Uncovering the Genetic Basis of Porcine Resilience Through GWAS of Feed Intake Data
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Data Collection
2.2. Quality Control
2.3. Resilience Traits and Production Traits
2.4. Statistical Analysis
2.5. Genotype Data
2.6. Population Structure Analysis
2.7. Single-Population GWAS
2.8. Bioinformatics Analysis
3. Results
3.1. Phenotype Statistics
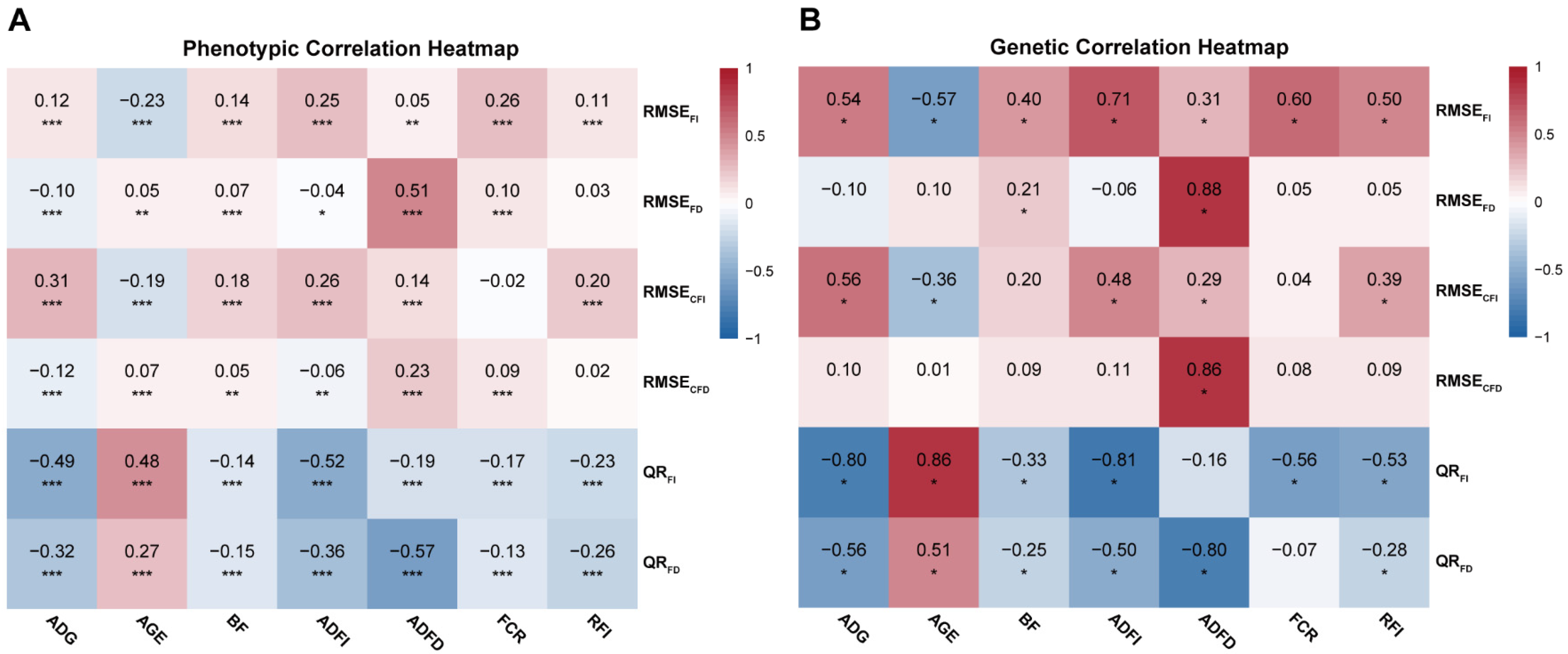
3.2. Correlation
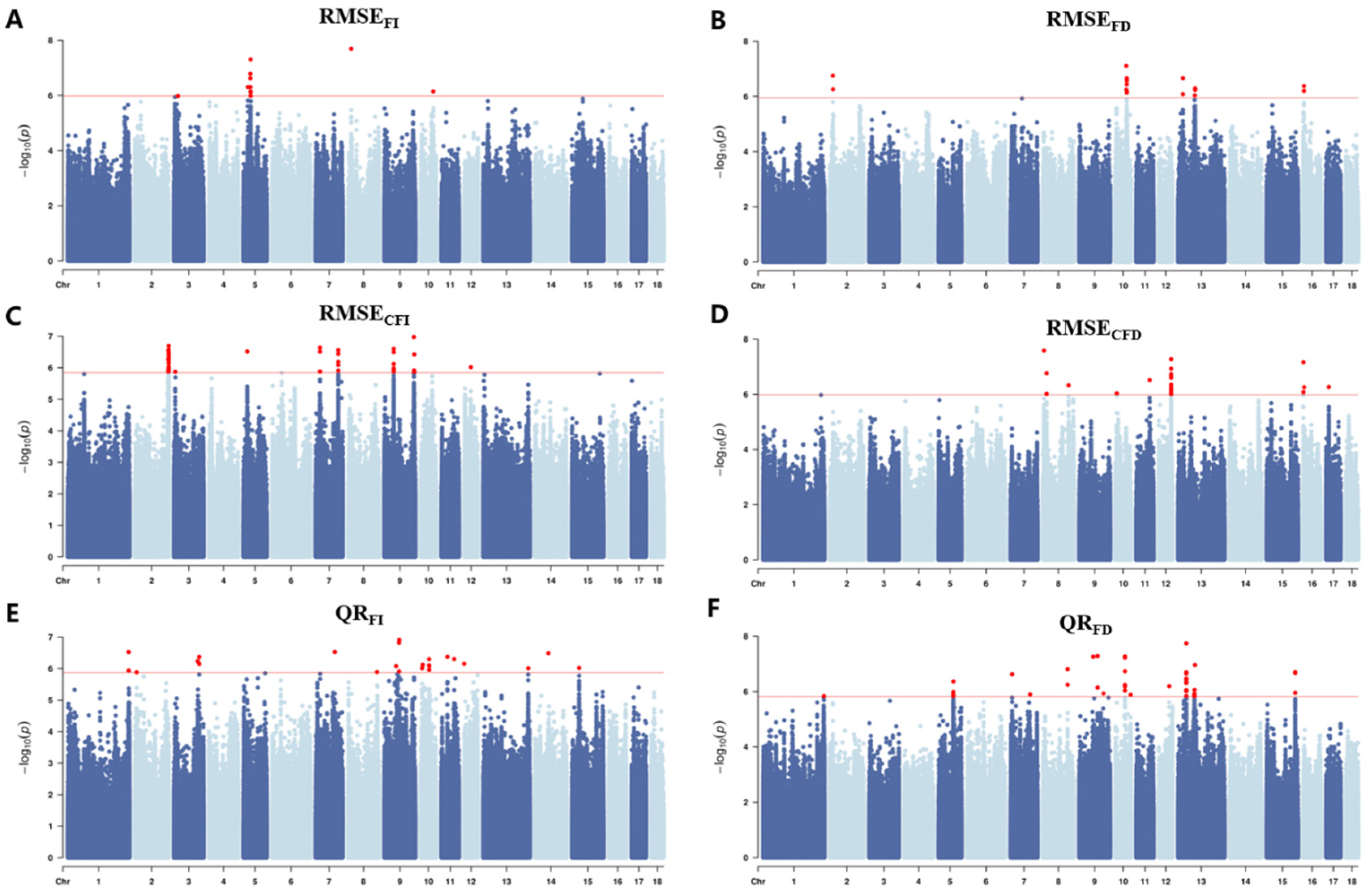
3.3. Population Genetic Structure and GWAS Results
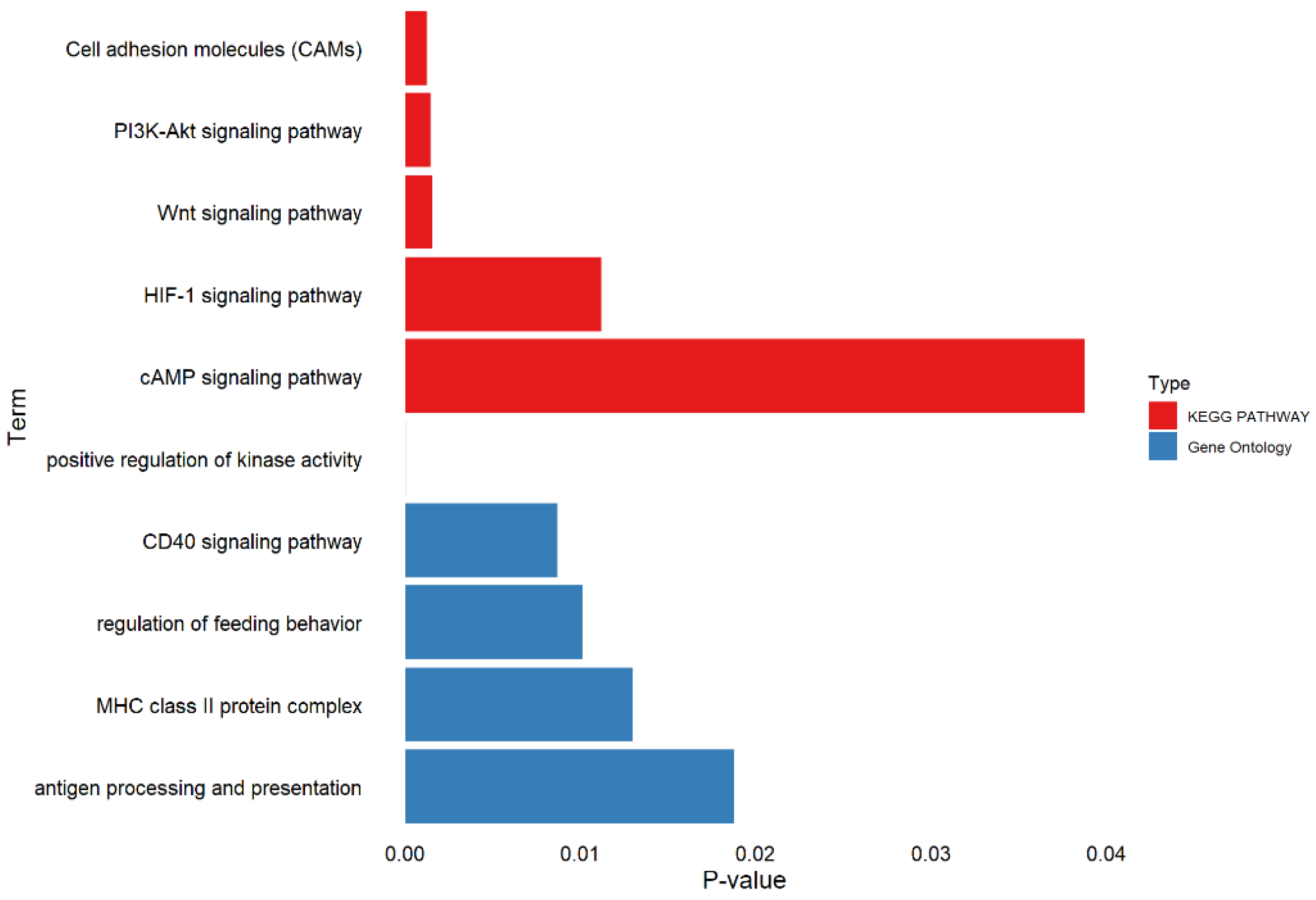
3.4. Functional Annotation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| RMSE | Mean square error roots |
| OLS | Ordinary least squares |
| QR | Quantile regression |
| FI | Daily feed intake |
| FD | Daily feed duration |
| CFI | Cumulative feed intake |
| CFD | Cumulative feed duration |
| ADG | Average daily gain |
| AGE | Adjust 100 kg age |
| BF | Adjust 100 kg backfat thickness |
| ADFI | Average occupation time in feeder per day |
| ADFD | Average number of visits to feeder per day |
| FCR | Feed conversion ratio |
| RFI | Residual feed intake |
| GWASs | Genome-wide association studies |
References
- Kim, S.W.; Gormley, A.; Jang, K.B.; Duarte, M.E. Invited Review—Current status of global pig production: An overview and research trends. Anim. Biosci. 2024, 37, 719–729. [Google Scholar] [CrossRef]
- Mote, B.E.; Rothschild, M.F. Modern genetic and genomic improvement of the pig. In Animal Agriculture; Elsevier: Amsterdam, The Netherlands, 2020; pp. 249–262. [Google Scholar]
- Kemp, B.; Da Silva, C.L.A.; Soede, N.M. Recent advances in pig reproduction: Focus on impact of genetic selection for female fertility. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 28–36. [Google Scholar] [CrossRef]
- Fablet, C.; Rose, N.; Grasland, B.; Robert, N.; Lewandowski, E.; Gosselin, M. Factors associated with the growing-finishing performances of swine herds: An exploratory study on serological and herd level indicators. Porc. Health Manag. 2018, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Iung, L.H.S.; Carvalheiro, R.; Neves, H.H.R.; Mulder, H.A. Genetics and genomics of uniformity and resilience in livestock and aquaculture species: A review. J. Anim. Breed. Genet. 2020, 137, 263–280. [Google Scholar] [CrossRef] [PubMed]
