1. Introduction
Bovine mastitis is a widespread and economically impactful disease that remains a persistent challenge due to the diverse and complex nature of its causative pathogens [
1]. Its etiology involves a broad spectrum of microorganisms, including
Staphylococcus aureus, non-aureus staphylococci,
Streptococcus spp.,
Escherichia coli, and
Klebsiella spp. [
2].
Nocardia spp., a group of filamentous, branching Gram-positive bacteria, have long been recognized as causative agents of localized or systemic suppurative diseases in both humans and animals [
3]. The predominant
Nocardia species associated with bovine mastitis include
N. asteroides,
N. farcinica,
N. nova,
N. otitidiscaviarum, and
N. cyriacigeorgica [
4]. Notably,
Nocardia spp. have also been identified as etiological agents of mastitis in several countries, including the United Kingdom [
5], Italy [
6], Brazil [
7], and China [
8], where infections frequently result in abscess formation, inflammation, and decreased milk production.
Widely distributed in soil, water, and both the internal and external environments of various animals and plants,
Nocardia species are generally regarded as environmental pathogens [
9,
10]. Although capable of causing severe invasive diseases,
Nocardia spp. are primarily opportunistic pathogens in both humans and dairy cattle. Notably, the incidence and clinical burden of human nocardiosis vary significantly across different geographical regions [
11].
The application of molecular epidemiology has greatly advanced our understanding of the population structure and diversity of pathogens associated with bovine mastitis [
12,
13,
14]. Analyzing bacterial genetic diversity is essential for characterizing epidemiological patterns and distinguishing strain-level variations [
15]. Various molecular typing methods have been employed to investigate the genetic diversity of
Nocardia species, including 16S rRNA sequencing and multilocus sequence analysis (MLSA) [
16,
17]. Among these, MLSA has proven to be a highly discriminative technique for identifying clinical
Nocardia isolates [
18]. These molecular approaches have played a critical role in clarifying genetic relationships and supporting epidemiological studies of bacterial pathogens [
11].
A comprehensive investigation of the microbiological agents and their pathogenic mechanisms underlying bovine mastitis is essential for developing effective prevention and control strategies. Although the pathogenicity of Nocardia species in humans and other animals has been documented, their presence, genetic diversity, and association with mastitis in dairy herds remain underexplored. In particular, the role of Nocardia cyriacigeorgica as a potential reservoir in extramammary and environmental niches on dairy farms has not been clearly established. Therefore, the objective of this study was to determine the phylogenetic characteristics of N. cyriacigeorgica isolated from cases of clinical mastitis (CM) and subclinical mastitis (SCM) on two large-scale dairy farms in China.
4. Discussion
Nocardia cyriacigeorgica, a pathogenic species within the genus
Nocardia, was initially classified as type VI of
N. asteroides [
29] and is widely recognized as an opportunistic pathogen in humans, where it causes severe infections. Although our study focuses on bovine mastitis, insights from human nocardiosis highlight its pathogenic potential. In recent years,
N. cyriacigeorgica has also been isolated from bovine milk samples associated with mastitis worldwide, contributing to significant economic losses in the dairy industry [
5,
7,
8]. During the mastitis outbreak investigated in this study,
N. cyriacigeorgica was detected at relatively high prevalence rates in milk samples from clinical mastitis cases (CM, 22.7%), subclinical mastitis (SCM, 11.2%), and even apparently healthy cows (HEAL, 28.0%), in agreement with previous reports from China [
30]. Notably, the distribution of isolates differed between the two farms: Farm A yielded more isolates, mainly from subclinical mastitis cases, whereas Farm B contributed mostly from clinical mastitis cases with only a single subclinical isolate. This difference may be explained by variations in management practices, diagnostic approaches, or sampling intensity, but could also reflect genuine epidemiological differences and distinct outbreak dynamics at each site. All samples were collected during an active outbreak, offering a unique opportunity to examine the genetic diversity of isolates under conditions of elevated pathogen pressure and environmental exposure. Whole genome sequencing showed closely related
N. cyriacigeorgica across clinical, subclinical, and healthy milk samples. This low divergence is compatible with exposure to shared farm sources (e.g., equipment, bedding, feed), but it does not identify the source or the direction of spread. Because we did not culture and sequence environmental isolates, we cannot distinguish direct environmental contamination, a single introduction with limited within-farm spread, from repeated reintroductions via external inputs (e.g., feed from the same fields). Given
Nocardia’s ecology as an environmental opportunist rather than a contagious mastitis pathogen, we view environmental exposure as plausible, not proven, in this dataset.
