis). Microorganisms 2024, 12, 216. [Google Scholar] [CrossRef]
- Jiang, F.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Cai, Z.; Gao, H.; Chi, X.; Zhang, T. Comparative Analysis of Gut Microbial Composition and Potential Functions in Captive Forest and Alpine Musk Deer. Appl. Microbiol. Biotechnol. 2022, 106, 1325–1339. [Google Scholar] [CrossRef]
- Yan, J.; Wu, X.; Wang, X.; Shang, Y.; Zhang, H. Uncovering the Fecal Bacterial Communities of Sympatric Sika Deer (Cervus nippon) and Wapiti (Cervus canadensis). Animals 2022, 12, 2468. [Google Scholar] [CrossRef]
- Saftien, A.; Puschhof, J.; Elinav, E. Fungi and Cancer. Gut 2023, 72, 1410–1425. [Google Scholar] [CrossRef]
- Sokol, H.; Leducq, V.; Aschard, H.; Pham, H.-P.; Jegou, S.; Landman, C.; Cohen, D.; Liguori, G.; Bourrier, A.; Nion-Larmurier, I.; et al. Fungal Microbiota Dysbiosis in IBD. Gut 2017, 66, 1039–1048. [Google Scholar] [CrossRef]
- Newbold, C.J.; Lassalas, B.; Jouany, J.P. The Importance of Methanogens Associated with Ciliate Protozoa in Ruminal Methane Production in Vitro. Lett. Appl. Microbiol. 1995, 21, 230–234. [Google Scholar] [CrossRef]
- Chabé, M.; Lokmer, A.; Ségurel, L. Gut Protozoa: Friends or Foes of the Human Gut Microbiota? Trends Parasitol. 2017, 33, 925–934. [Google Scholar] [CrossRef]
- Loke, P.; Lim, Y.A.L. A Commensal Protozoan Strikes a Balance in the Gut. Cell Host Microbe 2016, 20, 417–419. [Google Scholar] [CrossRef]
- Ranilla, M.J.; Jouany, J.-P.; Morgavi, D.P. Methane Production and Substrate Degradation by Rumen Microbial Communities Containing Single Protozoal Species in Vitro. Lett. Appl. Microbiol. 2007, 45, 675–680. [Google Scholar] [CrossRef]
- Veira, D.M. The Role of Ciliate Protozoa in Nutrition of the Ruminant. J. Anim. Sci. 1986, 63, 1547–1560. [Google Scholar] [CrossRef] [PubMed]
- Minas, K.; McEwan, N.R.; Newbold, C.J.; Scott, K.P. Optimization of a High-Throughput CTAB-Based Protocol for the Extraction of qPCR-Grade DNA from Rumen Fluid, Plant and Bacterial Pure Cultures. FEMS Microbiol. Lett. 2011, 325, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global Patterns of 16S rRNA Diversity at a Depth of Millions of Sequences per Sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Bachy, C.; Dolan, J.R.; López-García, P.; Deschamps, P.; Moreira, D. Accuracy of Protist Diversity Assessments: Morphology Compared with Cloning and Direct Pyrosequencing of 18S rRNA Genes and ITS Regions Using the Conspicuous Tintinnid Ciliates as a Case Study. ISME J. 2013, 7, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Cheung, M.K.; Au, C.H.; Chu, K.H.; Kwan, H.S.; Wong, C.K. Composition and Genetic Diversity of Picoeukaryotes in Subtropical Coastal Waters as Revealed by 454 Pyrosequencing. ISME J. 2010, 4, 1053–1059. [Google Scholar] [CrossRef]