- Berghof, T.V.L.; Poppe, M.; Mulder, H.A. Opportunities to Improve Resilience in Animal Breeding Programs. Front. Genet. 2018, 9, 692. [Google Scholar] [CrossRef] [PubMed]
- Gorssen, W.; Winters, C.; Meyermans, R.; Chapard, L.; Hooyberghs, K.; Janssens, S.; Huisman, A.; Peeters, K.; Mulder, H.; Buys, N. A promising resilience parameter for breeding: The use of weight and feed trajectories in growing pigs. J. Anim. Sci. Biotechnol. 2023, 14, 101. [Google Scholar] [CrossRef]
- Scheffer, M.; Bolhuis, J.E.; Borsboom, D.; Buchman, T.G.; Gijzel, S.M.W.; Goulson, D.; Kammenga, J.E.; Kemp, B.; van de Leemput, I.A.; Levin, S.; et al. Quantifying resilience of humans and other animals. Proc. Natl. Acad. Sci. USA 2018, 115, 11883–11890. [Google Scholar] [CrossRef]
- Bai, X.; Plastow, G.S. Breeding for disease resilience: Opportunities to manage polymicrobial challenge and improve commercial performance in the pig industry. CABI Agric. Biosci. 2022, 3, 6. [Google Scholar] [CrossRef]
- Taghipoor, M.; Pastell, M.; Martin, O.; Nguyen Ba, H.; van Milgen, J.; Doeschl-Wilson, A.; Loncke, C.; Friggens, N.C.; Puillet, L.; Munoz-Tamayo, R. Animal board invited review: Quantification of resilience in farm animals. Animal 2023, 17, 100925. [Google Scholar] [CrossRef]
- Chen, S.Y.; Gloria, L.S.; Pedrosa, V.B.; Doucette, J.; Boerman, J.P.; Brito, L.F. Unraveling the genomic background of resilience based on variability in milk yield and milk production levels in North American Holstein cattle through genome-wide association study and Mendelian randomization analyses. J. Dairy Sci. 2024, 107, 1035–1053. [Google Scholar] [CrossRef]
- Kessler, F.; Wellmann, R.; Chagunda, M.G.G.; Bennewitz, J. Toward a resilience selection index with indicator traits in German Holstein dairy cattle. J. Dairy Sci. 2025, 108, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Berghof, T.V.L.; Bedere, N.; Peeters, K.; Poppe, M.; Visscher, J.; Mulder, H.A. The genetics of resilience and its relationships with egg production traits and antibody traits in chickens. Genet. Sel. Evol. 2024, 56, 20. [Google Scholar] [CrossRef] [PubMed]
