Red Blood Cell Transcriptome Reflects Physiological Responses to Alternative Nutrient Sources in Gilthead Seabream (Sparus aurata)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
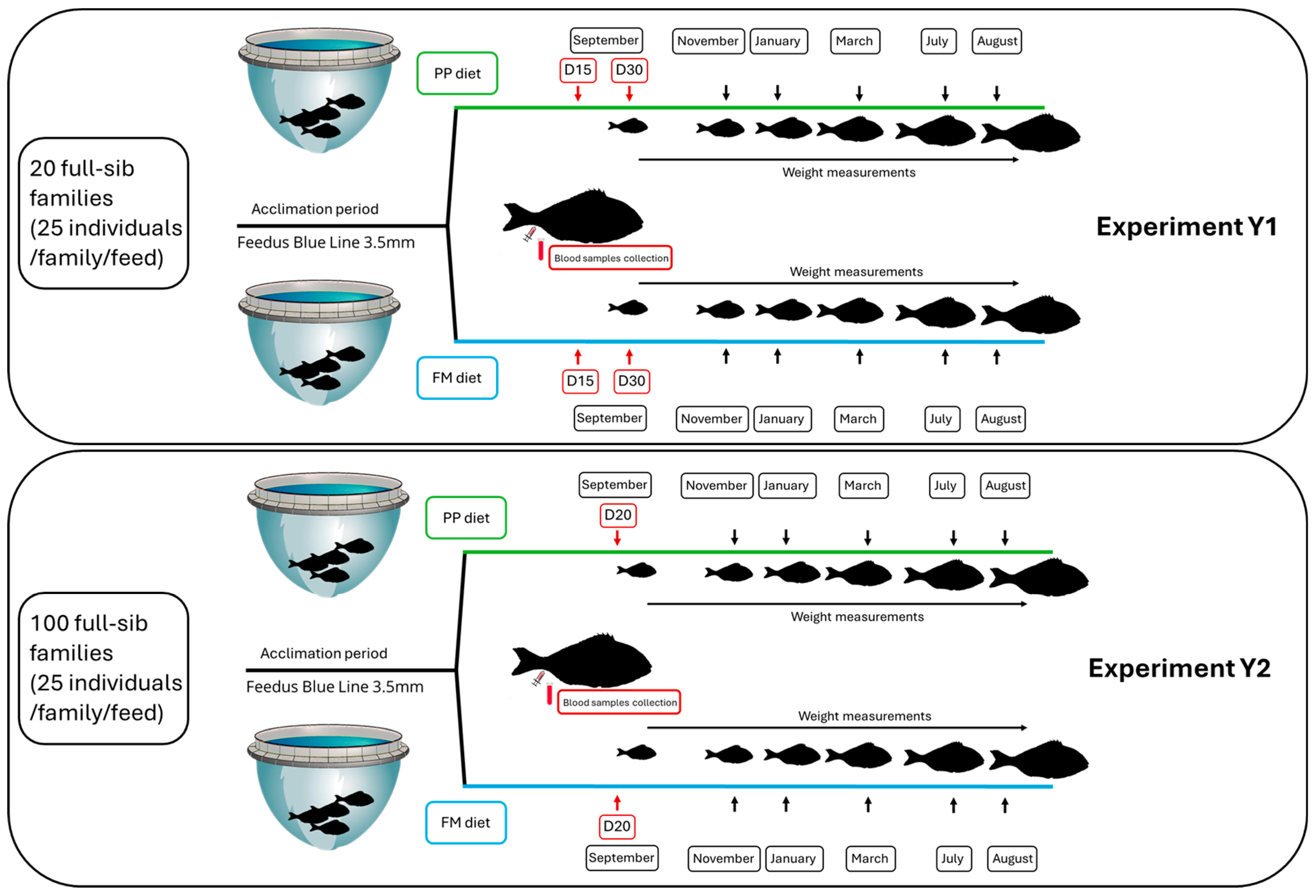
2.2. Experimental Fish
2.2.1. Experiment 1
2.2.2. Experiment 2
2.3. Blood Sampling
- Experiment 1: Approximately 200 µL blood was collected from 384 individuals (16 fish per family × 6 families × 2 sampling points (D15 and D30) × 2 diets (FM and PP diet)).
- Experiment 2: Approximately 200 µL blood was collected from 96 individuals (16 fish per family × 3 families × 1 sampling point (D20) × 2 diets (FM and PP diet)).
2.4. RNA Extraction
2.5. RNA Sequencing and Bioinformatics Analysis
- Experiment 1: A total of 24 pooled samples (6 families × 2 sampling points (D15 and D30) × 2 diets (FM and PP diet)) were sequenced.
- Experiment 2: A total of 6 pooled samples (3 families × 1 sampling point (D20) × 2 diets (FM and PP diet)) were sequenced.