- Mao, Y.; Wang, F.; Kong, W.; Wang, R.; Liu, X.; Ding, H.; Ma, Y.; Guo, Y. Dynamic Changes of Rumen Bacteria and Their Fermentative Ability in High-Producing Dairy Cows during the Late Perinatal Period. Front. Microbiol. 2023, 14, 1269123. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast Length Adjustment of Short Reads to Improve Genome Assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-Filtering Vastly Improves Diversity Estimates from Illumina Amplicon Sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME Improves Sensitivity and Speed of Chimera Detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Zhang, J.; Yang, Y.; Lei, X.; Wang, Y.; Li, Y.; Yang, Z.; Yao, J. Active Dry Yeast Supplementation Benefits Ruminal Fermentation, Bacterial Community, Blood Immunoglobulins, and Growth Performance in Young Dairy Goats, but Not for Intermittent Supplementation. Anim. Nutr. 2023, 13, 289–301. [Google Scholar] [CrossRef]
- Gao, Y.; Zhang, G.; Jiang, S.; Liu, Y. Wekemo Bioincloud: A User-friendly Platform for Meta-omics Data Analyses. Imeta 2024, 3, e175. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, Y.; Yang, B.; Yu, S.; Li, S.; Wright, A.-D.G.; Du, R.; Si, H.; Li, Z. Characteristics and Differences in the Antler Velvet Microbiota during Regeneration. Microorganisms 2024, 13, 36–49. [Google Scholar] [CrossRef]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An Open Source Software for Exploring and Manipulating Networks. In Proceedings of the 3rd International AAAI Conference on Web and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 3, pp. 361–362. [Google Scholar] [CrossRef]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial Degradation of Complex Carbohydrates in the Gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef]
- Guo, J.; Li, Z.; Jin, Y.; Sun, Y.; Wang, B.; Liu, X.; Yuan, Z.; Zhang, W.; Zhang, C.; Zhang, M. The Gut Microbial Differences between Pre-Released and Wild Red Deer: Firmicutes Abundance May Affect Wild Adaptation after Release. Front. Microbiol. 2024, 15, 1401373. [Google Scholar] [CrossRef]
- Huang, J.; Sheng, Y.; Xue, P.; Guan, P.; Ren, J.; Qian, W. Characteristics of Bacterial Community and Volatile Fatty Acids in the Gastrointestinal Tract of Tarim Wapiti. Ital. J. Anim. Sci. 2024, 23, 259–274. [Google Scholar] [CrossRef]
- Wang, B.; Deng, B.; Yong, F.; Zhou, H.; Qu, C.; Zhou, Z. Comparison of the Fecal Microbiomes of Healthy and Diarrheic Captive Wild Boar. Microb. Pathogen. 2020, 147, 104377. [Google Scholar] [CrossRef]
- Li, Y.; Hu, X.; Yang, S.; Zhou, J.; Qi, L.; Sun, X.; Fan, M.; Xu, S.; Cha, M.; Zhang, M.; et al. Comparison between the Fecal Bacterial Microbiota of Healthy and Diarrheic Captive Musk Deer. Front. Microbiol. 2018, 9, 300. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, M.; Xu, S.; Zhang, H.; Li, Y.; Hu, D. Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration. Animals 2023, 13, 3791. [Google Scholar] [CrossRef]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.C.; Jiang, Y.; et al. The Bamboo-Eating Giant Panda Harbors a Carnivore-like Gut Microbiota, with Excessive Seasonal Variations. mBio 2015, 6, e00022-15. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.E.; Brown, A.E.; Williams, C.L. The Role of Diet and Host Species in Shaping the Seasonal Dynamics of the Gut Microbiome. FEMS Microbiol. Ecol. 2023, 99, fiad156. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Zeng, D.; Zhou, Y.; Niu, L.; Deng, J.; Li, Y.; Pu, Y.; Lin, Y.; Xu, S.; Liu, Q.; et al. Microbial Biogeography along the Gastrointestinal Tract of a Red Panda. Front. Microbiol. 2018, 9, 1411. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Wen, X.; Jia, T.; Han, J.; Qin, X.; Zhang, Y.; Wang, Z. Comparative Study of the Gut Microbiota in Three Captive Rhinopithecus Species. BMC Genom. 2023, 24, 398. [Google Scholar] [CrossRef]