- Hine, B.; Acton, G.; Elks, D.; Niemeyer, D.; Bell, A.; Colditz, I.; Ingham, A.; Smith, J. Targeting improved resilience in Merino sheep–correlations between immune competence and health and fitness traits. Animal 2022, 16, 100544. [Google Scholar] [CrossRef]
- Putz, A.M.; Harding, J.C.S.; Dyck, M.K.; Fortin, F.; Plastow, G.S.; Dekkers, J.C.M.; PigGen, C. Novel Resilience Phenotypes Using Feed Intake Data From a Natural Disease Challenge Model in Wean-to-Finish Pigs. Front. Genet. 2018, 9, 660. [Google Scholar] [CrossRef]
- Casto-Rebollo, C.; Nunez, P.; Gol, S.; Reixach, J.; Ibanez-Escriche, N. Variability of daily feed intake as an indicator of resilience in Pietrain pigs. Animal 2025, 19, 101415. [Google Scholar] [CrossRef]
- Kavlak, A.T.; Uimari, P. Inheritance of feed intake-based resilience traits and their correlation with production traits in Finnish pig breeds. J. Anim. Sci. 2024, 102, skae037. [Google Scholar] [CrossRef]
- Homma, C.; Hirose, K.; Ito, T.; Kamikawa, M.; Toma, S.; Nikaido, S.; Satoh, M.; Uemoto, Y. Estimation of genetic parameter for feed efficiency and resilience traits in three pig breeds. Animal 2021, 15, 100384. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Tan, X.; He, Z.; Fahey, A.G.; Zhao, G.; Liu, R.; Wen, J. Research progress and applications of genome-wide association study in farm animals. Anim. Res. One Health 2023, 1, 56–77. [Google Scholar] [CrossRef]
- Chen, Y.; Tibbs-Cortes, L.E.; Ashley, C.; Putz, A.M.; Lim, K.S.; Dyck, M.K.; Fortin, F.; Plastow, G.S.; Dekkers, J.C.M.; Harding, J.C.S.; et al. The genetic basis of natural antibody titers of young healthy pigs and relationships with disease resilience. BMC Genom. 2020, 21, 648. [Google Scholar] [CrossRef]
- Walker, L.R.; Engle, T.B.; Vu, H.; Tosky, E.R.; Nonneman, D.J.; Smith, T.P.L.; Borza, T.; Burkey, T.E.; Plastow, G.S.; Kachman, S.D.; et al. Synaptogyrin-2 influences replication of Porcine circovirus 2. PLoS Genet. 2018, 14, e1007750. [Google Scholar] [CrossRef]