2.6. Quantitive Expression Analysis (qRT-PCR)
2.7. Statistical Analysis
3. Results
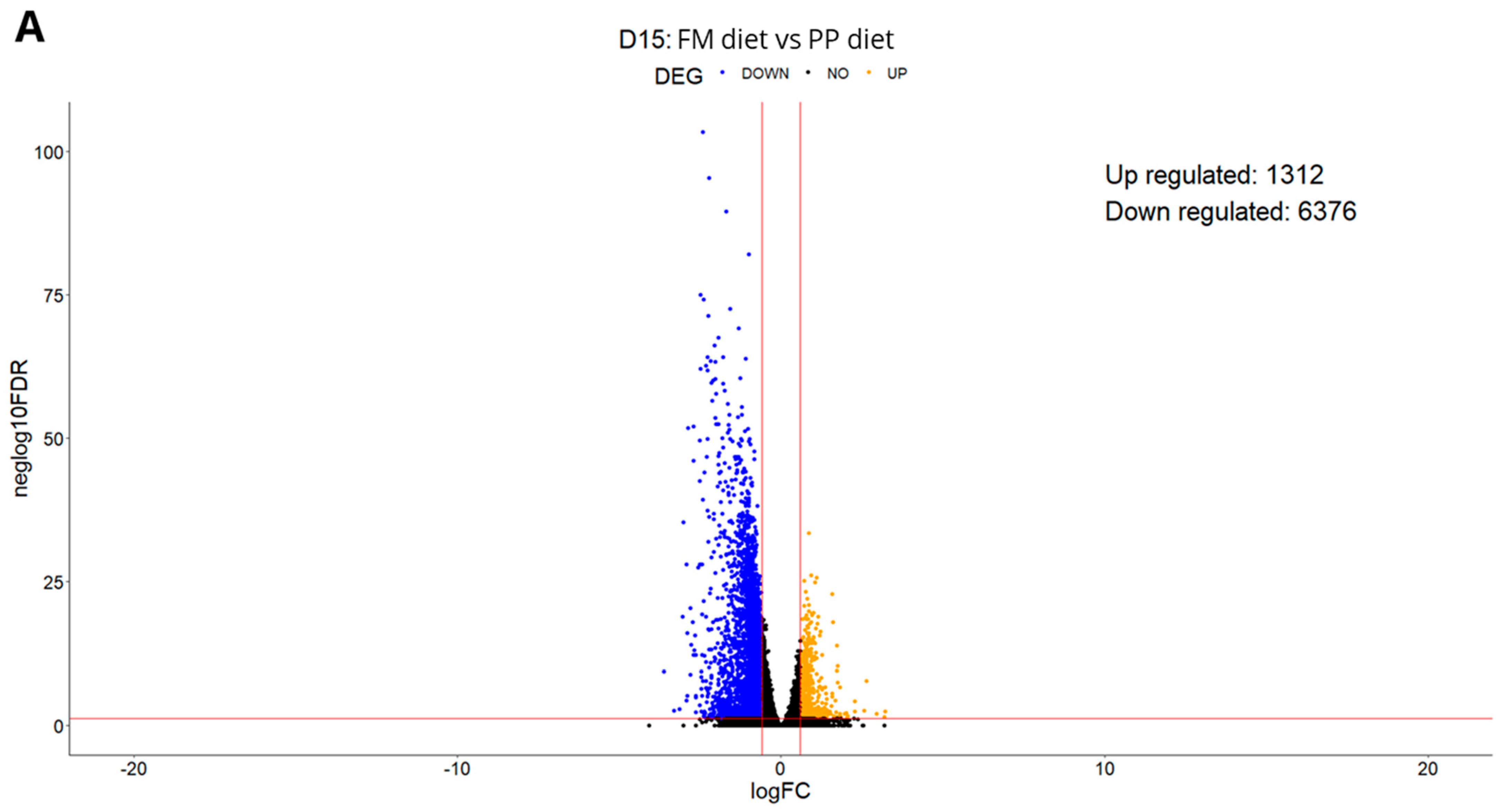
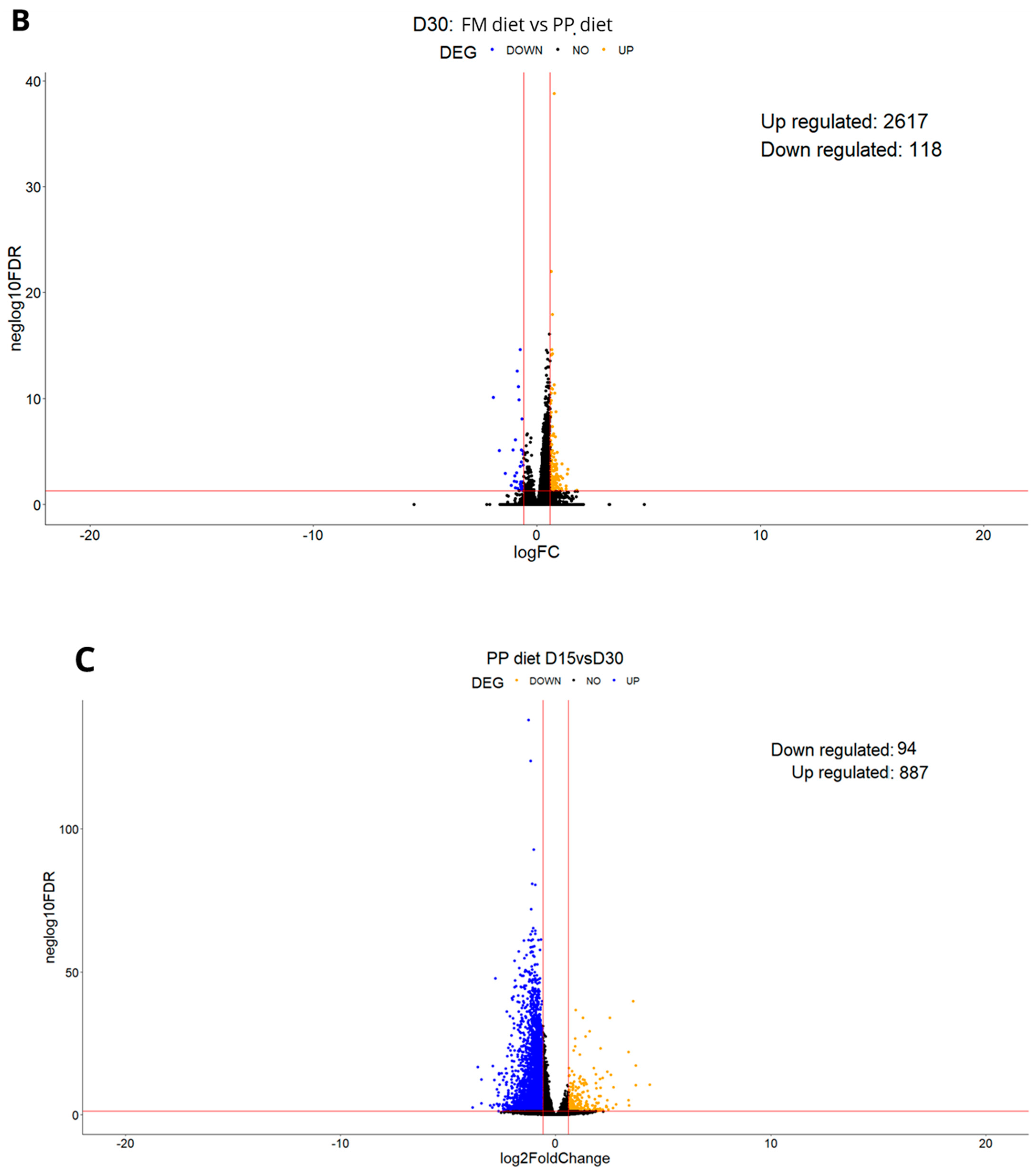
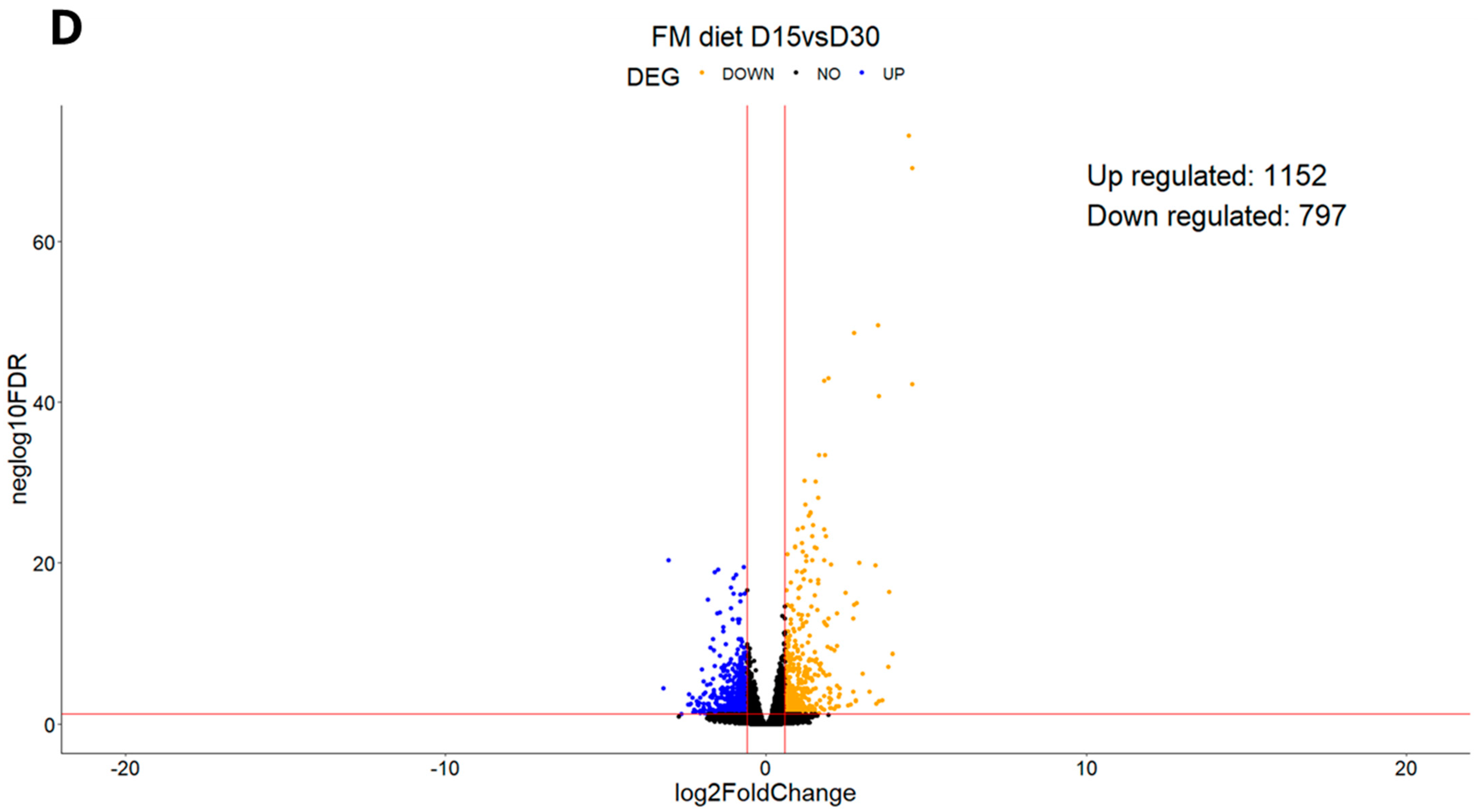
3.1. Experiment 1
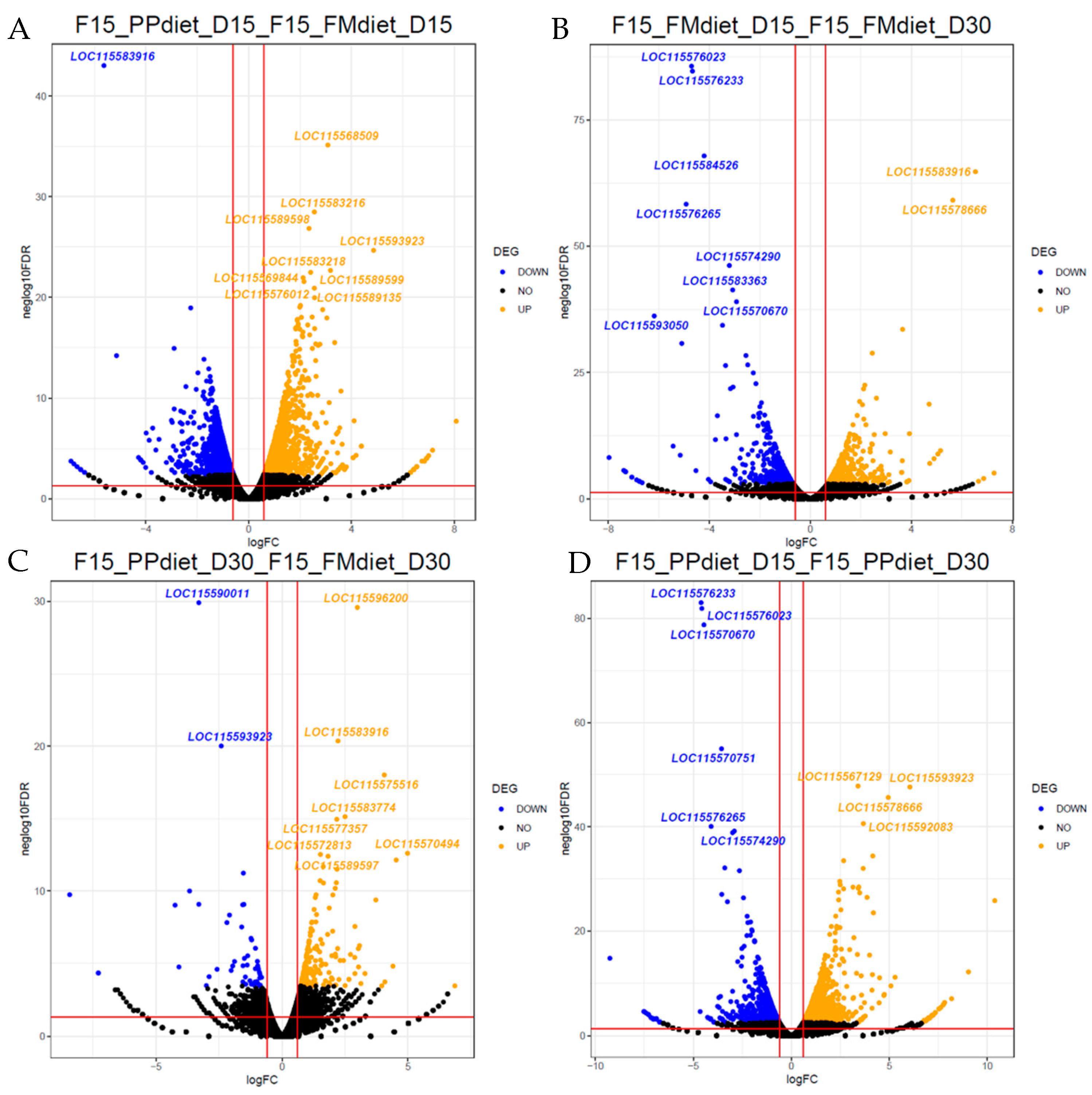
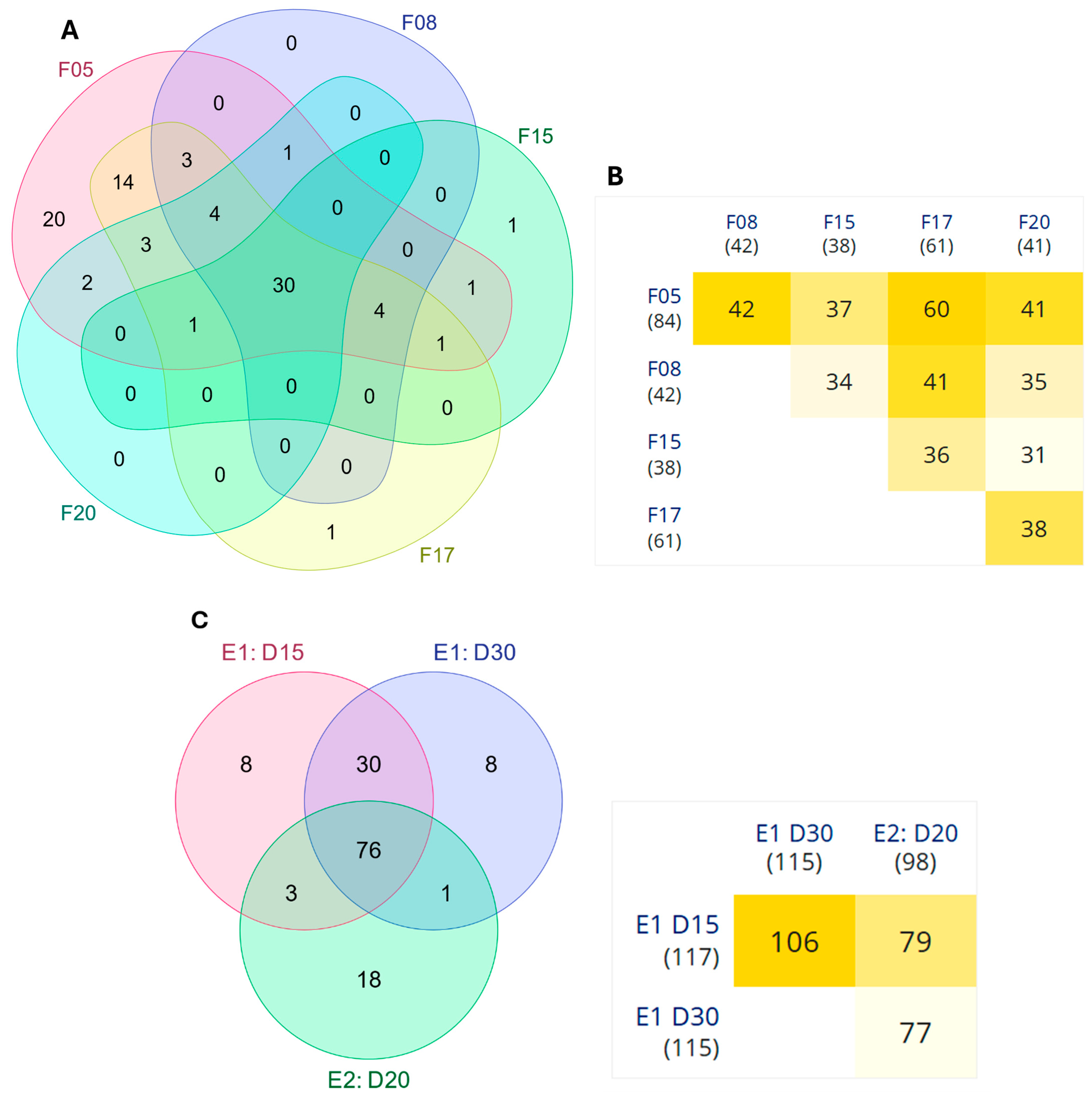
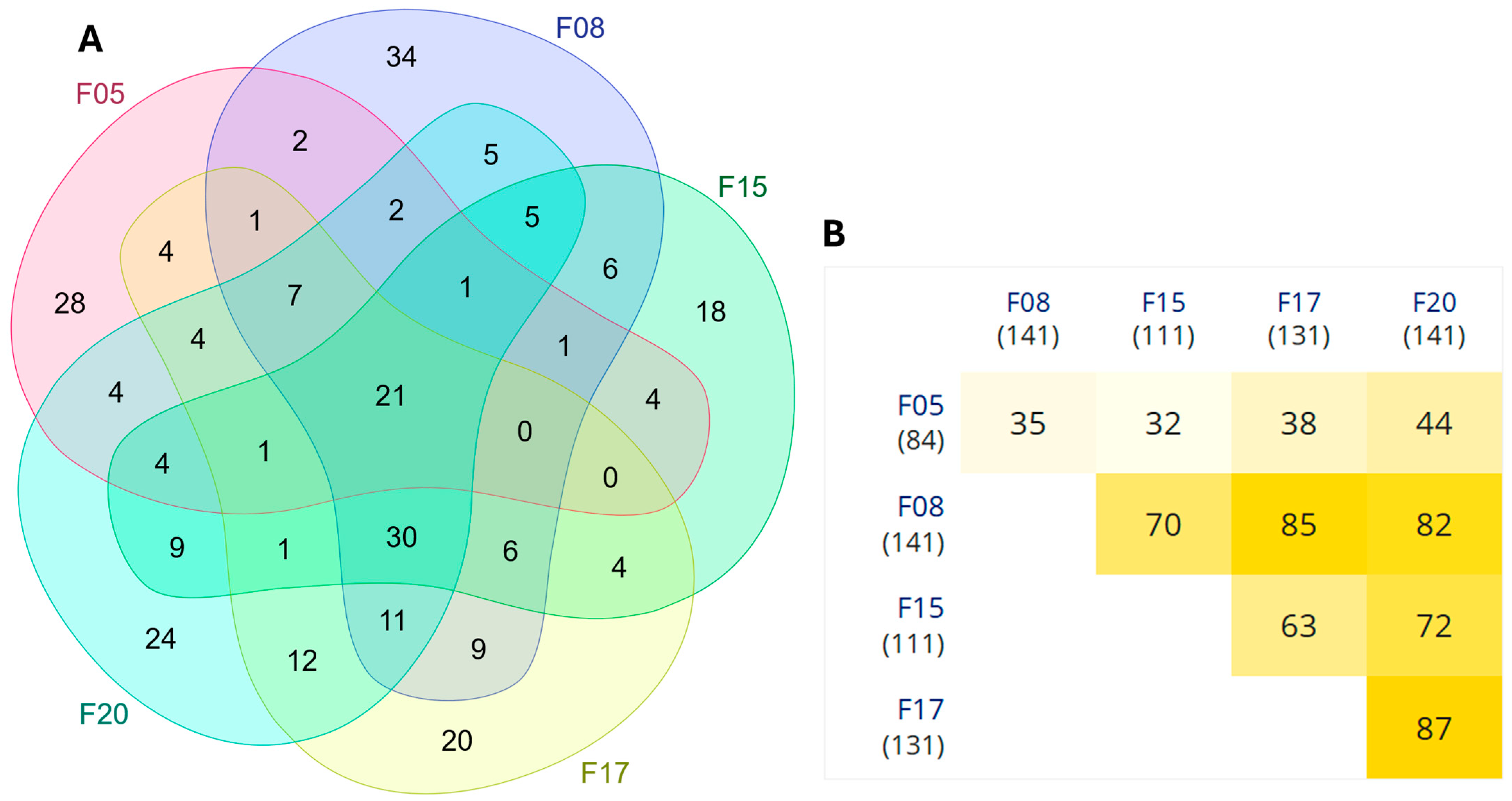
3.1.1. Differential Expression Analysis
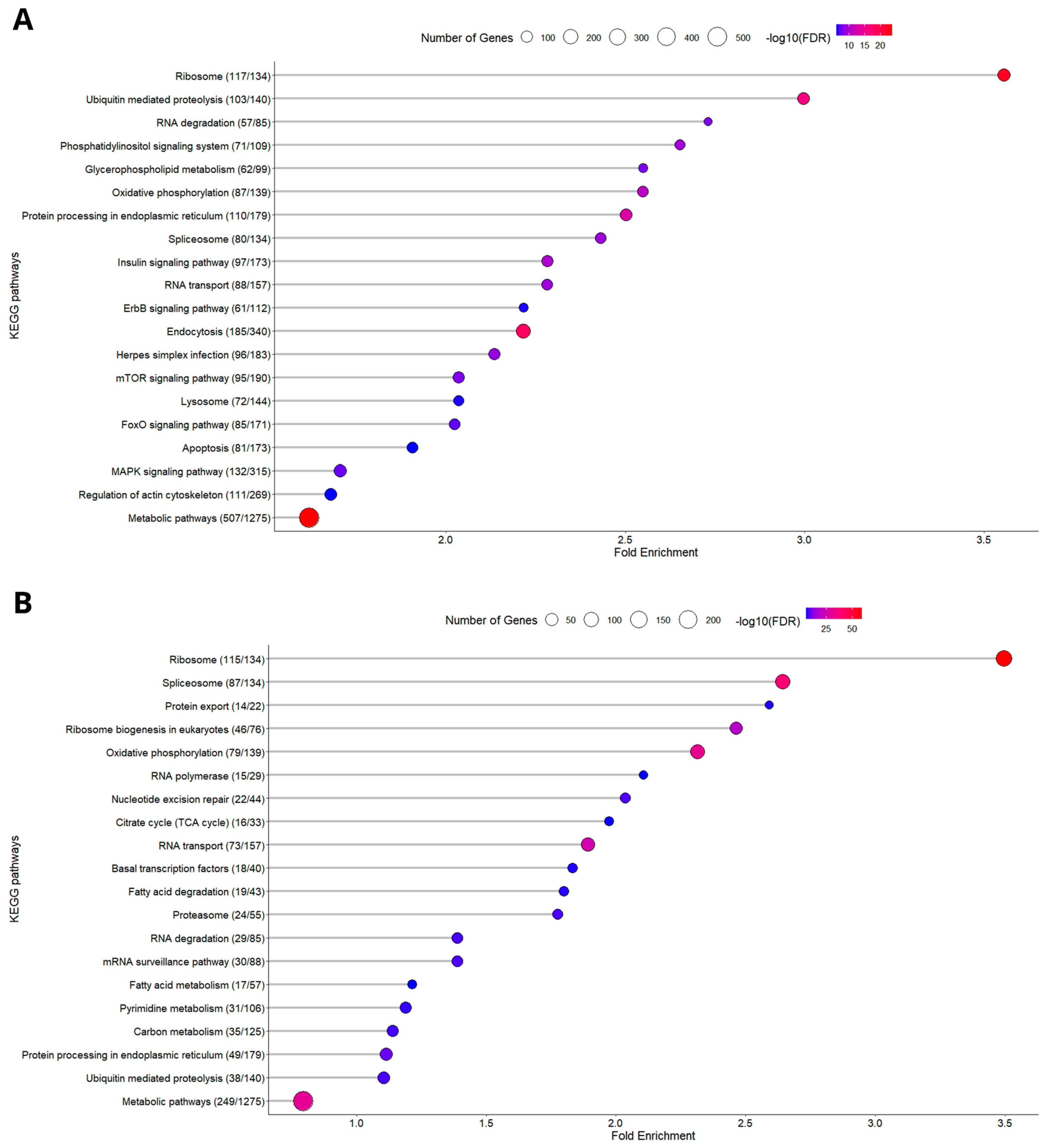
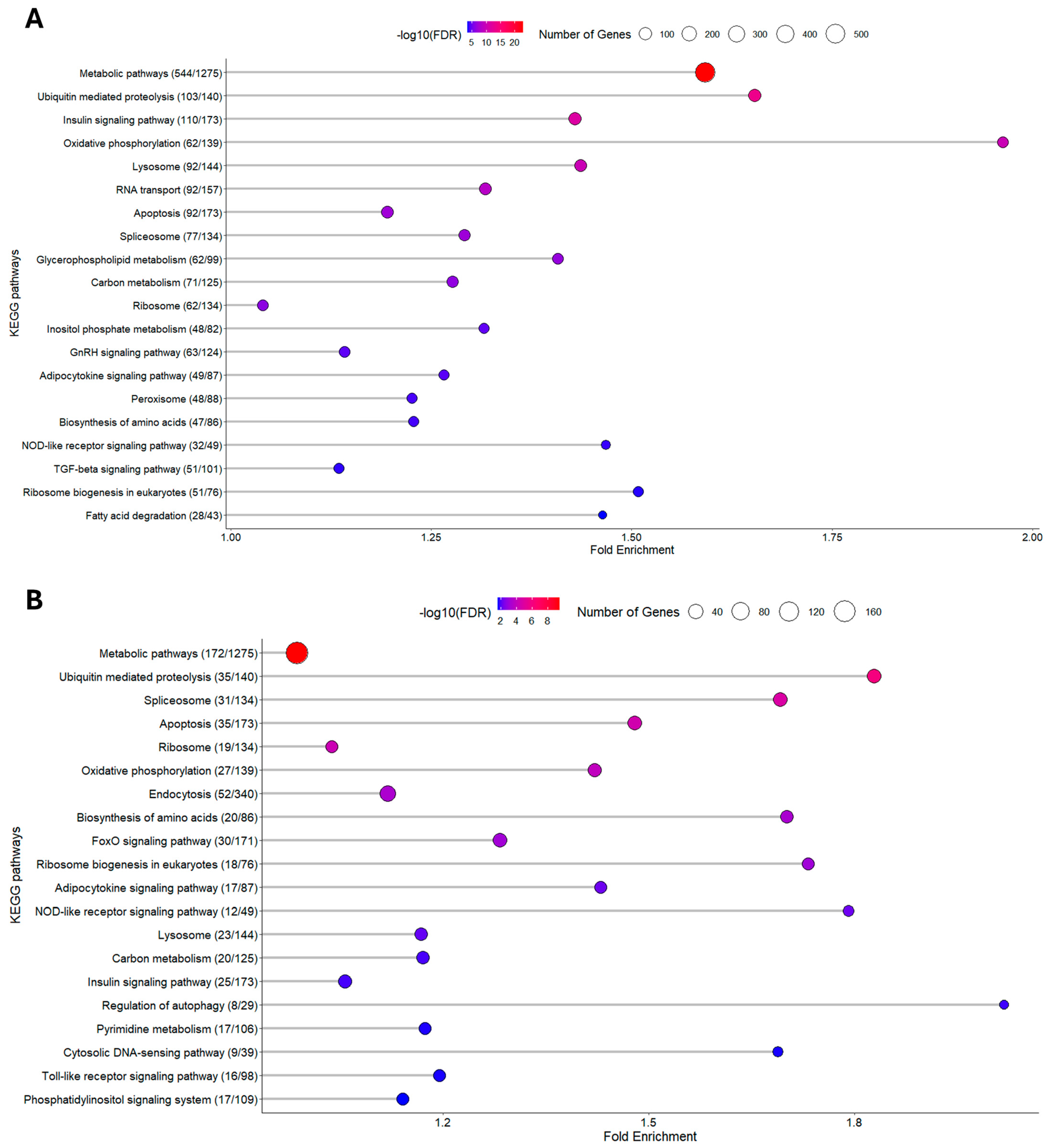
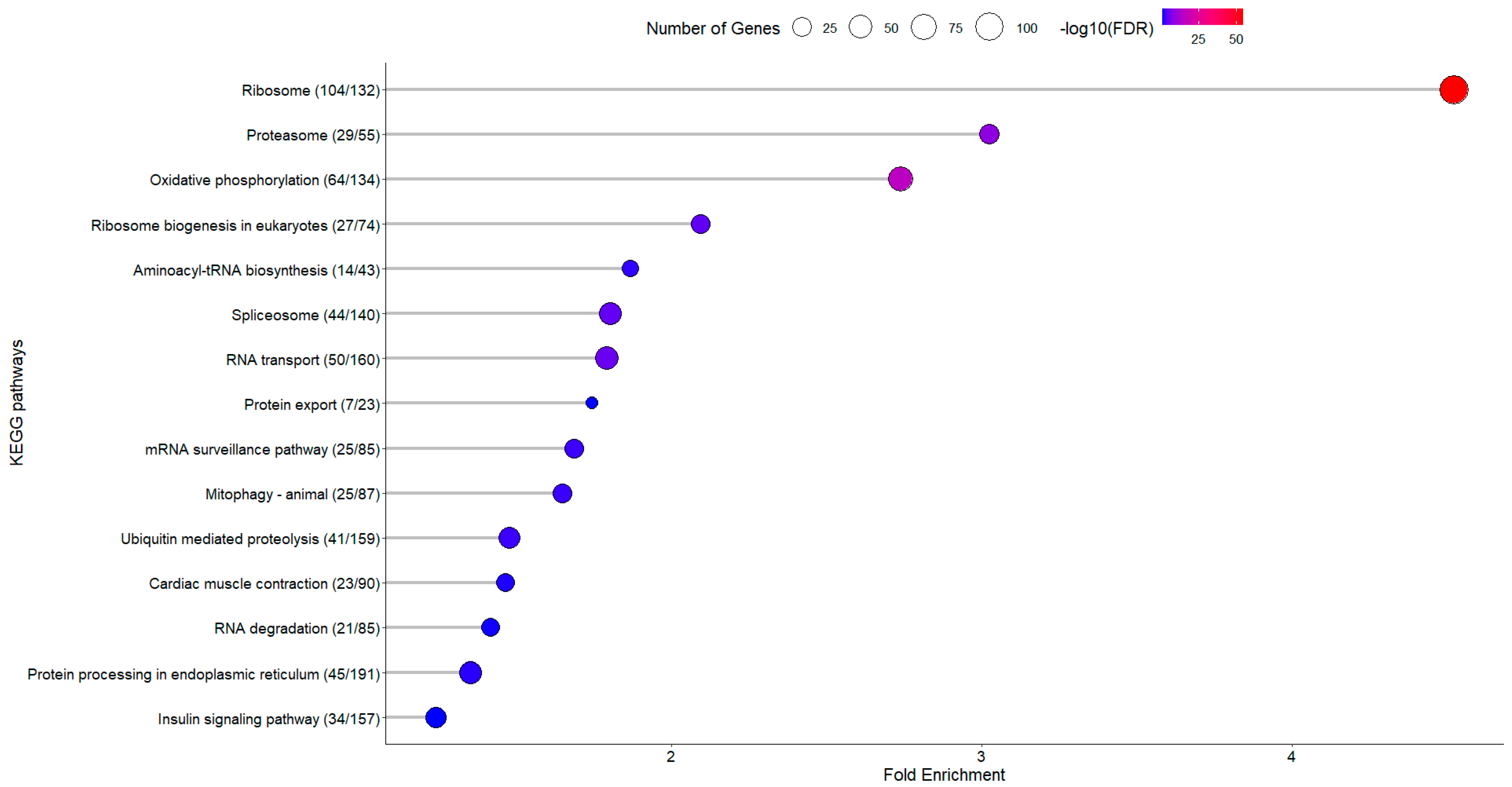
3.1.2. KEGG Enrichment Analysis
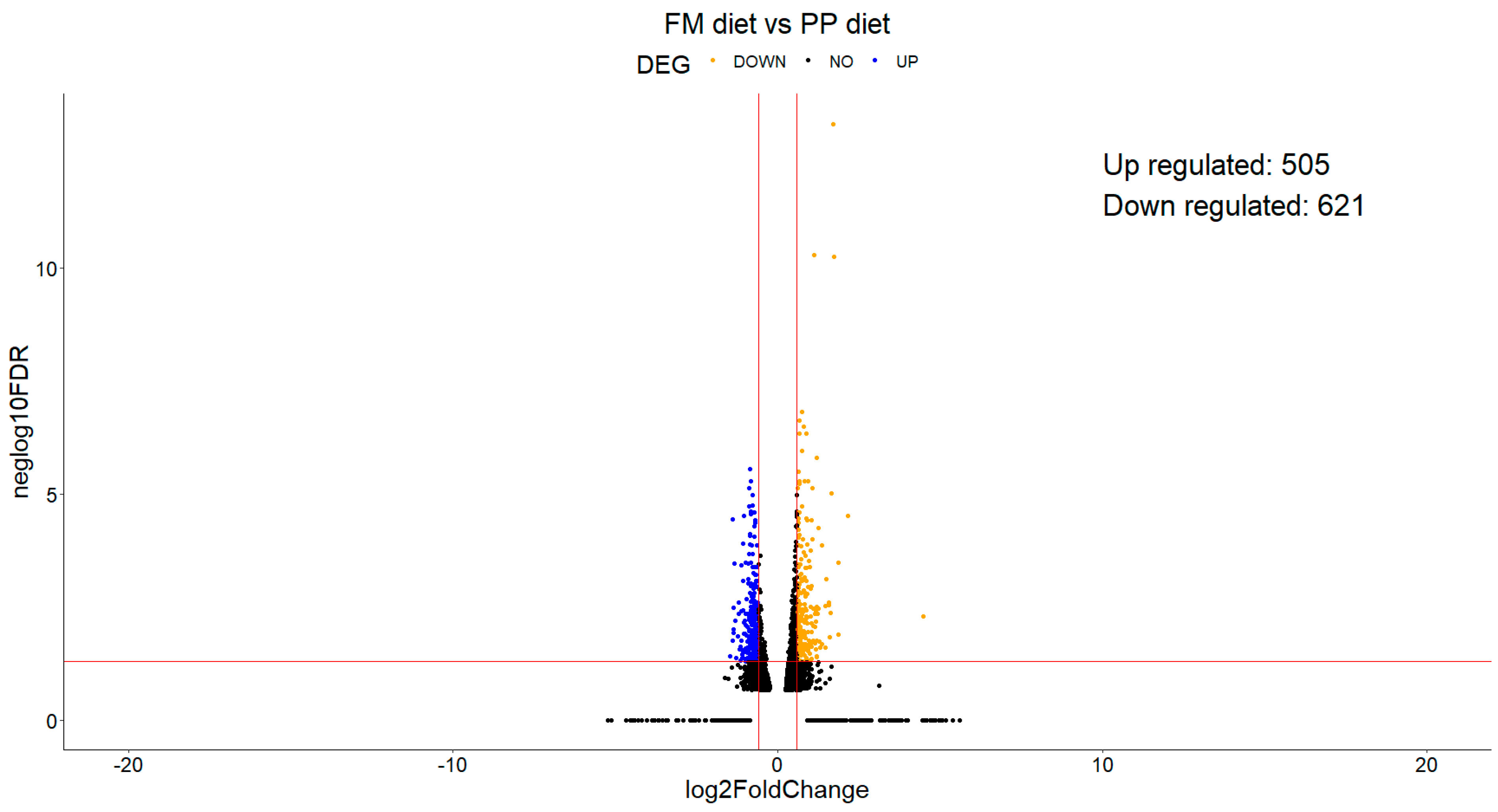
3.2. Experiment 2
3.2.1. Differential Expression Analysis
3.2.2. KEGG Enrichment Analysis
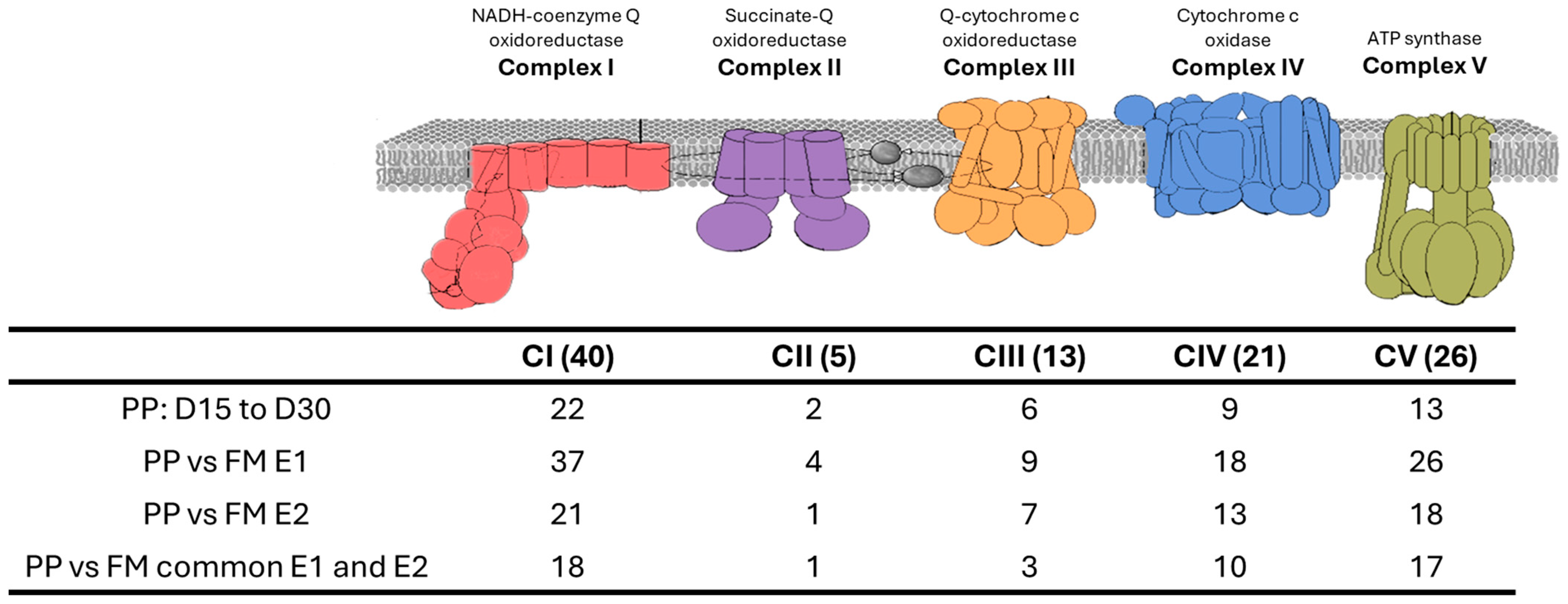
3.3. Oxidative Phosphorylation (OxPhos) Pathway
3.4. Ribosome Pathway
3.5. Metabolic Pathways
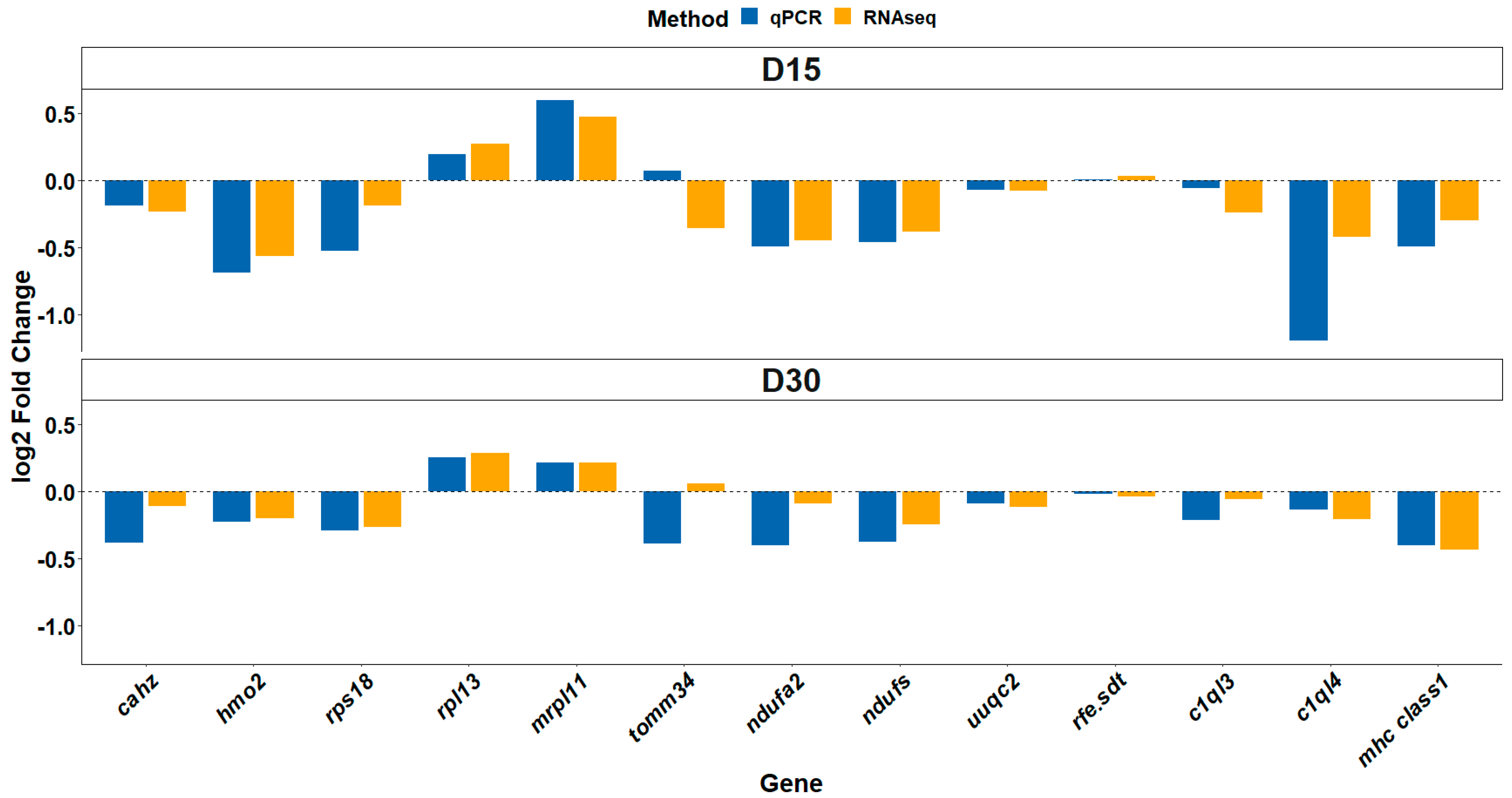
3.6. Differential Gene Expression Validation
4. Discussion
4.1. Oxidative Phosphorylation
4.2. Ribosomal KEGG Pathway