- Zhong, Z.; Sun, P.; Zhang, Y.; Li, L.; Han, D.; Pan, X.; Zhang, R. Differential Responses of Rumen and Fecal Fermentation and Microbiota of Liaoning Cashmere Goats after 2-Hydroxy-4-(Methylthio) Butanoic Acid Isopropyl Ester Supplementation. Sci. Rep. 2024, 14, 8505. [Google Scholar] [CrossRef]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial Cellulose Utilization: Fundamentals and Biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef]
- Sauer, U.; Eikmanns, B.J. The PEP-Pyruvate-Oxaloacetate Node as the Switch Point for Carbon Flux Distribution in Bacteria: We Dedicate This Paper to Rudolf K. Thauer, Director of the Max-Planck-Institute for Terrestrial Microbiology in Marburg, Germany, on the Occasion of His 65th Birthday. FEMS Microbiol. Rev. 2005, 29, 765–794. [Google Scholar] [CrossRef]
- Tang, L.; Gao, Y.; Yan, L.; Jia, H.; Chu, H.; Ma, X.; He, L.; Wang, X.; Li, K.; Hu, D.; et al. Comparative Analysis of Microbiome Metagenomics in Reintroduced Wild Horses and Resident Asiatic Wild Asses in the Gobi Desert Steppe. Microorganisms 2022, 10, 1166. [Google Scholar] [CrossRef]
- Clegg, S.R.; Carter, S.D.; Birtles, R.J.; Brown, J.M.; Hart, C.A.; Evans, N.J. Multilocus Sequence Typing of Pathogenic Treponemes Isolated from Cloven-Hoofed Animals and Comparison to Treponemes Isolated from Humans. Appl. Environ. Microbiol. 2016, 82, 4523–4536. [Google Scholar] [CrossRef]
- Qian, W.; Ao, W.; Jia, C.; Li, Z. Bacterial Colonisation of Reeds and Cottonseed Hulls in the Rumen of Tarim Red Deer (Cervus elaphus yarkandensis). Antonie Leeuwenhoek 2019, 112, 1283–1296. [Google Scholar] [CrossRef]
- Dias, D.; Cruz, A.; Fonseca, C.; Mendo, S.; Caetano, T.S. Antibiotic Resistance and Potential Bacterial Pathogens Identified in Red Deer’s Faecal DNA. Transbound. Emerg. Dis. 2022, 69, e3425–e3429. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Guan, L.L.; Liu, J.X. Assessment of Rumen Bacteria in Dairy Cows with Varied Milk Protein Yield. J. Dairy Sci. 2019, 102, 5031–5041. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhao, X.; Han, X.; Xu, S.; Zhao, L.; Hu, L.; Xu, T.; Zhao, N.; Zhang, X.; Chen, D.; et al. Comparative Study of Gut Microbiota in Tibetan Wild Asses (Equus kiang) and Domestic Donkeys (Equus asinus) on the Qinghai-Tibet Plateau. PeerJ 2020, 8, e9032. [Google Scholar] [CrossRef] [PubMed]
- Van Kuijk, S.J.A.; Sonnenberg, A.S.M.; Baars, J.J.P.; Hendriks, W.H.; Cone, J.W. Fungal Treated Lignocellulosic Biomass as Ruminant Feed Ingredient: A Review. Biotechnol. Adv. 2015, 33, 191–202. [Google Scholar] [CrossRef]