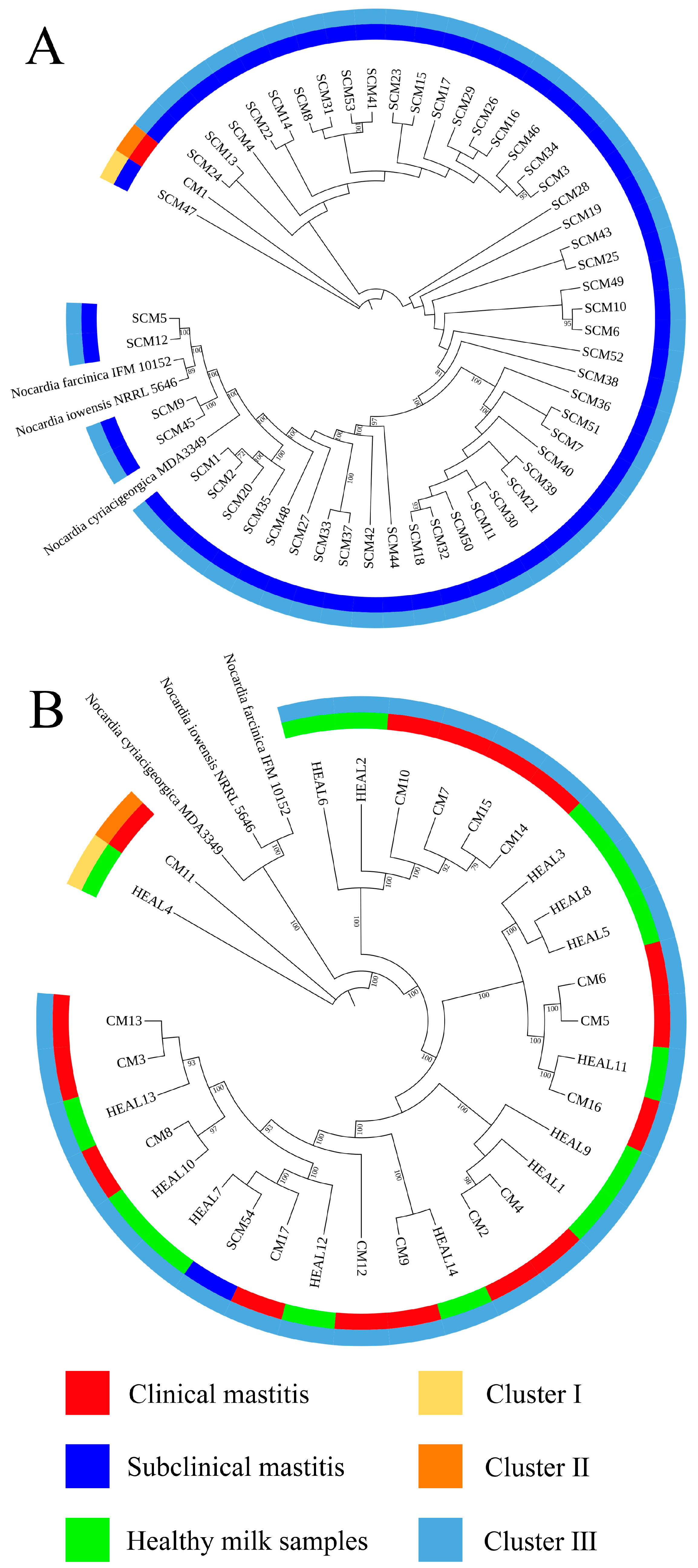
Accurate identification of
Nocardia species is essential for epidemiological investigations and clinical management, with full-length 16S rRNA gene sequencing being the gold standard for species-level confirmation [
31]. In this study, 16S rRNA sequencing and phylogenetic tree construction revealed a low level of genetic diversity among
N. cyriacigeorgica strains isolated from clinical mastitis (CM), subclinical mastitis (SCM), and healthy milk samples (HEAL), consistent with previous findings on the genetic relatedness between
N. cyriacigeorgica,
N. asteroides, and
N. abscessus [
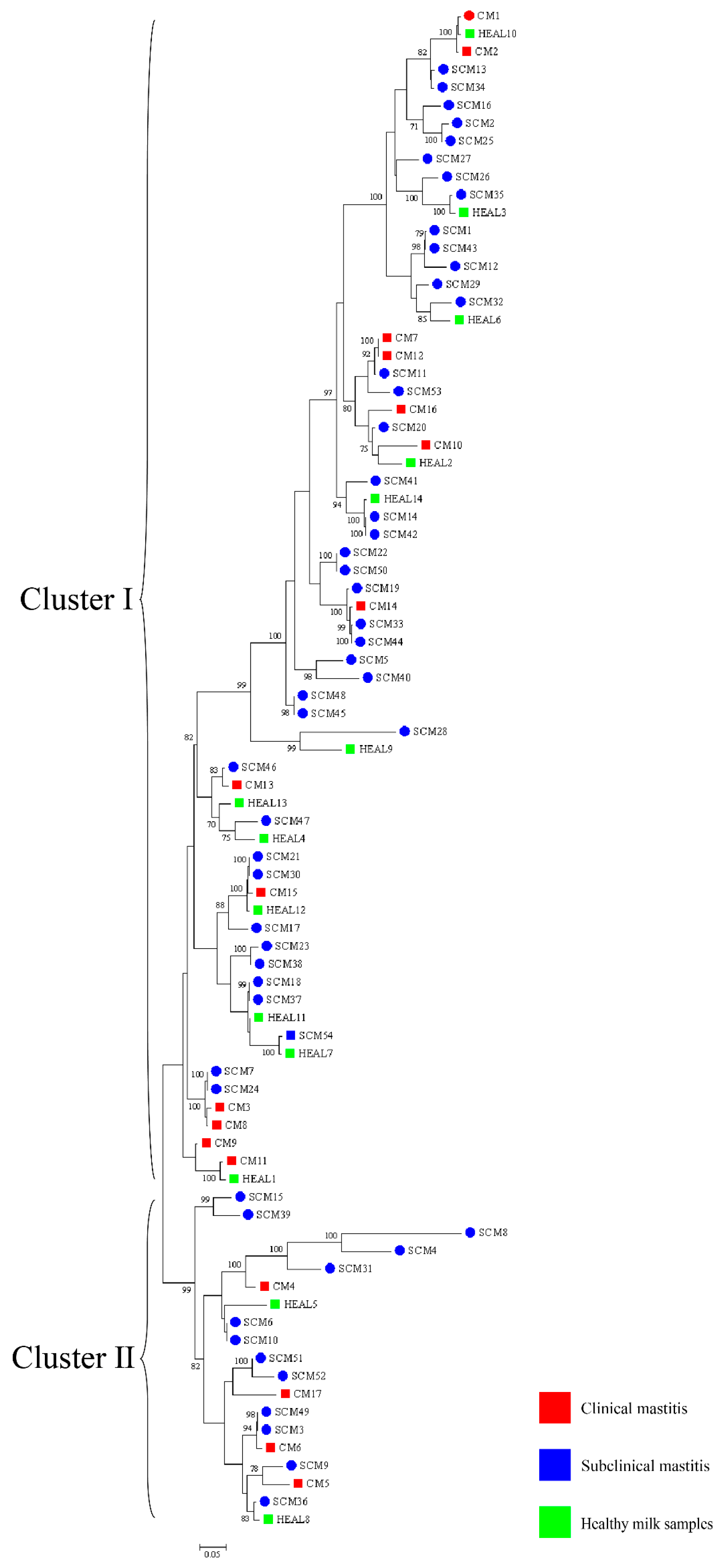
30]. To enhance phylogenetic resolution, multilocus sequence analysis (MLSA) based on three- and five-gene combinations was also performed. Notably, MLSA results demonstrated a high degree of homogeneity among isolates, particularly those obtained from mastitis cases, suggesting the