4.3. Metabolic Pathways
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Naylor, R.L.; Hardy, R.W.; Buschmann, A.H.; Bush, S.R.; Cao, L.; Klinger, D.H.; Little, D.C.; Lubchenco, J.; Shumway, S.E.; Troell, M. A 20-Year Retrospective Review of Global Aquaculture. Nature 2021, 591, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Egerton, S.; Wan, A.; Murphy, K.; Collins, F.; Ahern, G.; Sugrue, I.; Busca, K.; Egan, F.; Muller, N.; Whooley, J.; et al. Replacing Fishmeal with Plant Protein in Atlantic Salmon (Salmo salar) Diets by Supplementation with Fish Protein Hydrolysate. Sci. Rep. 2020, 10, 4194. [Google Scholar] [CrossRef] [PubMed]
- Turchini, G.M.; Trushenski, J.T.; Glencross, B.D. Thoughts for the Future of Aquaculture Nutrition: Realigning Perspectives to Reflect Contemporary Issues Related to Judicious Use of Marine Resources in Aquafeeds. N. Am. J. Aquac. 2019, 81, 13–39. [Google Scholar] [CrossRef]
- Pavan Kumar, B.; Ramudu, K.R.; Devi, B.C. Mini Review on Incorporation of Cotton Seed Meal, an Alternative to Fish Meal in Aquaculture Feeds. Int. J. Biol. Res. 2014, 2, 99–105. [Google Scholar] [CrossRef]
- Morais, S.; Pratoomyot, J.; Taggart, J.B.; Bron, J.E.; Guy, D.R.; Bell, J.G.; Tocher, D.R. Genotype-Specific Responses in Atlantic Salmon (Salmo salar) Subject to Dietary Fish Oil Replacement by Vegetable Oil: A Liver Transcriptomic Analysis. BMC Genom. 2011, 12, 255. [Google Scholar] [CrossRef]
- Hardy, R.W. Utilization of Plant Proteins in Fish Diets: Effects of Global Demand and Supplies of Fishmeal. Aquac. Res. 2010, 41, 770–776. [Google Scholar] [CrossRef]
- Hussain, S.M.; Bano, A.A.; Ali, S.; Rizwan, M.; Adrees, M.; Zahoor, A.F.; Sarker, P.K.; Hussain, M.; Arsalan, M.Z.-u.-H.; Yong, J.W.H.; et al. Substitution of Fishmeal: Highlights of Potential Plant Protein Sources for Aquaculture Sustainability. Heliyon 2024, 10, e26573. [Google Scholar] [CrossRef]