- Haitjema, C.H.; Solomon, K.V.; Henske, J.K.; Theodorou, M.K.; O’Malley, M.A. Anaerobic Gut Fungi: Advances in Isolation, Culture, and Cellulolytic Enzyme Discovery for Biofuel Production. Biotechnol. Bioeng. 2014, 111, 1471–1482. [Google Scholar] [CrossRef]
- Teunissen, M.J.; Smits, A.A.M.; Op Den Camp, H.J.M.; Huis In’T Veld, J.H.J.; Vogels, G.D. Fermentation of Cellulose and Production of Cellulolytic and Xylanolytic Enzymes by Anaerobic Fungi from Ruminant and Non-Ruminant Herbivores. Arch. Microbiol. 1991, 156, 290–296. [Google Scholar] [CrossRef]
- El-Zaiat, H.M.; Al-Marzooqi, W.; Al-Kharousi, K. Exploring Rumen Fermentation and Microbial Populations in Dhofari Goats Fed a Chitosan-Added Diet. Anim. Biotechnol. 2024, 35, 2337748. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Wu, Y.; Zhang, W. Effects of Fecal Microbiota Transplantation from Yaks on Weaning Diarrhea, Fecal Microbiota Composition, Microbial Network Structure and Functional Pathways in Chinese Holstein Calves. Front. Microbiol. 2022, 13, 898505. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Li, Q.; Liu, J.; Wang, X.; Gao, Q.; Yang, Y.; Zhao, X. Fecal Microbiota Reveal Adaptation of Herbivores to the Extreme Environment of the Qinghai-Tibet Plateau. Grassl. Res. 2024, 3, 155–170. [Google Scholar] [CrossRef]
- Butkovich, L.V.; Leggieri, P.A.; Lillington, S.P.; Navaratna, T.A.; Swift, C.L.; Malinov, N.G.; Zalunardo, T.R.; Vining, O.B.; Lipzen, A.; Wang, M.; et al. Separation of Life Stages within Anaerobic Fungi (Neocallimastigomycota) Highlights Differences in Global Transcription and Metabolism. Fungal Genet. Biol. 2025, 176, 103958. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Su, Y.; He, X.; Han, L.; Qu, H.; Zhou, L.; Huang, G. Membrane-Covered Composting Significantly Decreases Methane Emissions and Microbial Pathogens: Insight into the Succession of Bacterial and Fungal Communities. Sci. Total Environ. 2022, 845, 157343. [Google Scholar] [CrossRef]
- Enaud, R.; Cambos, S.; Viaud, E.; Guichoux, E.; Chancerel, E.; Marighetto, A.; Etchamendy, N.; Clark, S.; Mohammedi, K.; Cota, D.; et al. Gut Microbiota and Mycobiota Evolution Is Linked to Memory Improvement after Bariatric Surgery in Obese Patients: A Pilot Study. Nutrients 2021, 13, 4061. [Google Scholar] [CrossRef]
- Maas, E.; Penders, J.; Venema, K. Modelling the Gut Fungal-Community in TIM-2 with a Microbiota from Healthy Individuals. J. Fungi 2023, 9, 104. [Google Scholar] [CrossRef]
- Suhr, M.J.; Banjara, N.; Hallen-Adams, H.E. Sequence-Based Methods for Detecting and Evaluating the Human Gut Mycobiome. Lett. Appl. Microbiol. 2016, 62, 209–215. [Google Scholar] [CrossRef]
- Crous, P.W.; Cowan, D.A.; Maggs-Kölling, G.; Yilmaz, N.; Thangavel, R.; Wingfield, M.J.; Noordeloos, M.E.; Dima, B.; Brandrud, T.E.; Jansen, G.M.; et al. Fungal Planet Description Sheets: 1182–1283. Persoonia-Mol. Phylogeny Evol. Fungi 2021, 46, 313–528. [Google Scholar] [CrossRef]
- Du Preez, L.L.; Van Der Walt, E.; Valverde, A.; Rothmann, C.; Neser, F.W.C.; Cason, E.D. A Metagenomic Survey of the Fecal Microbiome of the African Savanna Elephant (Loxodonta africana). Anim. Genet. 2024, 55, 621–643. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, G.F.; Silva, M.R.L.; Hirata, D.B. Production of New Lipase from Preussia africana and Partial Characterization. Prep. Biochem. Biotechnol. 2022, 52, 942–949. [Google Scholar] [CrossRef]
