Conservation of Native Livestock Breeds in Russia: Current State and Promising Prospects
Simple Summary
Abstract
1. Introduction
2. Methodology
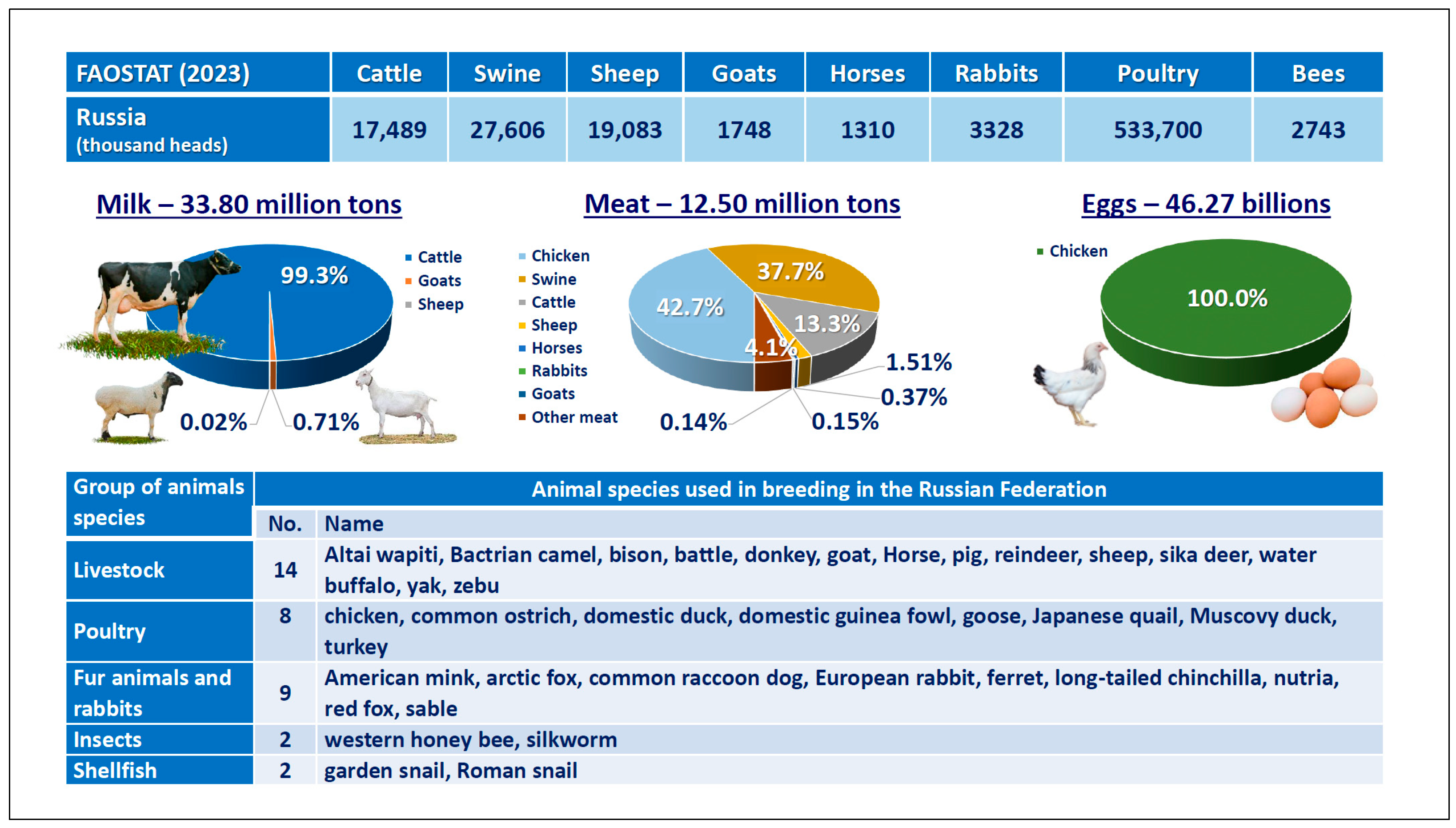
3. Overall Description of the State of the Art of AnGR in Russia
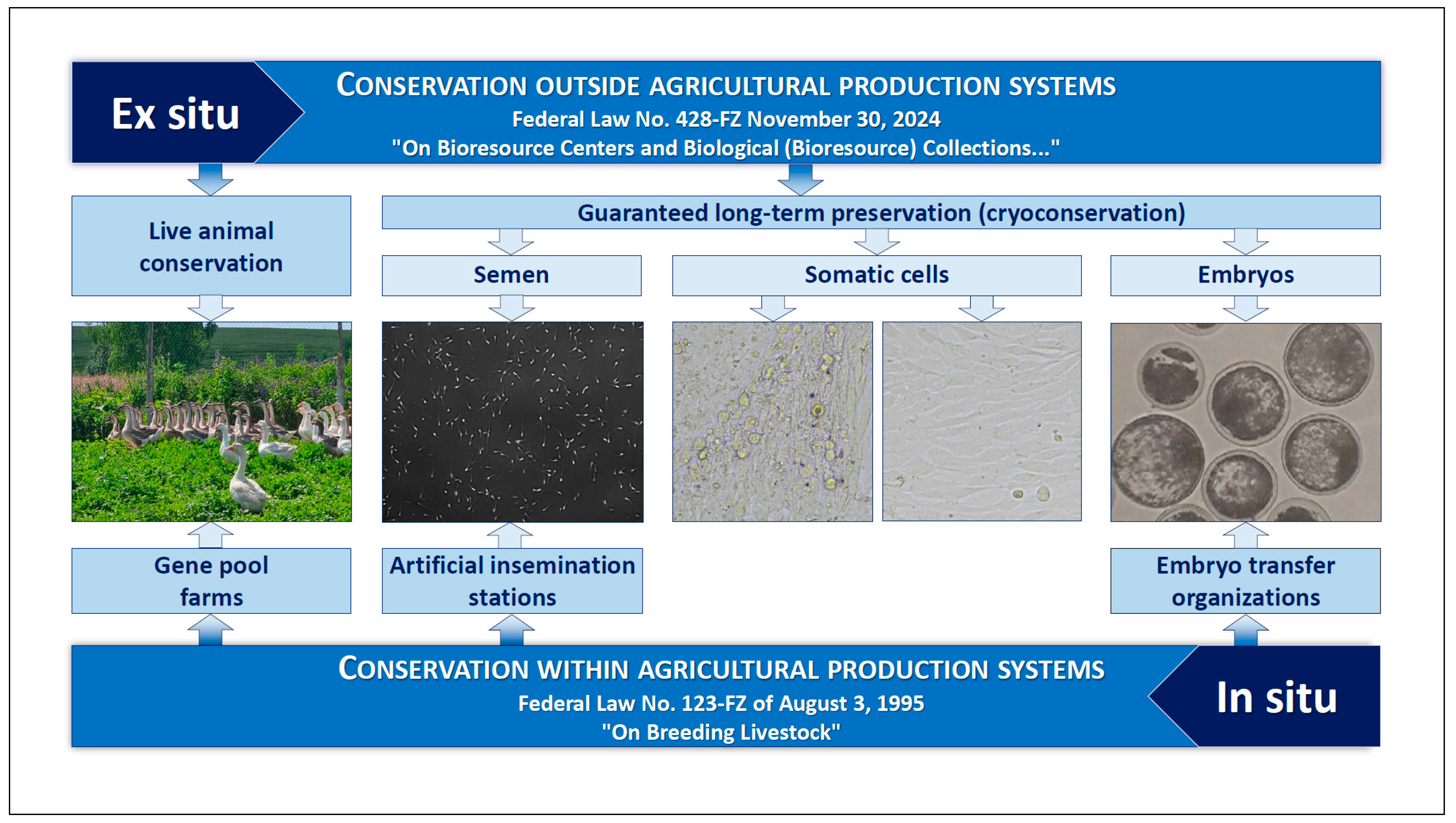
4. Preservation of AnGR in Russia
5. Conservation and Research of Russian AnGR
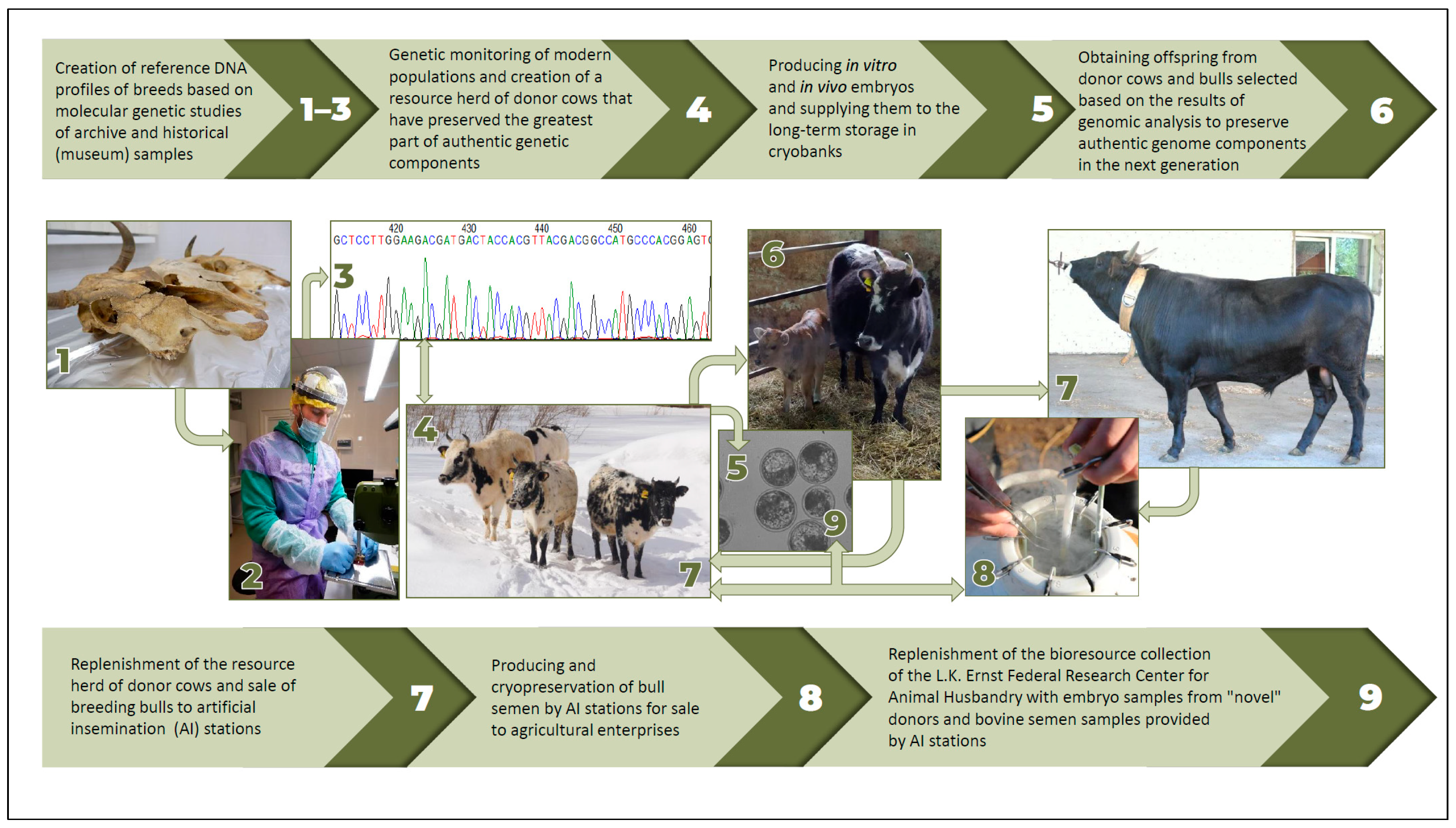
5.1. Key Conservation Strategy for AnGR
- Creation of reference DNA profiles of breeds based on molecular genetic studies of archival or historical (museum) samples [45];
- Conducting genetic monitoring of modern populations and creating a resource herd of donor females that have kept the largest proportion of authentic genetic components;
- Producing in vitro and in vivo embryos and placing them in a cryobank for long-term storage;
- Producing offspring from donor females and males selected based on the results of genomic analysis to preserve authentic genomic components in the next generation;
- Replenishment of the resource herd of donor females and sale of males to AI stations;
- Obtaining and cryopreserving semen at AI stations for sale to agricultural enterprises;
- Replenishment of the bioresource collection of the Russian National Center for AnGR with embryo samples from “new” donors and semen samples from males provided by AI stations.