- Perera, E.; Simó-Mirabet, P.; Shin, H.S.; Rosell-Moll, E.; Naya-Catalá, F.; de las Heras, V.; Martos-Sitcha, J.A.; Karalazos, V.; Armero, E.; Arizcun, M.; et al. Selection for Growth Is Associated in Gilthead Sea Bream (Sparus aurata) with Diet Flexibility, Changes in Growth Patterns and Higher Intestine Plasticity. Aquaculture 2019, 507, 349–360. [Google Scholar] [CrossRef]
- Xu, W.; Jin, J.; Han, D.; Liu, H.; Zhu, X.; Yang, Y.; Xie, S. Physiological and Transcriptomic Responses to Fishmeal-Based Diet and Rapeseed Meal-Based Diet in Two Strains of Gibel Carp (Carassius gibelio). Fish Physiol. Biochem. 2019, 45, 267–286. [Google Scholar] [CrossRef]
- Sabbagh, M.; Schiavone, R.; Brizzi, G.; Sicuro, B.; Zilli, L.; Vilella, S. Poultry By-Product Meal as an Alternative to Fish Meal in the Juvenile Gilthead Seabream (Sparus aurata) Diet. Aquaculture 2019, 511, 734220. [Google Scholar] [CrossRef]
- Aragão, C.; Gonçalves, A.T.; Costas, B.; Azeredo, R.; Xavier, M.J.; Engrola, S. Alternative Proteins for Fish Diets: Implications beyond Growth. Animals 2022, 12, 1211. [Google Scholar] [CrossRef] [PubMed]
- Liew, C.-C.; Ma, J.; Tang, H.-C.; Zheng, R.; Dempsey, A.A. The Peripheral Blood Transcriptome Dynamically Reflects System Wide Biology: A Potential Diagnostic Tool. J. Lab. Clin. Med. 2006, 147, 126–132. [Google Scholar] [CrossRef] [PubMed]
- Angelakopoulos, R.; Tsipourlianos, A.; Moutou, K.A.; Fytsili, A.E.; Tsingene, A.; Galliopoulou, E.; Papaharisis, L.; Mamuris, Z.; Giannoulis, T.; Dimitroglou, A. Selection of Nonlethal Early Biomarkers to Predict Gilthead Seabream (Sparus aurata) Growth. Aquac. Nutr. 2025, 2025, 9918595. [Google Scholar] [CrossRef]
- HAPO. Annual Report: Aquaculture in Greece. 2023. Available online: https://fishfromgreece.com/wp-content/uploads/2023/10/HAPO_AR23_WEB-NEW.pdf (accessed on 11 January 2025).
- FEAP. European Aquaculture Production Report; FEAP: Brussels, Belgium, 2024; Available online: https://feap.info (accessed on 11 January 2025).