- Li, M.; Pu, L.; MacHugh, D.E.; Tian, J.; Wang, X.; Zhao, Q.; Shi, L.; Gao, H.; Yu, Y.; Wang, L.; et al. Genome-wide Association Studies of Novel Resilience Traits Identify Important Immune QTL Regions and Candidate Genes in Duroc Pigs. J. Integr. Agric. 2024, 24, 4355–4369. [Google Scholar] [CrossRef]
- Casey, D.S.; Stern, H.S.; Dekkers, J.C. Identification of errors and factors associated with errors in data from electronic swine feeders. J. Anim. Sci. 2005, 83, 969–982. [Google Scholar] [CrossRef]
- Salgado, H.H.; Méthot, S.; Remus, A.; Létourneau-Montminy, M.P.; Pomar, C. A novel feeding behavior index integrating several components of the feeding behavior of finishing pigs. Animal 2021, 15, 100251. [Google Scholar] [CrossRef]
- Pham, L.M.; Le, D.T. High-performance simulation of disease outbreaks in growing-finishing pig herds raised by the precision feeding method. Comput. Electron. Agric. 2024, 225, 109335. [Google Scholar] [CrossRef]
- Berry, D.P.; Crowley, J.J. Residual intake and body weight gain: A new measure of efficiency in growing cattle. J. Anim. Sci. 2012, 90, 109–115. [Google Scholar] [CrossRef]
- Yin, L.; Zhang, H.; Tang, Z.; Yin, D.; Fu, Y.; Yuan, X.; Li, X.; Liu, X.; Zhao, S. HIBLUP: An integration of statistical models on the BLUP framework for efficient genetic evaluation using big genomic data. Nucleic Acids Res. 2023, 51, 3501–3512. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997. [Google Scholar] [CrossRef]
- Pedersen, B.S.; Quinlan, A.R. Mosdepth: Quick coverage calculation for genomes and exomes. Bioinformatics 2018, 34, 867–868. [Google Scholar] [CrossRef] [PubMed]
- Warr, A.; Affara, N.; Aken, B.; Beiki, H.; Bickhart, D.M.; Billis, K.; Chow, W.; Eory, L.; Finlayson, H.A.; Flicek, P.; et al. An improved pig reference genome sequence to enable pig genetics and genomics research. Gigascience 2020, 9, giaa051. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Browning, B.L.; Tian, X.; Zhou, Y.; Browning, S.R. Fast two-stage phasing of large-scale sequence data. Am. J. Hum. Genet. 2021, 108, 1880–1890. [Google Scholar] [CrossRef] [PubMed]
- R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 9 March 2025).