possible existence of a dominant or clonally derived strain circulating within the farms. Comparable patterns of clonal dissemination have been observed in recent molecular studies of mastitis pathogens. For example, Fu et al. reported extensive genetic diversity and antimicrobial resistance among
Klebsiella pneumoniae isolates from bovine mastitis using WGS and MLST [
13]. Wang et al. further characterized 108 isolates from dairy farms and identified multiple virulence and resistance gene clusters, with indications of local clonal expansion. These findings reinforce the hypothesis that under specific environmental and management conditions, certain strains may dominate through shared reservoirs and transmission pathways [
12]. These findings imply that
N. cyriacigeorgica may spread within farms via environmental routes under specific conditions, driven by shared environmental sources or management practices, rather than by direct transmission between cows. This highlights the need for targeted biosecurity and hygiene interventions to prevent within-farm spread, thereby safeguarding both animal health and farm productivity.
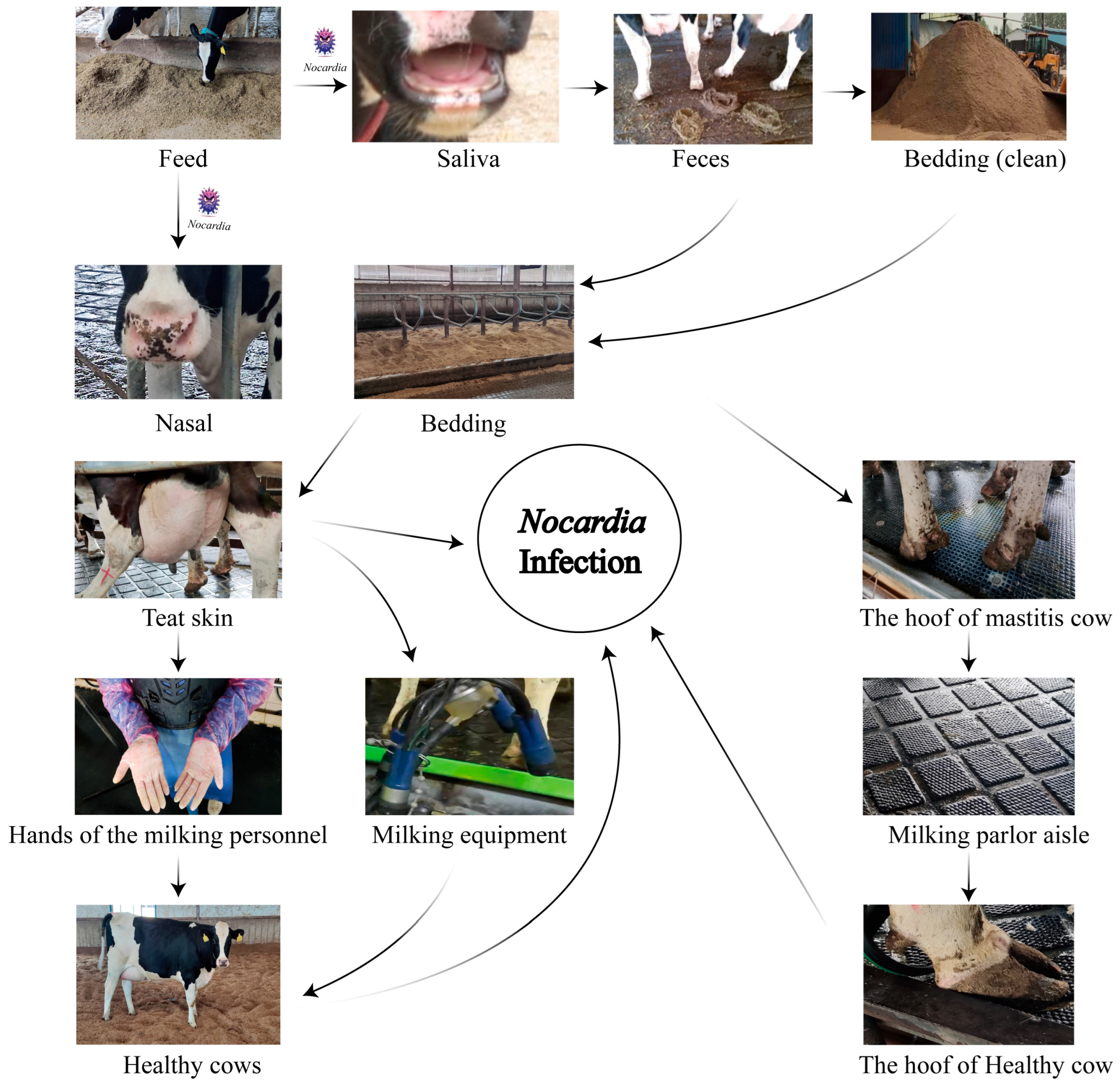
Nocardia spp., widely distributed in soil, water sources, and organic matter, demonstrates remarkable ecological adaptability [
32]. Its frequent detection in cattle farm environments, particularly in manure, bedding straw, and wastewater, reinforces its classification as an environmental pathogen [
33]. In this study,
N. cyriacigeorgica DNA was detected by qPCR at multiple on-farm sites on both farms, with the highest qPCR-positive frequencies in nasal swabs (75.0%), teat skin and interdigital spaces (75.0%), milking parlor aisles (50.0%), and equipment (41.7–50.0%). Lower qPCR-positive frequencies (8.3–25.0%) were observed in bedding, feed, feces, cow hooves, and personnel hands, while water, manure residue, and soil tested negative by qPCR in our assays. These findings suggest persistent contamination across multiple environmental sites, with repeated animal contact likely contributing to sustained infections. Previous studies have identified recycled manure solids as a major risk factor for environmental mastitis, due to their capacity to retain high bacterial loads and direct contact with the teat end [
34,
35]. Our results align with these findings, suggesting that recycled manure bedding may serve as a stable reservoir facilitating the persistence of
N. cyriacigeorgica within dairy herds. A likely cycle involves contaminated feed colonizing the nasal or oral cavities, subsequent fecal shedding, and recycled bedding, leading to re-exposure via teat skin and hooves. This highlights the importance of farm management practices, particularly bedding recycling and hygiene protocols, in sustaining or limiting pathogen dissemination. Comparable to