- Seddouk, L.; Jamai, L.; Tazi, K.; Ettayebi, M.; Alaoui-Mhamdi, M.; Aleya, L.; Janati-Idrissi, A. Isolation and Characterization of a Mesophilic Cellulolytic Endophyte Preussia africana from Juniperus oxycedrus. Environ. Sci. Pollut. Res. 2022, 29, 45589–45600. [Google Scholar] [CrossRef]
- Zaferanloo, B.; Bhattacharjee, S.; Ghorbani, M.M.; Mahon, P.J.; Palombo, E.A. Amylase Production by Preussia Minima, a Fungus of Endophytic Origin: Optimization of Fermentation Conditions and Analysis of Fungal Secretome by LC-MS. BMC Microbiol. 2014, 14, 55. [Google Scholar] [CrossRef]
- Leong, S.L.; Pettersson, O.V.; Rice, T.; Hocking, A.D.; Schnürer, J. The Extreme Xerophilic Mould Xeromyces Bisporus—Growth and Competition at Various Water Activities. Int. J. Food Microbiol. 2011, 145, 57–63. [Google Scholar] [CrossRef]
- Li, J.; Luo, Y.; Chen, D.; Yu, B.; He, J.; Huang, Z.; Mao, X.; Zheng, P.; Yu, J.; Luo, J.; et al. The Fungal Community and Its Interaction with the Concentration of Short-chain Fatty Acids in the Caecum and Colon of Weaned Piglets. J. Anim. Physiol. Anim. Nutr. 2020, 104, 616–628. [Google Scholar] [CrossRef]
- Chen, Y.; Meng, L.; Yuan, W.; Gao, Z.; Zhang, X.; Xie, B.; Song, J.; Li, J.; Zhong, J.; Liu, X. Gut Fungal Microbiota Alterations in Pulmonary Arterial Hypertensive Rats. Biomedicines 2024, 12, 298. [Google Scholar] [CrossRef]
- Jiang, H.; Pan, L.; Zhang, X.; Zhang, Z.; Zhou, Y.; Ruan, B. Altered Gut Bacterial-Fungal Interkingdom Networks in Patients with Current Depressive Episode. Brain Behav. 2020, 10, e01677. [Google Scholar] [CrossRef]
- Meng, P.; Zhang, G.; Ma, X.; Ding, X.; Song, X.; Dang, S.; Yang, R.; Xu, L. Characterization of Intestinal Fungal Community Diversity in People Living with HIV/AIDS (PLWHA). AIDS Res. Ther. 2024, 21, 10. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, A.E. Gibberella from A (venaceae) to Z (eae). Annu. Rev. Phytopathol. 2003, 41, 177–198. [Google Scholar] [CrossRef] [PubMed]
- Scherm, B.; Balmas, V.; Spanu, F.; Pani, G.; Delogu, G.; Pasquali, M.; Migheli, Q. Fusarium culmorum: Causal Agent of Foot and Root Rot and Head Blight on Wheat. Mol. Plant Pathol. 2013, 14, 323–341. [Google Scholar] [CrossRef] [PubMed]
- Gajęcka, M.; Majewski, M.S.; Zielonka, Ł.; Grzegorzewski, W.; Onyszek, E.; Lisieska-Żołnierczyk, S.; Juśkiewicz, J.; Babuchowski, A.; Gajęcki, M.T. Concentration of Zearalenone, Alpha-Zearalenol and Beta-Zearalenol in the Myocardium and the Results of Isometric Analyses of the Coronary Artery in Prepubertal Gilts. Toxins 2021, 13, 396. [Google Scholar] [CrossRef]
- Teunissen, M.J.; Op Den Camp, H.J.M. Anaerobic Fungi and Their Cellulolytic and Xylanolytic Enzymes. Antonie Leeuwenhoek 1993, 63, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Stincone, A.; Prigione, A.; Cramer, T.; Wamelink, M.M.C.; Campbell, K.; Cheung, E.; Olin-Sandoval, V.; Grüning, N.; Krüger, A.; Tauqeer Alam, M.; et al. The Return of Metabolism: Biochemistry and Physiology of the Pentose Phosphate Pathway. Biol. Rev. 2015, 90, 927–963. [Google Scholar] [CrossRef]