5.2. Selection of Animals for Conservation
5.3. Implementation Examples of the Strategy for AnGR Conservation
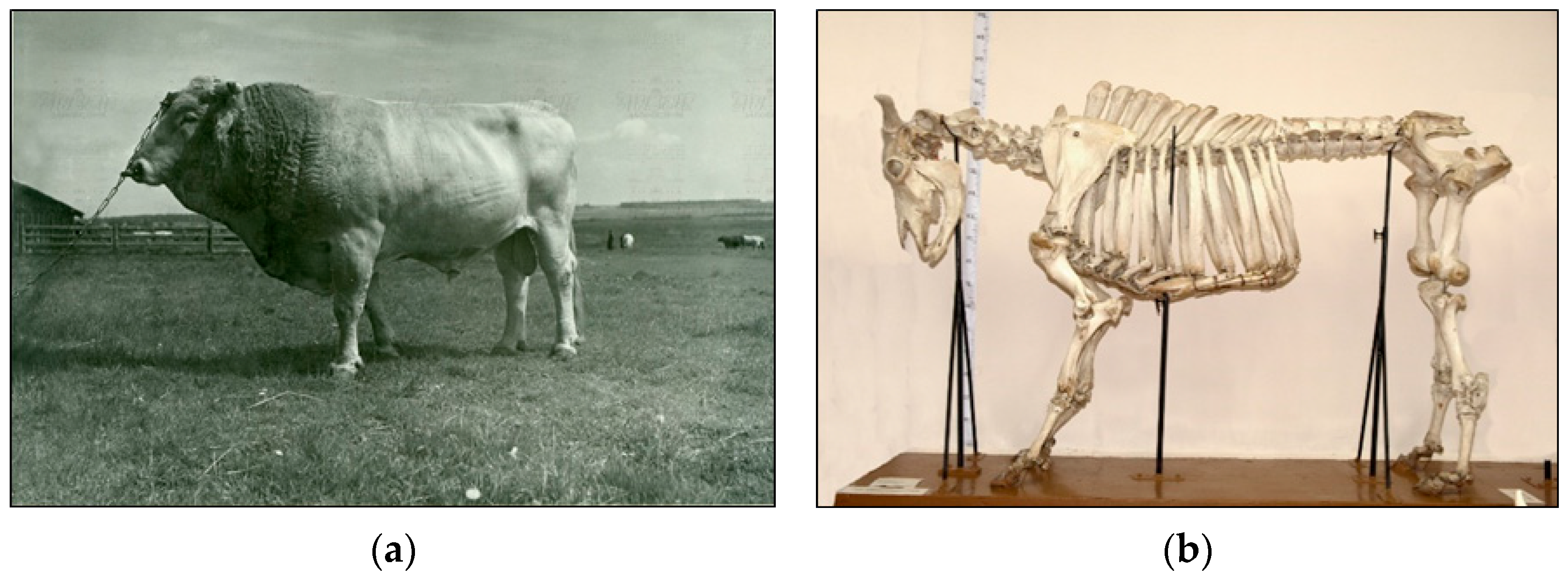
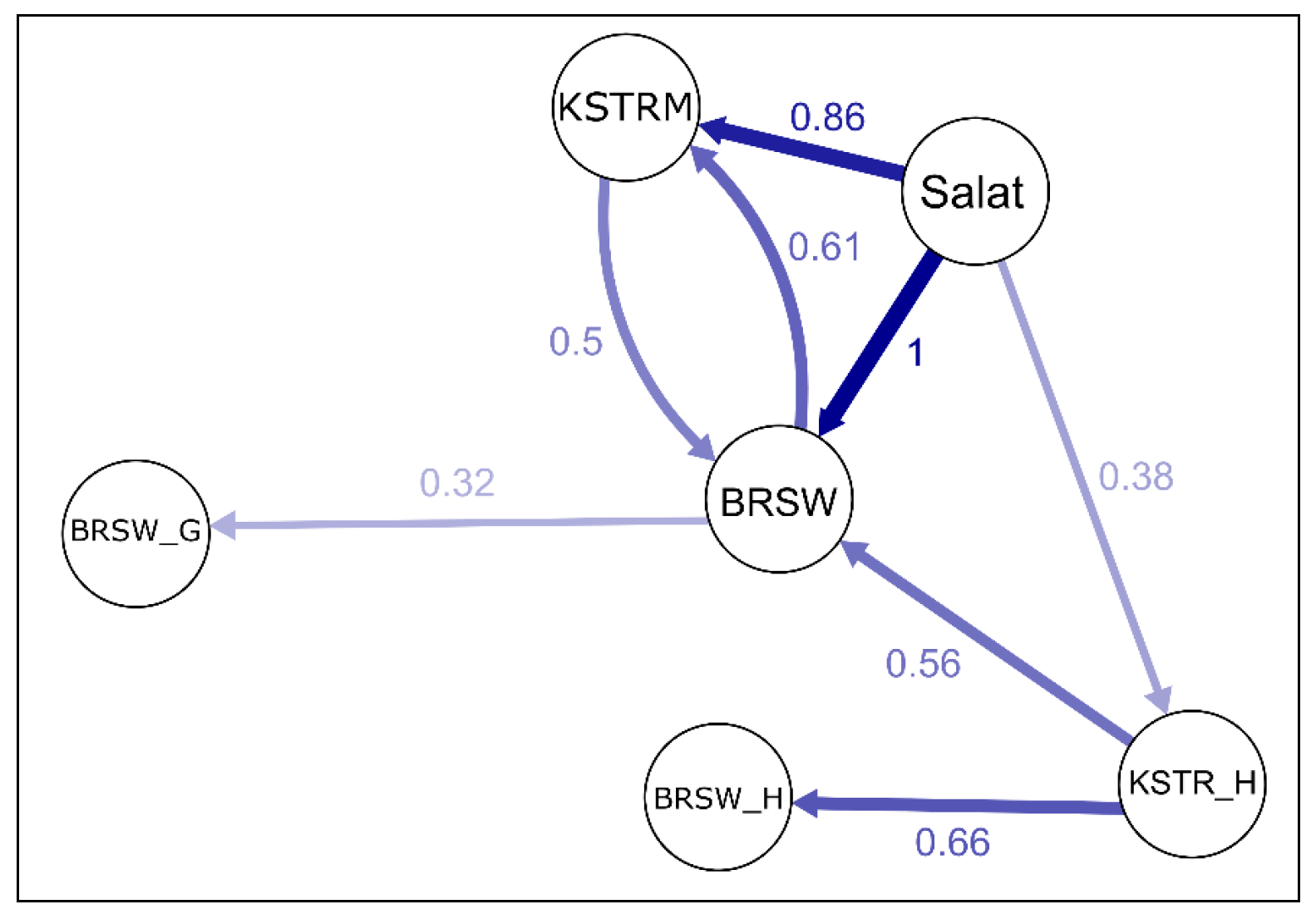
5.3.1. Tagil Cattle Breed
5.3.2. Kostroma Cattle Breed
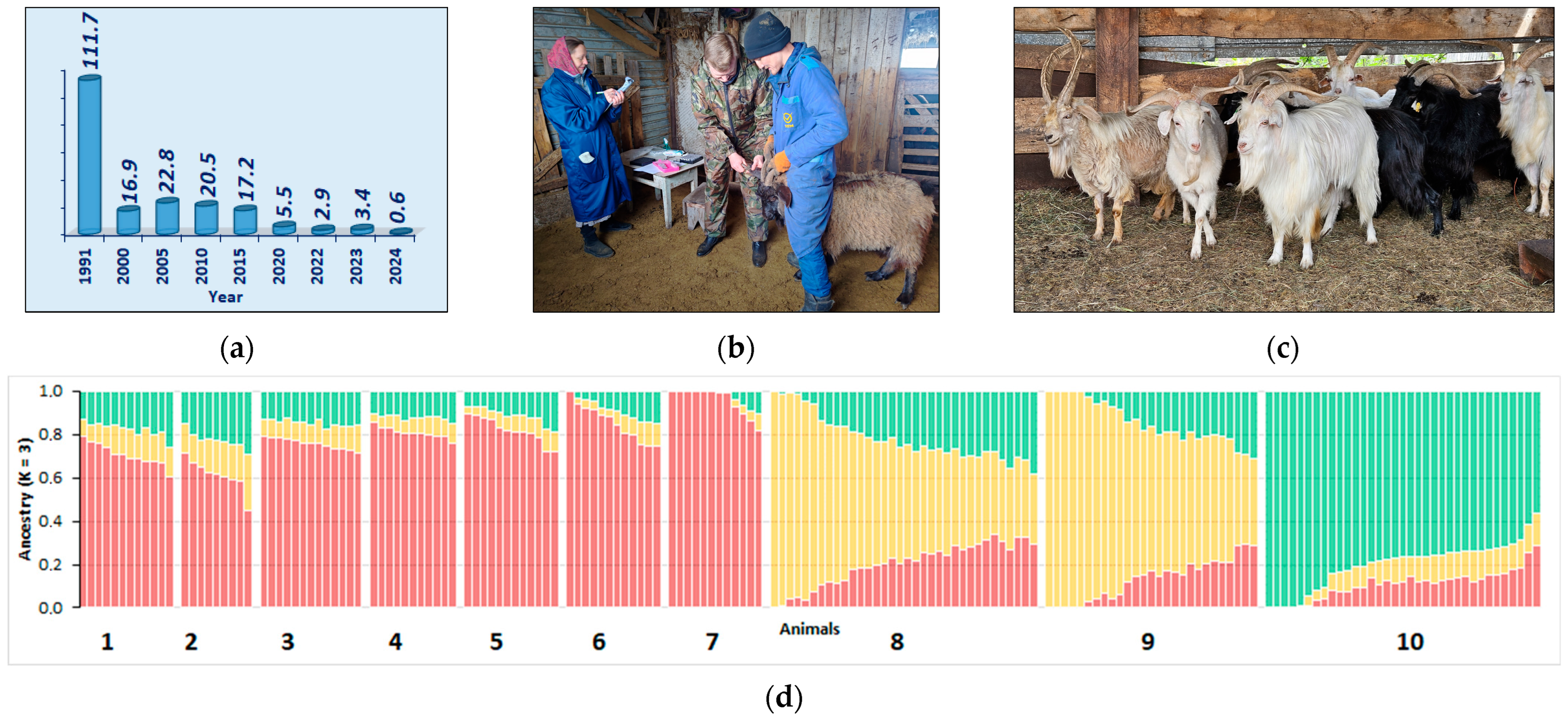
5.3.3. Orenburg Goats
5.4. Genetic Monitoring of Russian AnGR
5.4.1. Whole-Genome Resequencing
5.4.2. SNP Genotyping Arrays
5.4.3. Microsatellite Markers
5.4.4. New Prospects for Less Studied Species: A Case of Goose
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AI | Artificial insemination |
| AnGR | Animal genetic resources |
| DAD-IS | Domestic Animal Diversity Information System |
| DNA | Deoxyribonucleic acid |
| ET | Embryo transfer |
| FAO | Food and Agriculture Organization of the United Nations |
| SNP(s) | Single nucleotide polymorphism(s) |
| UN | United Nations |
| WGS | Whole-genome sequencing |
References
- FAO; Animal Production and Health Division (AGA). Shaping the Future of Livestock: Sustainably, Responsibly, Efficiently. In Proceedings of the 10th Global Forum for Food and Agriculture (GFFA), Berlin, Germany, 18–20 January 2018; FAO: Rome, Italy, 2018. Available online: https://openknowledge.fao.org/handle/20.500.14283/i8384en/ (accessed on 27 August 2025).