Klebsiella pneumoniae in Scottish dairy herds [
36],
N. cyriacigeorgica may establish stable environmental reservoirs with significant epidemiological impact.
During the milking process,
N. cyriacigeorgica present on the teat skin may invade the mammary gland, leading to intramammary infections. Contaminated teat skin can also transfer bacteria to the milking equipment liners, posing a risk of subsequent infection in healthy cows. Similarly, bacteria on the hooves may be deposited onto rubber mats in the milking parlor, increasing the risk of environmental transmission and hoof-related infections (
Figure 6). Notably, nocardial mastitis was predominantly diagnosed in clinical cases within dairy herds characterized by poor environmental hygiene between milking sessions and inappropriate intramammary therapy practices [
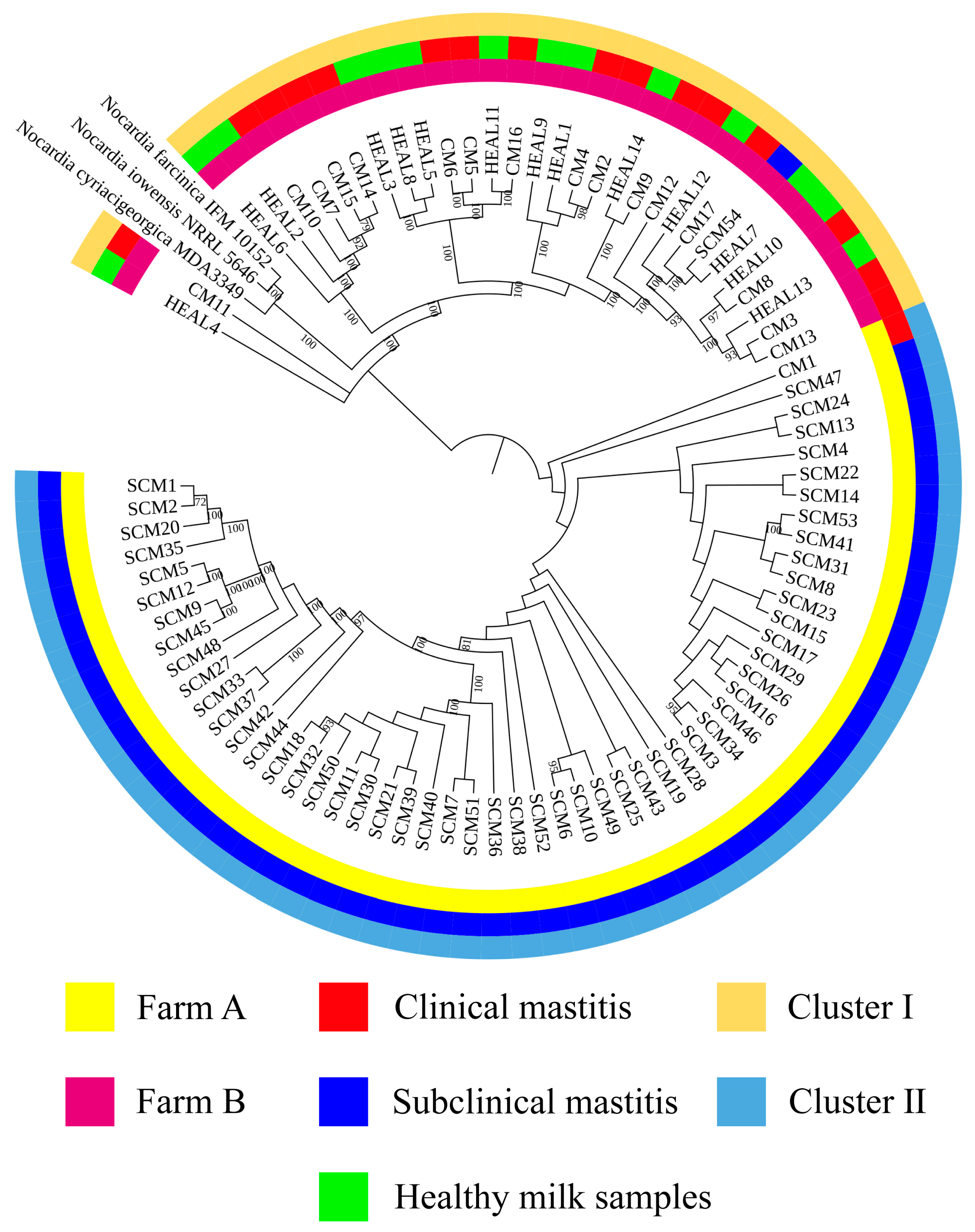
37]. Furthermore, phylogenetic analyses based on three-gene (16S rRNA–
gyrB–
hsp65), five-gene (
gyrB–16S rRNA–
secA1–
hsp65–
rpoB), and seven-gene (
gyrB–
hsp65–
secA1–
rpoB–
rpoA–
recA–
trpB) MLSA schemes revealed that isolates from farms A and B clustered separately, indicating genetic heterogeneity between farms. In contrast, isolates within each farm exhibited high genetic similarity over a short time frame, suggesting environmental clonal persistence, where specific lineages persist in farm environments and intermittently infect cows. Notably, the clustering results also varied among the three-gene, five-gene, and seven-gene MLSA schemes, reflecting the locus-dependent resolution of multilocus analyses. The three-gene set includes highly conserved loci, particularly 16S rRNA, which limits discriminatory power. The five-gene scheme improves resolution by incorporating additional housekeeping genes, while the seven-gene scheme captures even greater variation due to the inclusion of faster-evolving loci. In addition, locus-specific effects of recombination and horizontal gene transfer may contribute to conflicting topologies. These findings emphasize that MLSA outcomes depend strongly on gene selection, and whole genome sequencing provides the most robust framework for high-resolution phylogenetic inference. Previous studies have shown that the burden of human nocardiosis varies across geographic regions [
11]. Consistent with this, our findings suggest that the genetic diversity and persistence of
N. cyriacigeorgica are influenced by geographic location and farm-specific environmental conditions.
Currently, the antibiotic treatment of
N. cyriacigeorgica infections remains challenging. Therapy often requires prolonged courses, yet the clinical outcomes are frequently suboptimal, potentially resulting in chronic inflammation of the mammary gland, reduced milk yield, and compromised overall health of affected cows [
38,