- Yang, J.A.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A Tool for Genome-wide Complex Trait Analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef]
- Nakagawa, S. A farewell to Bonferroni: The problems of low statistical power and publication bias. Behav. Ecol. 2004, 15, 1044–1045. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Bu, D.C.; Luo, H.T.; Huo, P.P.; Wang, Z.H.; Zhang, S.; He, Z.H.; Wu, Y.; Zhao, L.H.; Liu, J.J.; Guo, J.C.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Niu, K.; Zhong, J.; Hu, X. Impacts of climate change-induced heat stress on pig productivity in China. Sci. Total Environ. 2024, 908, 168215. [Google Scholar] [CrossRef]
- Dervishi, E.; Yang, T.; Dyck, M.K.; Harding, J.C.S.; Fortin, F.; PigGen, C.; Cheng, J.; Dekkers, J.C.M.; Plastow, G. Heritability and genetic correlations of plasma metabolites of pigs with production, resilience and carcass traits under natural polymicrobial disease challenge. Sci. Rep. 2021, 11, 20628. [Google Scholar] [CrossRef] [PubMed]
- Laghouaouta, H.; Pena, R.N.; Ros-Freixedes, R.; Reixach, J.; Diaz, M.; Estany, J.; Armengol, R.; Bassols, A.; Fraile, L. A Methodology to Quantify Resilience in Growing Pigs. Animals 2021, 11, 2970. [Google Scholar] [CrossRef]
- Tong, X.K.; Chen, D.; Hu, J.C.; Lin, S.Y.; Ling, Z.Q.; Ai, H.S.; Zhang, Z.Y.; Huang, L.S. Accurate haplotype construction and detection of selection signatures enabled by high quality pig genome sequences. Nat. Commun. 2023, 14, 5126. [Google Scholar] [CrossRef]
- Wang, N.; Waghray, D.; Caveney, N.A.; Jude, K.M.; Garcia, K.C. Structural insights into human MHC-II association with invariant chain. Proc. Natl. Acad. Sci. USA 2024, 121, e2403031121. [Google Scholar] [CrossRef]
- Farr, L.; Ghosh, S.; Moonah, S. Role of MIF Cytokine/CD74 Receptor Pathway in Protecting Against Injury and Promoting Repair. Front. Immunol. 2020, 11, 1273. [Google Scholar] [CrossRef]
- Zimmerman, K.A.; Bentley, M.R.; Lever, J.M.; Li, Z.; Crossman, D.K.; Song, C.J.; Liu, S.; Crowley, M.R.; George, J.F.; Mrug, M.; et al. Single-Cell RNA Sequencing Identifies Candidate Renal Resident Macrophage Gene Expression Signatures across Species. J. Am. Soc. Nephrol. 2019, 30, 767–781. [Google Scholar] [CrossRef]
- Stanley, E.R.; Chitu, V. CSF-1 receptor signaling in myeloid cells. Cold Spring Harb. Perspect. Biol. 2014, 6, a021857. [Google Scholar] [CrossRef]
- Yadav, S.; Priya, A.; Borade, D.R.; Agrawal-Rajput, R. Macrophage subsets and their role: Co-relation with colony-stimulating factor-1 receptor and clinical relevance. Immunol. Res. 2023, 71, 130–152. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Zhang, P.; Lei, X.; Du, L.; Qu, B. Insights into CSF-1/CSF-1R signaling: The role of macrophage in radiotherapy. Front. Immunol. 2025, 16, 1530890. [Google Scholar] [CrossRef] [PubMed]