- Adams, E.; Frank, L. Metabolism of Proline and the Hydroxyprolines. Annu. Rev. Biochem. 1980, 49, 1005–1061. [Google Scholar] [CrossRef] [PubMed]
- Geisen, S.; Mitchell, E.A.D.; Adl, S.; Bonkowski, M.; Dunthorn, M.; Ekelund, F.; Fernández, L.D.; Jousset, A.; Krashevska, V.; Singer, D.; et al. Soil Protists: A Fertile Frontier in Soil Biology Research. FEMS Microbiol. Rev. 2018, 42, 293–323. [Google Scholar] [CrossRef]
- Gile, G.H. Protist Symbionts of Termites: Diversity, Distribution, and Coevolution. Biol. Rev. 2024, 99, 622–652. [Google Scholar] [CrossRef]
- Henriquez, F.L.; Mooney, R.; Bandel, T.; Giammarini, E.; Zeroual, M.; Fiori, P.L.; Margarita, V.; Rappelli, P.; Dessì, D. Paradigms of Protist/Bacteria Symbioses Affecting Human Health: Acanthamoeba Species and Trichomonas Vaginalis. Front. Microbiol. 2021, 11, 616213. [Google Scholar] [CrossRef]
- Mattison, R.G.; Harayama, S. The Predatory Soil Flagellate Heteromita globosa Stimulates Toluene Biodegradation by a Pseudomonas sp. FEMS Microbiol. Lett. 2001, 194, 39–45. [Google Scholar] [CrossRef]
- Quispe-Rodríguez, G.H.; Wankewicz, A.A.; Luis Málaga Granda, J.; Lewis, B.; Stockert, K.; Clinton White, A. ‘Entamoeba Histolytica’ Identified by Stool Microscopy from Children with Acute Diarrhoea in Peru Is Not E. histolytica. Trop. Doct. 2020, 50, 19–22. [Google Scholar] [CrossRef]
- Burrell, A.; Tomley, F.M.; Vaughan, S.; Marugan-Hernandez, V. Life Cycle Stages, Specific Organelles and Invasion Mechanisms of Eimeria Species. Parasitology 2020, 147, 263–278. [Google Scholar] [CrossRef]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The Hidden World within Plants: Ecological and Evolutionary Considerations for Defining Functioning of Microbial Endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Ley, R.E.; Hamady, M.; Lozupone, C.; Turnbaugh, P.J.; Ramey, R.R.; Bircher, J.S.; Schlegel, M.L.; Tucker, T.A.; Schrenzel, M.D.; Knight, R.; et al. Evolution of Mammals and Their Gut Microbes. Science 2008, 320, 1647–1651. [Google Scholar] [CrossRef]
- Azad, E.; Fehr, K.B.; Derakhshani, H.; Forster, R.; Acharya, S.; Khafipour, E.; McGeough, E.; McAllister, T.A. Interrelationships of Fiber-Associated Anaerobic Fungi and Bacterial Communities in the Rumen of Bloated Cattle Grazing Alfalfa. Microorganisms 2020, 8, 1543. [Google Scholar] [CrossRef]
- Tasnim, N.; Abulizi, N.; Pither, J.; Hart, M.M.; Gibson, D.L. Linking the Gut Microbial Ecosystem with the Environment: Does Gut Health Depend on Where We Live? Front. Microbiol. 2017, 8, 1935. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet Rapidly and Reproducibly Alters the Human Gut Microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Mishra, P.; Tulsani, N.J.; Jakhesara, S.J.; Dafale, N.A.; Patil, N.V.; Purohit, H.J.; Koringa, P.G.; Joshi, C.G. Exploring the Eukaryotic Diversity in Rumen of Indian Camel (Camelus dromedarius) Using 18S rRNA Amplicon Sequencing. Arch. Microbiol. 2020, 202, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, Y.; Zhao, Z.; Deng, F.; Jiang, H.; Liu, C.; Li, Y.; Chai, J. Bacterial-Fungal Interactions: Mutualism, Antagonism, and Competition. Life 2025, 15, 1242. [Google Scholar] [CrossRef] [PubMed]