- Sponenberg, D.P.; Martin, A.; Couch, C.; Beranger, J. Conservation strategies for local breed biodiversity. Diversity 2019, 11, 177. [Google Scholar] [CrossRef]
- Ortiz, A.M.D.; Outhwaite, C.L.; Dalin, C.; Newbold, T. A review of the interactions between biodiversity, agriculture, climate change, and international trade: Research and policy priorities. One Earth 2021, 4, 88–101. [Google Scholar] [CrossRef]
- Bittante, G. Biodiversity and genetics of beef quality, a review. Ital. J. Anim. Sci. 2023, 22, 867–884. [Google Scholar] [CrossRef]
- Solodneva, E.; Svishcheva, G.; Smolnikov, R.; Bazhenov, S.; Konorov, E.; Mukhina, V.; Stolpovsky, Y. Genetic structure analysis of 155 transboundary and local populations of cattle (Bos taurus, Bos indicus and Bos grunniens) based on STR markers. Int. J. Mol. Sci. 2023, 24, 5061. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Sermyagin, A.A.; Dotsev, A.V.; Boronetslaya, O.I.; Petrikeeva, L.V.; Abdelmanova, A.S.; Brem, G. Animal genetic resources: Developing the research of allele pool of Russian cattle breeds—Minireview. Sel’skokhozyaistvennaya Biol. 2019, 54, 631–641. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Kharzinova, V.R.; Volkova, V.V.; Dotsev, A.V.; Sermyagin, A.A.; Boronetskaya, O.I.; Chinarov, R.Y.; Lutshikhina, E.M.; Sölkner, J.; Brem, G.; et al. Comparative study of the genetic diversity of local steppe cattle breeds from Russia, Kazakhstan and Kyrgyzstan by microsatellite analysis of museum and modern samples. Diversity 2021, 13, 351. [Google Scholar] [CrossRef]
- Restoux, G.; Rognon, X.; Vieaud, A.; Guemene, D.; Petitjean, F.; Rouger, R.; Brard-Fudulea, S.; Lubac-Paye, S.; Chiron, G.; Tixier-Boichard, M. Managing genetic diversity in breeding programs of small populations: The case of French local chicken breeds. Genet. Sel. Evol. 2022, 54, 56. [Google Scholar] [CrossRef]
- United Nations. Convention on Biological Diversity of 5 June 1992 (1760 U.N.T.S. 69); United Nations: New York, NY, USA, 1992; Available online: https://www.cbd.int/convention (accessed on 27 August 2025).
- Scherf, B.D. (Ed.) World Watch List for Domestic Animal Diversity, 2nd ed.; FAO; UNEP: Rome, Italy, 1995; Available online: https://www.fao.org/3/x6197e/x6197e.pdf (accessed on 27 August 2025).
- FAO. Global Plan of Action for Animal Genetic Resources and the Interlaken Declaration. In Proceedings of the Adopted by the International Technical Conference on Animal Genetic Resources for Food and Agriculture, Interlaken, Switzerland, 3–7 September 2007; FAO: Rome, Italy, 2007. Available online: https://openknowledge.fao.org/items/5038d1cf-73a7-4215-bbb0-bf0c4f5797ca (accessed on 27 August 2025).
- FAO. The State of the World’s Animal Genetic Resources for Food and Agriculture; Rischkowsky, B., Pilling, D., Eds.; FAO: Rome, Italy, 2007; Available online: https://books.google.com/books?id=Skpj197tU0oC (accessed on 27 August 2025).
- FAO. DAD-IS: Domestic Animal Diversity Information System; FAO: Rome, Italy, 2025; Available online: https://www.fao.org/dad-is/en (accessed on 27 August 2025).
- Bodó, I. Methods and experiences with in situ preservation of farm animals. In Animal Genetic Resources—A Global Programme for Sustainable Development; FAO Animal Production and Health Paper 80; Wiener, G., Ed.; FAO: Rome, Italy, 1990; pp. 85–102. Available online: http://www.fao.org/3/a-t0284e.pdf (accessed on 27 August 2025).
- FAO. Vivo Conservation of Animal Genetic Resources; FAO Animal Production and Health Guidelines No. 14; Commission on Genetic Resources for Food and Agriculture, FAO: Rome, Italy, 2013; Available online: https://www.fao.org/4/i3327e/i3327e00.htm (accessed on 27 August 2025).
- Loftus, R.; Scherf, B. (Eds.) World Watch List for Domestic Animal Diversity; FAO: Rome, Italy, 1993; Available online: https://books.google.com/books?id=wJEYkNNxllAC (accessed on 27 August 2025).
- Hilton, M.; Walsh, J.C.; Liddell, E.; Cook, C.N. Lessons from other disciplines for setting management thresholds for biodiversity conservation. Conserv. Biol. 2022, 36, e13865. [Google Scholar] [CrossRef]
- Dmitriev, N.G.; Ernst, L.K. (Eds.) Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper 65; FAO: Rome, Italy, 1989; Available online: https://www.fao.org/4/ah759e/AH759E00.htm (accessed on 27 August 2025).
- PubMed. National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2025. Available online: https://pubmed.ncbi.nlm.nih.gov/ (accessed on 27 August 2025).
- eLIBRARY.RU. Scientific Electronic Library LLC.: Moscow, Russia, 2000–2025. Available online: https://elibrary.ru/defaultx.asp? (accessed on 27 August 2025).
- Scientific Electronic Library. CyberLeninka: Moscow, Russia, 2025. Available online: https://cyberleninka.ru/ (accessed on 27 August 2025).
- ResearchGate. ResearchGate GmbH: Berlin, Germany, 2008–2024. Available online: https://www.researchgate.net/ (accessed on 27 August 2025).
- FAO. Food and Agriculture Organization of the United Nations: Rome, Italy, 2025. Available online: https://www.fao.org/home/en/ (accessed on 27 August 2025).
- Scientific Library. L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2005–2025. Available online: https://www.vij.ru/biblioteka/o-podrazdelenii (accessed on 27 August 2025).
- News: Conferences. L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2005–2025. Available online: https://www.vij.ru/novosti2/konferentsii (accessed on 27 August 2025).