- Sikorska, A.; Stachowiak, M.; Flisikowska, T.; Stachecka, J.; Flisikowski, K.; Switonski, M. Polymorphisms of CSF1R and WISP1 genes are associated with severity of familial adenomatous polyposis in APC(1311) pigs. Gene 2020, 759, 144988. [Google Scholar] [CrossRef] [PubMed]
- Faba, L.; de Groot, N.; Ramis, G.; Cabrera-Gomez, C.G.; Doelman, J. Serotonin receptors and their association with the immune system in the gastrointestinal tract of weaning piglets. Porc. Health Manag. 2022, 8, 8. [Google Scholar] [CrossRef]
- Wen, H.; Johnson, J.S.; Mulim, H.A.; Araujo, A.C.; De Carvalho, F.E.; Rocha, A.O.; Huang, Y.; Tiezzi, F.; Maltecca, C.; Schinckel, A.P.; et al. Genomic regions and biological mechanisms underlying climatic resilience traits derived from automatically-recorded vaginal temperature in lactating sows under heat stress conditions. Front. Genet. 2024, 15, 1498380. [Google Scholar] [CrossRef] [PubMed]
- Danilowicz, E.; Martinez-Arias, R.; Dolf, G.; Singh, M.; Probst, I.; Tummler, B.; Holtig, D.; Waldmann, K.H.; Gerlach, G.F.; Stanke, F.; et al. Characterization of the porcine transferrin gene (TF) and its association with disease severity following an experimental Actinobacillus pleuropneumoniae infection. Anim. Genet. 2010, 41, 424–427. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Huang, S.; Chen, Z.; Liu, W.; Zhou, X.; Zhang, D. Hypoxia-induced autophagy contributes to the invasion of salivary adenoid cystic carcinoma through the HIF-1alpha/BNIP3 signaling pathway. Mol. Med. Rep. 2015, 12, 6467–6474. [Google Scholar] [CrossRef] [PubMed]
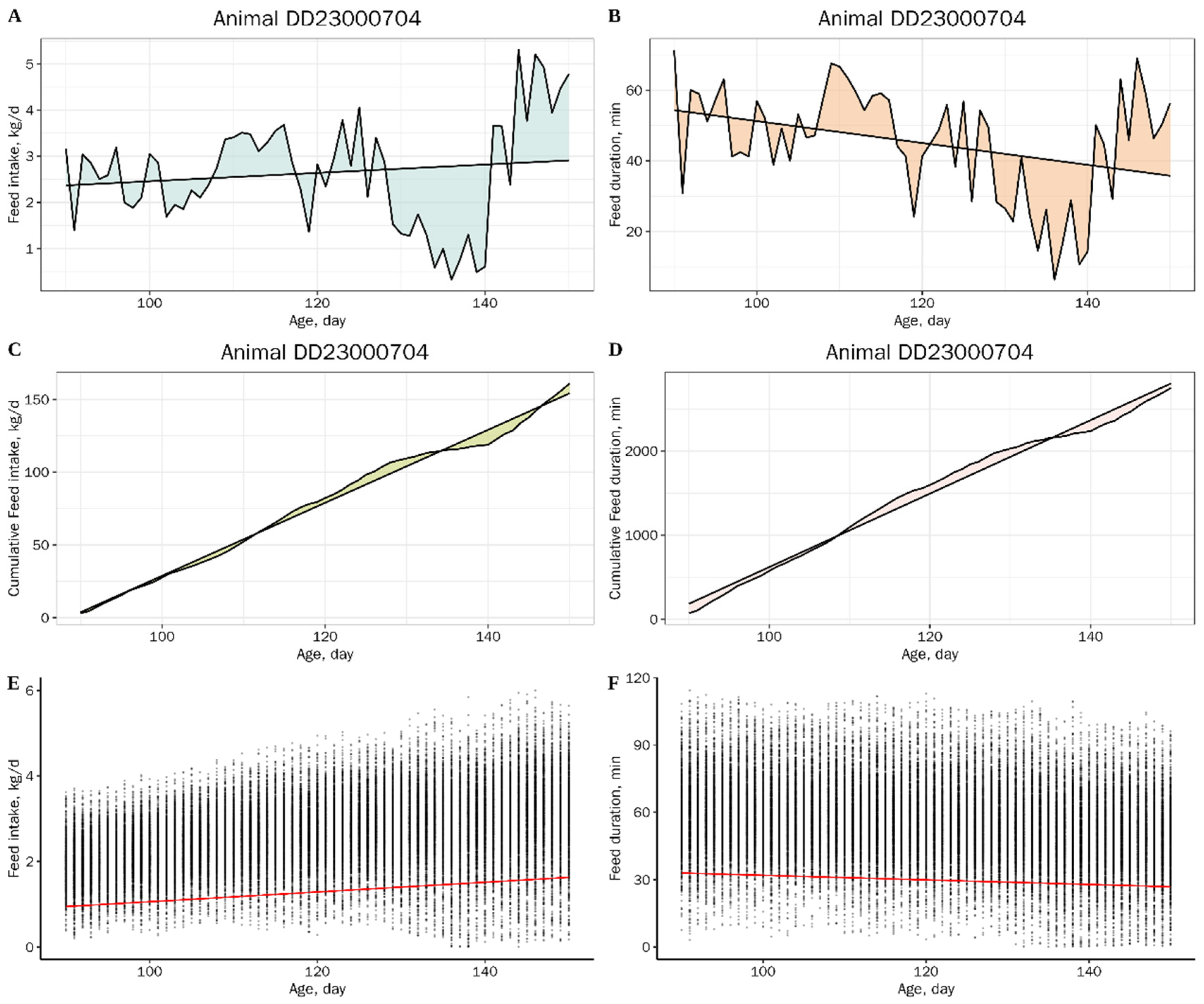
| Trait | Abbreviations 1 | Unit | N | Mean | SD | Min | Max | h2 (SE) |
|---|---|---|---|---|---|---|---|---|
| Resilience trait | ||||||||
| RMSE of daily feed intake | RMSEFI | 3437 | 0.56 | 0.15 | 0.18 | 1.29 | 0.237 (0.05) | |
| RMSE of daily feed duration | RMSEFD | 3437 | 11.38 | 3.05 | 4.43 | 26.30 | 0.267 (0.06) | |
| RMSE of cumulative feed intake | RMSECFI | 3437 | 3.55 | 1.29 | 0.80 | 8.92 | 0.230 (0.05) | |
| RMSE of cumulative feed duration | RMSECFD | 3437 | 41.84 | 22.71 | 5.69 | 182.85 | 0.103 (0.04) | |
| Quantile regression of daily feed intake | QRFI | 3437 | 5.00 | 6.19 | 0 | 77.05 | 0.160 (0.04) | |
| Quantile regression of daily feed duration | QRFD | 3437 | 5.00 | 6.06 | 0 | 59.02 | 0.171 (0.04) | |
| Growth trait | ||||||||
| Average daily gain from 90 to 150 d | ADG | kg | 3437 | 1.05 | 0.14 | 0.50 | 1.54 | 0.293 (0.06) |