- Andrew, S.C.; Primmer, C.R.; Debes, P.V.; Erkinaro, J.; Verta, J.P. The Atlantic Salmon Whole Blood Transcriptome and How It Relates to Major Locus Maturation Genotypes and Other Tissues. Mar. Genom. 2021, 56, 100809. [Google Scholar] [CrossRef]
- Jégou, M.; Gondret, F.; Vincent, A.; Tréfeu, C.; Gilbert, H.; Louveau, I. Whole Blood Transcriptomics Is Relevant to Identify Molecular Changes in Response to Genetic Selection for Feed Efficiency and Nutritional Status in the Pig. PLoS ONE 2016, 11, e0146550. [Google Scholar] [CrossRef]
- Götting, M.; Nikinmaa, M.J. Transcriptomic Analysis of Young and Old Erythrocytes of Fish. Front. Physiol. 2017, 8, 1046. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Čikoš, Š.; Bukovská, A.; Koppel, J. Relative Quantification of MRNA: Comparison of Methods Currently Used for Real-Time PCR Data Analysis. BMC Mol. Biol. 2007, 8, 113. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate Normalization of Real-Time Quantitative RT-PCR Data by Geometric Averaging of Multiple Internal Control Genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 13 December 2024).
- RStudio Team. RStudio: Integrated Development for R. RStudio; PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 13 December 2024).
- Shapiro, S.S.; Wilk, M.B. An Analysis of Variance Test for Normality (Complete Samples). Biometrika 1965, 52, 591–611. [Google Scholar] [CrossRef]
- Dodge, Y. Kruskal-Wallis Test. In The Concise Encyclopedia of Statistics; Springer: New York, NY, USA, 2008. [Google Scholar]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Kassambara, A. ggpubr: ‘ggplot2’ Based Publication Ready Plots. R Package Version 0.6.0. 2023. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 13 December 2024).
- Leduc, A.; Zatylny-Gaudin, C.; Robert, M.; Corre, E.; Corguille, G.L.; Castel, H.; Lefevre-Scelles, A.; Fournier, V.; Gisbert, E.; Andree, K.B.; et al. Dietary Aquaculture By-Product Hydrolysates: Impact on the Transcriptomic Response of the Intestinal Mucosa of European Seabass (Dicentrarchus labrax) Fed Low Fish Meal Diets. BMC Genom. 2018, 19, 396. [Google Scholar] [CrossRef] [PubMed]
- Francis, G.; Makkar, H.P.S.; Becker, K. Antinutritional Factors Present in Plant-Derived Alternate Fish Feed Ingredients and Their Effects in Fish. Aquaculture 2001, 199, 197–227. [Google Scholar] [CrossRef]
- Harter, T.S.; Brauner, C.J. Teleost Red Blood Cells Actively Enhance the Passive Diffusion of Oxygen That Was Discovered by August Krogh. Comp. Biochem. Physiol.-Part A Mol. Integr. Physiol. 2021, 253, 110855. [Google Scholar] [CrossRef]
- Gilmour, K.M.; Perry, S.F. Carbonic Anhydrase and Acid-Base Regulation in Fish. J. Exp. Biol. 2009, 212, 1647–1661. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.; Egée, S. Fish Red Blood Cells: Characteristics and Physiological Role of the Membrane Ion Transporters. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 1998, 119, 79–86. [Google Scholar] [CrossRef]
- Dichiera, A.M.; Khursigara, A.J.; Esbaugh, A.J. The Effects of Warming on Red Blood Cell Carbonic Anhydrase Activity and Respiratory Performance in a Marine Fish. Comp. Biochem. Physiol.-Part A Mol. Integr. Physiol. 2021, 260, 111033. [Google Scholar] [CrossRef]
- Quyet, D.H.; Dung, P.T.; Le Na, N.T.; Dung, M.B.; Huong, N.T.M.; Vuong, T.P. Hematological Profile of Red Drum Sciaenops ocellatus (Linnaeus, 1766) under Natural and Commercial Feed Nutritional Conditions. Isr. J. Aquac.-Bamidgeh 2025, 77, 91–103. [Google Scholar] [CrossRef]
- Tsipourlianos, A.; Cardoso, J.C.R.; Angelakopoulos, R.; Kotoula, A.; Power, D.M.; Mamuris, Z.; Moutou, K.A. Regulatory Subfunctionalization Drives OXPHOS Evolution in Teleosts. bioRxiv 2024. [Google Scholar] [CrossRef]
- Silva-Marrero, J.I.; Sáez, A.; Caballero-Solares, A.; Viegas, I.; Almajano, M.P.; Fernández, F.; Baanante, I.V.; Metón, I. A Transcriptomic Approach to Study the Effect of Long-Term Starvation and Diet Composition on the Expression of Mitochondrial Oxidative Phosphorylation Genes in Gilthead Sea Bream (Sparus aurata). BMC Genom. 2017, 18, 768. [Google Scholar] [CrossRef]
- Torricelli, M.; Felici, A.; Branciari, R.; Trabalza-Marinucci, M.; Galarini, R.; Biagetti, M.; Manfrin, A.; Boriani, L.; Radicchi, E.; Sebastiani, C.; et al. Gene Expression Study in Gilthead Seabream (Sparus aurata): Effects of Dietary Supplementation with Olive Oil Polyphenols on Immunity, Metabolic, and Oxidative Stress Pathways. Int. J. Mol. Sci. 2024, 25, 12185. [Google Scholar] [CrossRef]
- Vera, L.M.; Metochis, C.; Taylor, J.F.; Clarkson, M.; Skjærven, K.H.; Migaud, H.; Tocher, D.R. Early Nutritional Programming Affects Liver Transcriptome in Diploid and Triploid Atlantic Salmon, Salmo salar. BMC Genom. 2017, 18, 886. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.K.; Xu, Y.C.; Luo, Z.; Zhao, T.; Zheng, H.; Tan, X.Y. Fish Meal Replacement by Mixed Plant Protein in the Diets for Juvenile Yellow Catfish Pelteobagrus gulvidraco: Effects on Growth Performance and Health Status. Aquac. Nutr. 2022, 2022, 2677885. [Google Scholar] [CrossRef] [PubMed]
- Calduch-Giner, J.A.; Sitjà-Bobadilla, A.; Davey, G.C.; Cairns, M.T.; Kaushik, S.; Pérez-Sánchez, J. Dietary Vegetable Oils Do Not Alter the Intestine Transcriptome of Gilthead Sea Bream (Sparus aurata), but Modulate the Transcriptomic Response to Infection with Enteromyxum Leei. BMC Genom. 2012, 13, 470. [Google Scholar] [CrossRef]
- Xu, H.; Liao, Z.; Wang, C.; Wei, Y.; Liang, M. Hepatic Transcriptome of the Euryhaline Teleost Japanese Seabass (Lateolabrax japonicus) Fed Diets Characterized by α-Linolenic Acid or Linoleic Acid. Comp. Biochem. Physiol. Part D Genom. Proteom. 2019, 29, 106–116. [Google Scholar] [CrossRef]
- de Magalhães, C.R.; Sandoval, K.; Kagan, F.; McCormack, G.; Schrama, D.; Carrilho, R.; Farinha, A.P.; Cerqueira, M.; Rodrigues, P.M. Transcriptomic Changes behind Sparus aurata Hepatic Response to Different Aquaculture Challenges: An RNA-Seq Study and Multiomics Integration. PLoS ONE 2024, 19, e0300472. [Google Scholar] [CrossRef]
- Geay, F.; Ferraresso, S.; Zambonino-Infante, J.L.; Bargelloni, L.; Quentel, C.; Vandeputte, M.; Kaushik, S.; Cahu, C.L.; Mazurais, D. Effects of the Total Replacement of Fish-Based Diet with Plant-Based Diet on the Hepatic Transcriptome of Two European Sea Bass (Dicentrarchus labrax) Half-Sibfamilies Showing Different Growth Rates with the Plant-Based Diet. BMC Genom. 2011, 12, 522. [Google Scholar] [CrossRef]
- De Santis, C.; Crampton, V.O.; Bicskei, B.; Tocher, D.R. Replacement of Dietary Soy- with Air Classified Faba Bean Protein Concentrate Alters the Hepatic Transcriptome in Atlantic Salmon (Salmo salar) Parr. Comp. Biochem. Physiol. Part D Genom. Proteom. 2015, 16, 48–58. [Google Scholar] [CrossRef]