- Order of the Ministry of Agriculture of the Russian Federation No. 196 of 31 March 2025 “On Approval of the List of Species and Breeds (Types, Crosses of Lines) of Animals Used in Breeding Breeder Animals”. Available online: http://publication.pravo.gov.ru/document/0001202505070019 (accessed on 27 August 2025).
- FAOSTAT. Food and Agriculture Data; Statistics Division, FAO: Rome, Italy, 2025; Available online: https://www.fao.org/faostat/en/#home (accessed on 27 August 2025).
- Zinovieva, N.A. Synergy of Genetic and Reproductive Technologies Is the Key to the Development of Highly Efficient Animal Husbandry. In Proceedings of the 16th Vavilov Readings, Moscow, Russia, 15 November 2024; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2024. Available online: https://www.vij.ru/images/news-24/Vavilov_Readings/2024_Zinovieva_Vavilov.pdf (accessed on 27 August 2025).
- On Approval of the Procedure for Testing New Breeds, Types, Lines and Crosses of Farm Animals in the Member States of the Eurasian Economic Union. Decision of the Board of the Eurasian Economic Commission No. 113 of 22 September 2020. Available online: https://faolex.fao.org/docs/pdf/kaz204509.pdf (accessed on 27 August 2025).
- Voskresensky, S.B.; Lukonina, O.N.; Safina, G.F. (Eds.) Yearbook on Breeding Work in Dairy Cattle Breeding in Farms of the Russian Federation (2024); All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2025; Available online: https://vniiplem.ru/wp-content/uploads/2025/%D0%95%D0%B6%D0%B5%D0%B3%D0%BE%D0%B4%D0%BD%D0%B8%D0%BA.%D0%9C%D0%BE%D0%BB.%D0%A1%D0%BA%D0%BE%D1%82.2025.pdf (accessed on 27 August 2025).
- Ernst, L.K.; Dmitriev, N.G. Cattle (excluding zebus). In Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper 65; Dmitriev, N.G., Ernst, L.K., Eds.; FAO: Rome, Italy, 1989; pp. 1–94. Available online: https://www.fao.org/4/ah759e/AH759E07.htm (accessed on 27 August 2025).
- L.K. Ernst Federal Research Center for Animal Husbandry. Science News on the Conservation of the Red Gorbatov Cattle by Scientists of the Center on the Website Gazeta.ru.; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2024; Available online: https://www.vij.ru/fits-v-smi/smi-kras-gorb-skot (accessed on 29 September 2025).
- Voskresensky, S.B.; Lukonina, O.N.; Safina, G.F. (Eds.) Yearbook on Breeding Work in Sheep and Goat Breeding in Farms of the Russian Federation (2024); All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2025; Available online: https://vniiplem.ru/wp-content/uploads/2025/%D0%95%D0%B6%D0%B5%D0%B3%D0%BE%D0%B4%D0%BD%D0%B8%D0%BA.%D0%9E%D0%B2%D1%86%D0%B5%D0%B2%D0%BE%D0%B4%D1%81%D1%82%D0%B2%D0%BE.%D0%9A%D0%BE%D0%B7%D0%BE%D0%B2%D0%BE%D0%B4%D1%81%D1%82%D0%B2%D0%BE.2025.pdf (accessed on 27 August 2025).
- Federal Law “On Breeding Livestock” No. 123-FZ of 3 August 1995 (Last Edition No. 232-FZ of 8 August 2024). Available online: http://pravo.gov.ru/proxy/ips/?docbody=&nd=102036917 (accessed on 27 August 2025).
- Federal Law “On Bioresource Centers and Biological (Bioresource) Collections and Amendments to Article 29 of the Federal Law “On Wildlife”” No. 428-FZ of 30 November 2024. Available online: http://publication.pravo.gov.ru/Document/View/0001202411300018 (accessed on 27 August 2025).
- Viana, J.H.M. 2022 Statistics of embryo production and transfer in domestic farm animals: The main trends for the world embryo industry still stand. Embryo Technol. Newsl. 2023, 41, 20–38. [Google Scholar]
- Shichkin, G.I.; Butusov, D.V. (Eds.) Yearbook on Breeding Work in Dairy Cattle Breeding in Farms of the Russian Federation (2022); All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2023; Available online: https://www.elibrary.ru/item.asp?id=54228742 (accessed on 27 August 2025).
- Lukonina, O.N.; Safina, G.F. (Eds.) Yearbook on Breeding Work in Dairy Cattle Breeding in Farms of the Russian Federation (2023); All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2024; Available online: https://vniiplem.ru/wp-content/uploads/%D0%95%D0%B6%D0%B5%D0%B3%D0%BE%D0%B4%D0%BD%D0%B8%D0%BA.%D0%9C%D0%BE%D0%BB.%D0%A1%D0%BA%D0%BE%D1%82.2024-1.pdf (accessed on 27 August 2025).
- Decree of the President of the Russian Federation No. 195 of 19 March 2024 “On the National Center for Genetic Resources of Agricultural Animals”. Available online: http://publication.pravo.gov.ru/document/0001202403190018 (accessed on 27 August 2025).
- Zinovieva, N.A. (Ed.) Report on the Progress of the Implementation of the Program for the Development of the National Center for Genetic Resources of Agricultural Animals for 2024; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2025; Available online: https://www.vij.ru/novosti2/nc-mvk-1 (accessed on 27 August 2025).
- Pizzi, F.; Turri, F.; Gliozzi, T.M.; Gandini, G. Implementation and cost analysis of a regional farm animal cryobank: An Italian case study. Ital. J. Anim. Sci. 2016, 15, 207–210. [Google Scholar] [CrossRef]
- Leroy, G.; Boettcher, P.; Besbes, B.; Danchin-Burge, C.; Baumung, R.; Hiemstra, S.J. Cryoconservation of animal genetic resources in Europe and two African countries: A gap analysis. Diversity 2019, 11, 240. [Google Scholar] [CrossRef]
- Silversides, F.G.; Purdy, P.H.; Blackburn, H.D. Comparative costs of programmes to conserve chicken genetic variation based on maintaining living populations or storing cryopreserved material. Br. Poult. Sci. 2012, 53, 599–607. [Google Scholar] [CrossRef]
- Program for the Development of the National Center for Genetic Resources of Agricultural Animals for 2024–2030. Available online: http://publication.pravo.gov.ru/document/0001202410250065 (accessed on 27 August 2025).