| Adjust 100 kg age | AGE | day | 3295 | 143.97 | 9.73 | 119.91 | 180.28 | 0.357 (0.06) |
| Adjust 100 kg backfat thickness | BF | mm | 3293 | 10.75 | 2.06 | 5.22 | 22.20 | 0.560 (0.07) |
| Feeding behaviour traits | ||||||||
| Average occupation time in feeder per day | ADFI | kg | 3437 | 2.51 | 0.39 | 1.12 | 4.02 | 0.342 (0.06) |
| Average number of visits to feeder per day | ADFD | min | 3437 | 55.94 | 9.62 | 29.23 | 97.3 | 0.324 (0.06) |
| Feed efficiency trait | ||||||||
| Feed conversion ratio | FCR | 3437 | 2.40 | 0.27 | 1.24 | 3.95 | 0.324 (0.061) | |
| Residual feed intake | RFI | kg | 3437 | 0 | 0.23 | −1.14 | 0.88 | 0.343 (0.061) |
| Trait | RMSEFI (SE) | RMSEFD (SE) | RMSECFI (SE) | RMSECFD (SE) | QRFI (SE) | QRFD (SE) |
|---|---|---|---|---|---|---|
| RMSEFI | - | 0.521 (0.099) * | 0.301 (0.135) * | 0.417 (0.167) * | −0.258 (0.152) | −0.030 (0.147) |
| RMSEFD | 0.588 (0.014) *** | - | 0.216 (0.132) | 0.823 (0.101) * | 0.345 (0.125) * | −0.347 (0.137) * |
| RMSECFI | 0.119 (0.017) *** | 0.106 (0.017) *** | - | 0.288 (0.185) | −0.266 (0.145) | −0.288 (0.143) * |
| RMSECFD | 0.308 (0.017) *** | 0.477 (0.015) *** | 0.030 (0.017) | - | 0.179 (0.185) | −0.426 (0.179) * |
| QRFI | 0.265 (0.017) *** | 0.366 (0.016) *** | 0.001 (0.017) | 0.273 (0.017) *** | - | 0.546 (0.121) * |
| QRFD | 0.262 (0.017) *** | 0.026 (0.017) | −0.035 (0.017) * | 0.091 (0.017) *** | 0.510 (0.015) *** | - |
| Trait | SSC | N | Position (Mb) | Top SNP | |
|---|---|---|---|---|---|
| SNP | p-Value | ||||
| RMSEFI | 3 | 1 | 18.24 | 3_18244026 | 1.04 × 10−6 |
| 5 | 8 | 32.13 | 5_32134126 | 4.98 × 10−8 | |
| 8 | 1 | 14.94 | 8_14938763 | 2.02 × 10−8 | |
| 10 | 1 | 56.18 | 10_56176108 | 7.14 × 10−7 | |
| RMSEFD | 2 | 2 | 13.78 | 2_13777555 | 1.78 × 10−7 |
| 10 | 8 | 47.61–49.68 | 10_49631485 | 2.16 × 10−7 | |
| 13 | 5 | 74.96–76.14 | 13_22269276 | 5.17 × 10−7 | |
| 16 | 3 | 3.98 | 16_3980191 | 4.18 × 10−7 | |
| QRFI | 1 | 3 | 270.39 | 1_270386804 | 1.16 × 10−6 |
| 2 | 1 | 8.96 | 2_8960899 | 1.30 × 10−6 | |
| 3 | 3 | 106.22–112.56 | 3_112556064 | 4.28 × 10−7 | |
| 7 | 1 | 86.34 | 7_86344231 | 2.96 × 10−7 | |
| 8 | 1 | 129.80 | 8_129801413 | 1.28 × 10−6 | |
| 9 | 4 | 52.97 66.05–66.91 | 9_66914339 | 1.22 × 10−7 | |
| 10 | 6 | 6.46–8.93 37.71–38.17 | 10_37759538 | 5.01 × 10−7 | |
| 11 | 2 | 26.92 | 11_26916442 | 4.23 × 10−7 | |
| 12 | 1 | 0.01 | 12_10921 | 6.98 × 10−7 | |
| 13 | 1 | 200.95 | 13_200945969 | 9.73 × 10−7 | |
| 14 | 1 | 59.23 | 14_59225961 | 3.28 × 10−7 | |
| 15 | 1 | 31.74 | 15_31744440 | 9.46 × 10−7 | |
| QRFD | 1 | 1 | 270.91 | 1_270916635 | 1.48 × 10−6 |
| 5 | 5 | 6.71 | 5_67126971 | 4.30 × 10−7 | |
| 7 | 2 | 89.69 | 7_88693147 | 1.25 × 10−6 | |