- Estruch, G.; Collado, M.C.; Monge-Ortiz, R.; Tomás-Vidal, A.; Jover-Cerdá, M.; Peñaranda, D.S.; Pérez Martínez, G.; Martínez-Llorens, S. Long-Term Feeding with High Plant Protein Based Diets in Gilthead Seabream (Sparus aurata, L.) Leads to Changes in the Inflammatory and Immune Related Gene Expression at Intestinal Level. BMC Vet. Res. 2018, 14, 302. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Alvarez, M.J.; Arzel, J.; Corraze, G.; Diez, A.; Bautista, J.M.; Kaushik, S.J. Dietary Protein Source Affects Lipid Metabolism in the European Seabass (Dicentrarchus labrax). Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2005, 142, 19–31. [Google Scholar] [CrossRef]
- Kortner, T.M.; Björkhem, I.; Krasnov, A.; Timmerhaus, G.; Krogdahl, Å. Dietary Cholesterol Supplementation to a Plant-Based Diet Suppresses the Complete Pathway of Cholesterol Synthesis and Induces Bile Acid Production in Atlantic Salmon (Salmo salar L.). Br. J. Nutr. 2014, 111, 2089–2103. [Google Scholar] [CrossRef] [PubMed]
- Kemski, M.M.; Rappleye, C.A.; Dabrowski, K.; Bruno, R.S.; Wick, M. Transcriptomic Response to Soybean Meal-Based Diets as the First Formulated Feed in Juvenile Yellow Perch (Perca flavescens). Sci. Rep. 2020, 10, 3998. [Google Scholar] [CrossRef] [PubMed]
- Krogdahl, Å.; Penn, M.; Thorsen, J.; Refstie, S.; Bakke, A.M. Important Antinutrients in Plant Feedstuffs for Aquaculture: An Update on Recent Findings Regarding Responses in Salmonids. Aquac. Res. 2010, 41, 333–344. [Google Scholar] [CrossRef]
- Bermejo-Nogales, A.; Calduch-Giner, J.A.; Pérez-Sánchez, J. Unraveling the Molecular Signatures of Oxidative Phosphorylation to Cope with the Nutritionally Changing Metabolic Capabilities of Liver and Muscle Tissues in Farmed Fish. PLoS ONE 2015, 10, e0122889. [Google Scholar] [CrossRef]
- Kristoffersen, S.; Tobiassen, T.; Steinsund, V.; Olsen, R.L. Slaughter Stress, Postmortem Muscle PH and Rigor Development in Farmed Atlantic Cod (Gadus morhua L.). Int. J. Food Sci. Technol. 2006, 41, 861–864. [Google Scholar] [CrossRef]
- Cai, W.; Liu, H.; Han, D.; Zhu, X.; Jin, J.; Yang, Y.; Xie, S. Complete Replacement of Fishmeal With Plant Protein Ingredients in Gibel Carp (Carassius auratus gibelio) Diets by Supplementation With Essential Amino Acids Without Negative Impact on Growth Performance and Muscle Growth-Related Biomarkers. Front. Mar. Sci. 2022, 8, 759086. [Google Scholar] [CrossRef]
| Ingredients | FM Diet | PP Diet |
|---|---|---|
| Fish meal standard % | 24.6 | |
| Fish meal LT % | 8.1 | |
| Fish oil % | 4.6 | 5.0 |
| Salmon oil % | 6.5 | |
| Rapeseed oil % | 6.5 | |
| Soya bean meal % | 9.9 | 21.0 |
| Rapeseed meal % | 14.0 | |
| Soya protein concentrate % | 17.0 | 7.0 |
| Corn gluten % | 14.5 | 9.6 |
| Plant premix * % | 6.6 | 8.0 |
| Sunflower meal % | 5.0 | 4.1 |
| Wheat % | 9.6 | 10.7 |
| Amino acid premix ** % | 3.2 | |
| Vitamin and mineral premix % | 1.0 | 1.0 |
| Phospholipids *** % | 0.3 | 0.8 |
| Ca & P source **** % | 0.4 | 1.0 |
| Gene ID | Gene Description | Gene Name | Forward Primer | Reverse Primer | Product Size (bp) |
|---|---|---|---|---|---|
| ENSSAUG00010018560/XM_030411990.1 | elongation factor 1-alpha, somatic form | ef1a | TCAAGGGATGGAAGGTTGAG | AGTTCCAATACCGCCGAT | 152 |
| ENSSAUG00010007010 | hemoglobin, beta adult 2 | hbba2 | GCAAGGGTGCTGATCGTCTA | GGGCTGCCACTTTGGAGTTA | 103 |
| ENSSAUG00010003114 | ribosomal protein L13a | rpl13 | TCTGGAGGACTGTCAGGGGCATGC | AGACGCACAATCTTAAGAGCAG | 197 |
| ENSSAUG00010000811 | 40S ribosomal protein S18 | rps18 | AGGGTGTTGGCAGACGTTAC | GAGGACCTGGCTGTATTTGC | 148 |
| ENSSAUG00010012992 | complement C1q-like protein 3 | c1ql3 | TTTGGAGACGGAGCGAAGAC | CCATACGCCTCACCACCTTT | 121 |
| ENSSAUG00010012990 | complement C1q-like protein 4 | c1ql4 | AGGTTGACACAGCCTTCCATA | CACTCATGTTGGGTTTGCAGG | 111 |
| ENSSAUG00010011851 | Carbonic anhydrase | cahz | AGGTGGACTTTGTGGACGAC | AAGCTCACAGGGGAACTTGA | 155 |
| ENSSAUG00010003856 | Rieske (Fe-S) domain containing | rfe.sdt | AGATGTGCATCGTTTGTCCA | TAGACATCCCCGTTGGTCTC | 166 |
| ENSSAUG00010006080 | NADH:ubiquinone oxidoreductase core subunit S1 | ndufs | CCCACTCTTCAACGCCAGAA | TCCCAGGTGGTCATACGAGT | 106 |
| ENSSAUG00010002150 | NADH:ubiquinone oxidoreductase subunit A2 | ndufa2 | CAGTAAGGGGGCCAGAGATT | GTTGTCCACCATGACACTGC | 157 |
| ENSSAUG00010006606 | mitochondrial ribosomal protein L11 | mrpl11 | ACGAGATCGCAAGGGTTAAA | GCTGCTCCAGGAAGATTTTG | 159 |
| ENSSAUG00010007642 | translocase of outer mitochondrial membrane 34 | tomm34 | CCTGTCGGTGAAGCAGTACA | AGGTTGTTCAGGTCGTCCAC | 150 |
| ENSSAUG00010025859 | heme oxygenase-like | hmo2 | CGCCTACACCCGTTATCTGG | GCTGTTCATCCTGCTCCTGT | 166 |
| ENSSAUG00010025665 | heat shock protein HSP 90-alpha | hsp90aa | TGACCCTCAGACACACTCCA | GTCGTCATCGTCCCCTTCAA | 139 |
| ENSSAUG00010000727 | major histocompatibility complex classI-related gene protein -like | mhc class 1 | AGATCGGATCGGAACCAACG | CGATGAATCCAACAGCACCG | 107 |
| Families | IBW (g) | FBW (g) | SGR1 (%) | SGR2 (%) | SGR3 (%) | SGR4 (%) | SGR5 (%) | Final SGR (%) | Survival Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| F05 | 50/46 | 449/343 | 0.54/0.46 | 0.14/0.14 | 0.11/0.09 | 0.24/0.21 | 0.14/0.15 | 1.77/1.58 | 87/86 |
| F06 | 56/54 | 556/512 | 0.55/0.51 | 0.17/0.18 | 0.1/0.11 | 0.27/0.27 | 0.13/0.12 | 1.85/1.81 | 89/86 |
| F08 | 54/51 | 533/436 | 0.53/0.4 | 0.16/0.16 | 0.1/0.12 | 0.29/0.31 | 0.14/0.15 | 1.79/1.66 | 87/80 |
| F15 | 53/49 | 439/343 | 0.51/0.44 | 0.15/0.12 | 0.1/0.08 | 0.22/0.24 | 0.14/0.14 | 1.75/1.62 | 85/78 |
| F17 | 52/43 | 423/320 | 0.52/0.46 | 0.14/0.12 | 0.08/0.08 | 0.23/0.24 | 0.14/0.15 | 1.73/1.6 | 88/85 |
| F20 | 37/34 | 479/407 | 0.63/0.52 | 0.17/0.22 | 0.13/0.11 | 0.3/0.35 | 0.19/0.15 | 2.03/1.99 | 91/92 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Angelakopoulos, R.; Tsipourlianos, A.; Fytsili, A.E.; Papaharisis, L.; Dimitroglou, A.; Barkas, D.; Mamuris, Z.; Giannoulis, T.; Moutou, K.A. Red Blood Cell Transcriptome Reflects Physiological Responses to Alternative Nutrient Sources in Gilthead Seabream (Sparus aurata). Animals 2025, 15, 1279. https://doi.org/10.3390/ani15091279
Angelakopoulos R, Tsipourlianos A, Fytsili AE, Papaharisis L, Dimitroglou A, Barkas D, Mamuris Z, Giannoulis T, Moutou KA. Red Blood Cell Transcriptome Reflects Physiological Responses to Alternative Nutrient Sources in Gilthead Seabream (Sparus aurata). Animals. 2025; 15(9):1279. https://doi.org/10.3390/ani15091279
Chicago/Turabian StyleAngelakopoulos, Rafael, Andreas Tsipourlianos, Alexia E. Fytsili, Leonidas Papaharisis, Arkadios Dimitroglou, Dimitrios Barkas, Zissis Mamuris, Themistoklis Giannoulis, and Katerina A. Moutou. 2025. "Red Blood Cell Transcriptome Reflects Physiological Responses to Alternative Nutrient Sources in Gilthead Seabream (Sparus aurata)" Animals 15, no. 9: 1279. https://doi.org/10.3390/ani15091279
APA StyleAngelakopoulos, R., Tsipourlianos, A., Fytsili, A. E., Papaharisis, L., Dimitroglou, A., Barkas, D., Mamuris, Z., Giannoulis, T., & Moutou, K. A. (2025). Red Blood Cell Transcriptome Reflects Physiological Responses to Alternative Nutrient Sources in Gilthead Seabream (Sparus aurata). Animals, 15(9), 1279. https://doi.org/10.3390/ani15091279