- Abdelmanova, A.S.; Mishina, A.I.; Volkova, V.V.; Chinarov, R.Y.; Sermyagin, A.A.; Dotsev, A.V.; Boronetskaya, O.I.; Petrikeeva, L.V.; Kostyunina, O.V.; Brem, G.; et al. Comparative study of different methods of DNA extraction from cattle bones specimens maintained in a craniological collection. Sel’skokhozyaistvennaya Biol. 2019, 54, 1110–1121. [Google Scholar] [CrossRef]
- Matyukov, V.S.; Tyrina, Y.O.; Kantanen, Y.; Stolpovskii, Y.A. About features and selective value of the gene pool in local cattle (for Kholmogory breed as an example). Sel’skokhozyaistvennaya Biol. 2013, 2, 19–30. [Google Scholar] [CrossRef]
- Rowe, K.C.; Singhal, S.; Macmanes, M.D.; Ayroles, J.F.; Morelli, T.L.; Rubidge, E.M.; Bi, K.; Moritz, C.C. Museum genomics: Low-cost and high-accuracy genetic data from historical specimens. Mol. Ecol. Resour. 2011, 11, 1082–1092. [Google Scholar] [CrossRef]
- Billerman, S.M.; Walsh, J. Historical DNA as a tool to address key questions in avian biology and evolution: A review of methods, challenges, applications, and future directions. Mol. Ecol. Resour. 2019, 19, 1115–1130. [Google Scholar] [CrossRef]
- McHugo, G.P.; Dover, M.J.; MacHugh, D.E. Unlocking the origins and biology of domestic animals using ancient DNA and paleogenomics. BMC Biol. 2019, 17, 98. [Google Scholar] [CrossRef] [PubMed]
- Card, D.C.; Shapiro, B.; Giribet, G.; Moritz, C.; Edwards, S.V. Museum genomics. Annu. Rev. Genet. 2021, 55, 633–659. [Google Scholar] [CrossRef]
- Edwards, C.J.; Connellan, J.; Wallace, P.F.; Park, S.D.E.; McCormick, F.M.; Olsaker, I.; Eythórsdóttir, E.; MacHugh, D.E.; Bailey, J.F.; Bradley, D.G. Feasibility and utility of microsatellite markers in archaeological cattle remains from a Viking Age settlement in Dublin. Anim. Genet. 2003, 34, 410–416. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gargani, M.; Pariset, L.; Lenstra, J.A.; De Minicis, E.; European Cattle Genetic Diversity Consortium; Valentini, A. Microsatellite genotyping of medieval cattle from central Italy suggests an old origin of Chianina and Romagnola cattle. Front. Genet. 2015, 6, 68. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef]
- Parejo, M.; Wragg, D.; Henriques, D.; Charrière, J.D.; Estonba, A. Digging into the genomic past of Swiss honey bees by whole-genome sequencing museum specimens. Genome Biol. Evol. 2020, 12, 2535–2551. [Google Scholar] [CrossRef]
- Kvist, L.; Honka, J.; Salazar, D.; Kirkinen, T.; Hemmann, K. Memories, museum artefacts and excavations in resolving the history of maternal lineages in the Finnhorse. Anim. Genet. 2022, 53, 821–828. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Kharzinova, V.R.; Volkova, V.V.; Mishina, A.I.; Dotsev, A.V.; Sermyagin, A.A.; Boronetskaya, O.I.; Petrikeeva, L.V.; Chinarov, R.Y.; Brem, G.; et al. Genetic diversity of historical and modern populations of Russian cattle breeds revealed by microsatellite analysis. Genes 2020, 11, 940. [Google Scholar] [CrossRef]
- Abdelmanova, A.A.; Deniskova, T.E.; Kharzinova, V.R.; Chinarov, R.Y.; Boronetskaya, O.I.; Sölkner, J.; Brem, G.; Ai, H.; Huang, L.; Trukhachev, V.I.; et al. Tracing the dynamical genetic diversity changes of Russian Livni pigs during the last 50 years with the museum, old, and modern samples. Animals 2024, 14, 1629. [Google Scholar] [CrossRef]
- Middendorf, A.F.A. Middendorf’s report. On the breed of cattle of northern Russia and its improvement. In Research of the Current State of Cattle Breeding in Russia. Horned Cattle, Issue 1; Ministry of State Property, Department of Agriculture and Rural Industry, Printing House of M.N. Lavrov and Ko: Moscow, Russia, 1884; pp. 226–260. Available online: https://elibrary.tambovlib.ru/?ebook=4383#n=226 (accessed on 27 August 2025).
- Chinarov, R.Y. Developing the Ovum Pick-Up technology in cattle: State-of-the-art and research directions (review). Sel’skokhozyaistvennaya Biol. 2024, 59, 194–220. [Google Scholar] [CrossRef]
- Zinovieva, N.A. Using OPU/IVP Technology to Implement the Developmental Program of the National Center for Animal Genetic Resources. In Proceedings of the International Agro-Industrial Exhibition AgroRus, St.-Petersburg, Russia, 15 October 2025; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2025. Available online: https://www.vij.ru/images/conf-25/AgroRus_25/2025_agrorus_NA.pdf (accessed on 20 October 2025).
- Sabaneev, L.P. Essays on the Trans-Urals and Steppe Economy on the Bashkir Lands; Printing House of V. Gautier: Moscow, Russia, 1873; Available online: https://elis.psu.ru/node/77462 (accessed on 27 August 2025).
- Romanov, A.I. On Tagil Cattle in Connection with Its History and the Establishment of a Zemstvo Nursery Based on Crossbreeding with Dutch Cattle. In Proceedings of the Conference of Veterinarians and Representatives of District Zemstvos of Perm Governorate, Perm, Russia, 18–24 March 1908; Governorate Zemstvo Printing House: Perm, Russia, 1913; pp. 11–129. Available online: https://archives.permkrai.ru/library/view/8940 (accessed on 27 August 2025).