| 8 | 2 | 110.7 | 8_110707400 | 1.55 × 10−7 | |
| 9 | 4 | 62.88 82.53 | 9_82536133 | 5.18 × 10−8 | |
| 10 | 7 | 42.74 | 10_42741896 | 5.34 × 10−8 | |
| 12 | 1 | 45.18 | 12_45187896 | 6.38 × 10−7 | |
| 13 | 19 | 37.07–37.09 72.99–75.57 | 13_37080425 | 1.81 × 10−8 | |
| 15 | 3 | 127.9 | 15_127864633 | 1.98 × 10−7 | |
| RMSECFI | 2 | 31 | 149.29–151.10 | 2_151078512 | 2.67 × 10−7 |
| 3 | 1 | 6.2 | 3_6201890 | 1.33 × 10−6 | |
| 5 | 1 | 17.66 | 5_17660757 | 3.06 × 10−7 | |
| 7 | 9 | 20.27–20.28 101.71–101.73 | 7_20275252 | 2.31 × 10−7 | |
| 9 | 13 | 42.35–42.49 131.76–132.83 | 9_131763229 | 1.05 × 10−7 | |
| 12 | 2 | 29.58 | 12_29583861 | 9.51 × 10−7 | |
| RMSECFD | 8 | 5 | 5.46 17.33–17.47 115.67 | 8_17477285 | 1.74 × 10−7 |
| 10 | 1 | 6.07 | 6073366 | 9.16 × 10−7 | |
| 11 | 1 | 61.07 | 61073632 | 3.00 × 10−7 | |
| 12 | 21 | 55.01–55.08 | 55.67 | 5.31 × 10−8 | |
| 16 | 4 | 0.955–0.961 4.92 | 16_955767 | 6.79 × 10−8 | |
| 17 | 1 | 11.29 | 11294396 | 5.42 × 10−7 | |
| SSC 1 | Candidate Genes | Position (Mb) 2 | N 3 | Consequence |
|---|---|---|---|---|
| 1 | HMCN2 | 270.36–270.52 | 2 | intron_variant |
| QRFP | 270.91 | 1 | upstream_gene_variant | |
| FIBCD1 | 270.91–270.95 | 1 | intron_variant | |
| 2 | STX5 | 8.93–8.96 | 1 | intron_variant |
| HTR4 | 149.73–149.91 | 7 | intron_variant | |
| CSF1R | 151.10–151.14 | 1 | intron_variant | |
| TCOF1 | 151.36–151.40 | 2 | downstream_gene_variant | |
| CD74 | 151.40 | 1 | downstream_gene_variant | |
| 13 | TF | 74.93–74.97 | 1 | intron_variant |
| 16 | CTNND2 | 0.50–1.52 | 3 | intron_variant downstream_gene_variant |
| 16 | FBXL7 | 4.78–5.19 | 1 | upstream_gene_variant |
| 17 | IKBKB | 11.28–11.34 | 1 | intron_variant |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Xin, W.; Li, M.; Duan, D.; Han, J.; Wang, M.; Zhou, S.; Li, X. Uncovering the Genetic Basis of Porcine Resilience Through GWAS of Feed Intake Data. Animals 2025, 15, 3269. https://doi.org/10.3390/ani15223269
Wang Z, Xin W, Li M, Duan D, Han J, Wang M, Zhou S, Li X. Uncovering the Genetic Basis of Porcine Resilience Through GWAS of Feed Intake Data. Animals. 2025; 15(22):3269. https://doi.org/10.3390/ani15223269
Chicago/Turabian StyleWang, Zhenyu, Wenshui Xin, Mengyu Li, Dongdong Duan, Jinyi Han, Mingyu Wang, Shenping Zhou, and Xinjian Li. 2025. "Uncovering the Genetic Basis of Porcine Resilience Through GWAS of Feed Intake Data" Animals 15, no. 22: 3269. https://doi.org/10.3390/ani15223269
APA StyleWang, Z., Xin, W., Li, M., Duan, D., Han, J., Wang, M., Zhou, S., & Li, X. (2025). Uncovering the Genetic Basis of Porcine Resilience Through GWAS of Feed Intake Data. Animals, 15(22), 3269. https://doi.org/10.3390/ani15223269