39]. In this study, antimicrobial susceptibility testing (AST) showed universal resistance to clarithromycin and rifampin and high resistance to ciprofloxacin, confirming the limited utility of these agents. In contrast, all isolates were susceptible to amikacin and tobramycin, suggesting aminoglycosides as the most reliable therapeutic option. Moderate resistance to doxycycline and moxifloxacin highlights variable efficacy and reinforces the need for susceptibility-guided therapy. These patterns accord with Brazilian reports [
37,
40] where environmental lapses have been implicated and multidrug-resistant
Nocardia have emerged, underscoring the need for environmental control and cautious antimicrobial stewardship. To prevent further spread, affected cows should be promptly isolated or removed, while future research should integrate AST profiling with studies on virulence and host–pathogen interactions to inform more effective control strategies.
In this study, we investigated the genetic diversity of N. cyriacigeorgica within two dairy herds under outbreak conditions. The close clustering of milk isolates on whole genome sequencing and multilocus sequence analysis, together with qPCR detection at shared contact sites, is consistent with indirect exposure involving milking parlor aisles, shared equipment, and high animal density. However, environmental samples were not cultured and environmental isolates were not sequenced, so the data do not allow attribution of source or inference about transmission direction. In the absence of established interpretive standards for bovine nocardial mastitis, we used homology-based comparisons to describe relatedness among clinical, subclinical, and healthy milk isolates, and these comparisons require further methodological validation before they can inform attribution. The findings generate hypotheses regarding environmental exposure pathways, but repeated acquisition from environmental reservoirs cannot be concluded from the present dataset. Alternative explanations remain plausible, including a single introduction followed by limited within-farm spread or repeated reintroductions through external inputs such as feed from the same fields. From a practical standpoint, the results support prioritizing control at shared exposure points, including teat-skin hygiene, sanitation of liners and other equipment, management of bedding, and secure storage of feed, while confirmation should rely on longitudinal environmental culture with parallel sequencing of environmental and milk isolates. This investigation was conducted on two farms in Hebei and Shandong over a short interval; therefore, the findings should be considered preliminary and context specific, and multi-farm longitudinal studies with environmental culture and sequencing are warranted.