- Bhandari, R.; Wong, A.C.-N.; Lee, J.C.; Boyd, A.; Shelby, K.; Ringbauer, J.; Kang, D.S. Microbiome Composition and Co-Occurrence Dynamics in Wild Drosophila suzukii Are Influenced by Host Crop, Fly Sex, and Sampling Location. Microbiol. Spectr. 2025, 13, e02608-24. [Google Scholar] [CrossRef] [PubMed]
- Moraïs, S.; Winkler, S.; Zorea, A.; Levin, L.; Nagies, F.S.P.; Kapust, N.; Lamed, E.; Artan-Furman, A.; Bolam, D.N.; Yadav, M.P.; et al. Cryptic Diversity of Cellulose-Degrading Gut Bacteria in Industrialized Humans. Science 2024, 383, eadj9223. [Google Scholar] [CrossRef]
- Martínez-Romero, E.; Aguirre-Noyola, J.L.; Bustamante-Brito, R.; González-Román, P.; Hernández-Oaxaca, D.; Higareda-Alvear, V.; Montes-Carreto, L.M.; Martínez-Romero, J.C.; Rosenblueth, M.; Servín-Garcidueñas, L.E. We and Herbivores Eat Endophytes. Microb. Biotechnol. 2021, 14, 1282–1299. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.H.; Chen, Z.D.; Zhou, S.; Song, X.Z.; Ouyang, K.H.; Pan, K.; Xu, L.J.; Liu, C.J.; Qu, M.R. Effects of Daidzein on Performance, Serum Metabolites, Nutrient Digestibility, and Fecal Bacterial Community in Bull Calves. Anim. Feed Sci. Technol. 2017, 225, 87–96. [Google Scholar] [CrossRef]
- Pitta, D.W.; Kumar, S.; Veiccharelli, B.; Parmar, N.; Reddy, B.; Joshi, C.G. Bacterial Diversity Associated with Feeding Dry Forage at Different Dietary Concentrations in the Rumen Contents of Mehshana Buffalo (Bubalus bubalis) Using 16S Pyrotags. Anaerobe 2014, 25, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Barko, P.C.; McMichael, M.A.; Swanson, K.S.; Williams, D.A. The Gastrointestinal Microbiome: A Review. J. Vet. Intern. Med. 2018, 32, 9–25. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhou, Y.; Wu, Y.; Ma, C.; Ruan, X.; Cha, M.; Zhou, Y.; Li, T.; Sun, W.; Liu, H. Comparative Profiling of the Fecal Bacteriome, Mycobiome, and Protist Community in Wild Versus Captive (Cervus canadensis). Animals 2026, 16, 44. https://doi.org/10.3390/ani16010044
Zhou Y, Wu Y, Ma C, Ruan X, Cha M, Zhou Y, Li T, Sun W, Liu H. Comparative Profiling of the Fecal Bacteriome, Mycobiome, and Protist Community in Wild Versus Captive (Cervus canadensis). Animals. 2026; 16(1):44. https://doi.org/10.3390/ani16010044
Chicago/Turabian StyleZhou, Yalin, Yan Wu, Cuiliu Ma, Xingzhou Ruan, Muha Cha, Yulei Zhou, Tao Li, Weili Sun, and Hanlu Liu. 2026. "Comparative Profiling of the Fecal Bacteriome, Mycobiome, and Protist Community in Wild Versus Captive (Cervus canadensis)" Animals 16, no. 1: 44. https://doi.org/10.3390/ani16010044
APA StyleZhou, Y., Wu, Y., Ma, C., Ruan, X., Cha, M., Zhou, Y., Li, T., Sun, W., & Liu, H. (2026). Comparative Profiling of the Fecal Bacteriome, Mycobiome, and Protist Community in Wild Versus Captive (Cervus canadensis). Animals, 16(1), 44. https://doi.org/10.3390/ani16010044