- Stolpovsky, Y.A.; Beketov, S.V.; Solodneva, E.V.; Absalikov, V.M.; Abdelmanova, A.S.; Gladyr, E.A.; Zinovieva, N.A. The population-genetic structure of native Tagil cattle by STR- and SNP-markers. Sel’skokhozyaistvennaya Biol. 2021, 56, 1123–1133. [Google Scholar] [CrossRef]
- Ernst, L.K.; Zinovieva, N.A. Biological Problems of Livestock in the 21st Century; Russian Academy of Agricultural Sciences, All-Russian Research Institute of Animal Husbandry: Moscow, Russia, 2008; Available online: https://www.agrobiology.ru/ernstbook.html (accessed on 27 August 2025).
- Abdelmanova, A.S.; Sermyagin, A.A.; Dotsev, A.V.; Rodionov, A.N.; Stolpovsky, Y.A.; Zinovieva, N.A. Whole-genomic studies of the population structure of Russian local black-pied breeds. Russ. J. Genet. 2022, 58, 804–813. [Google Scholar] [CrossRef]
- L.K. Ernst Federal Research Center for Animal Husbandry. Science News. In Viable Offspring Were Obtained After Transplantation of OPU/IVPembryos of the Tagil Cattle Breed; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2025; Available online: https://www.vij.ru/novosti2/spc-tagilki (accessed on 27 August 2025).
- Zinovieva, N.A. Ex Situ Conservation of Genetic Resources of Native Cattle Breeds. In Proceedings of the Scientific and Production Conference “Problems of Dairy Cattle Breed Diversity Conservation”, Dubrovitsy, Russia, 20–21 April 2023; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2023. Available online: https://www.vij.ru/images/conf-23/Conf_VIZH_21.04.23/Zinovieva_NA.pdf (accessed on 27 August 2025).
- Zinovieva, N.A. Scientific and Methodological Approaches to Increasing the Efficiency of Using Donor Cows in Programs for Obtaining IVP Embryos. In Round Table “Theoretical and Practical Aspects of the Use of Assisted Reproductive Technologies in Animal Husbandry”, Proceedings of the 33rd International Agro-Industrial Exhibition AgroRus, St. Petersburg, Russia, 28–30 August 2024; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2024; Available online: https://www.vij.ru/images/conf-24/Agrorus/2024_.pdf (accessed on 27 August 2025).
- L.K. Ernst Federal Research Center for Animal Husbandry. Science News. In Russian Geneticists Have More Than 100 Thousand Samples for the National Center for Farm Animals; TASS Publication: Moscow, Russia; L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2025; Available online: https://www.vij.ru/novosti2/tass-nauka-nc (accessed on 27 August 2025).
- Korolev, A.A.; Baranova, N.S.; Koroleva, E.A. Improvement of Kostroma Breed Cattle with the Use of Domestic and Imported Breeding Bulls; INFRA-M: Moscow, Russia. Available online: https://www.elibrary.ru/item.asp?id=50239756 (accessed on 27 August 2025).
- Boronetskaya, O.I. (Ed.) Catalogue of the Craniological Collection of the E.F. Liskun State Museum of Animal Husbandry; Russian State Agrarian University—Moscow Timiryazev Agricultural Academy: Moscow, Russia, 2012; Available online: https://museum.timacad.ru/%D0%BA%D0%BE%D0%BB%D0%BB%D0%B5%D0%BA%D1%86%D0%B8%D0%B8-%D0%BC%D1%83%D0%B7%D0%B5%D1%8F/#ZCranyo (accessed on 27 August 2025).
- OGBUK “Kostroma State Historical, Architectural and Art Museum-Reserve”. Kostroma Breed. In Museum on Connection.ru; OGBUK “Kostroma State Historical, Architectural and Art Museum-Reserve”: Kostroma, Russia, 2025; Available online: https://xn--80aehhdgdvm5bq5m.xn--p1ai/muzeynyy_predmet/estestvennye_nauki_i_priroda/kostromskaya_poroda (accessed on 27 August 2025).
- Orekhov, A.A. Goats. In Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper 65; FAO: Rome, Italy, 1989; pp. 344–365. Available online: https://www.fao.org/4/ah759e/AH759E14.htm (accessed on 27 August 2025).
- Tarasova, E.I.; Frolov, A.N.; Lebedev, S.V.; Romanov, M.N. History, Breeding, Selection and Genetics of the Orenburg Goat Breed. In Molecular Genetic Technologies for Analysis of Gene Expression Related to Animal Productivity and Disease Resistance, Proceedings of the Materials of the 3rd International Scientific and Practical Conference, Moscow, Russia, 30 September 2021; Sel’skokhozyaistvennye Tekhnologii: Moscow, Russia, 2021; pp. 450–454. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Abdelmanova, A.S.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Gladyr, E.A.; Gusev, I.V.; Selionova, M.I.; Rodionov, A.N.; et al. Genetic diversity in the Orenburg goat breed revealed by single-nucleotide polymorphism (SNP) analysis: Initial steps in saving a threatened population. Genes 2024, 15, 1375. [Google Scholar] [CrossRef]
- Tarasova, E.I.; Frolov, A.N.; Lebedev, S.V.; Romanov, M.N. Landmark native breed of the Orenburg goats: Progress in its breeding and genetics and future prospects. Anim. Biotechnol. 2023, 34, 5139–5154. [Google Scholar] [CrossRef] [PubMed]
- Tyurin, A.N. Orenburg downy shawl. In Orenburg Oblast: Geography, Economy, Ecology: Collection of Scientific Articles; Orenburg State Pedagogical University: Orenburg, Russia, 2014; pp. 122–143. Available online: https://ospu.ru/assets/resources/izdatelstvo/Sb_geo_2014/122_143.pdf (accessed on 27 August 2025).
- Grigoryan, L.N.; Khatataev, S.A.; Sverchkova, S.V. The state of goat breeding in the Russian Federation and its breeding base. In Yearbook on Breeding Work in Sheep and Goat Breeding in Farms of the Russian Federation (2005); Desyatov, V.G., Ed.; All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2006; pp. 312–313. Available online: https://www.elibrary.ru/item.asp?id=24656156 (accessed on 27 August 2025).
- Desyatov, V.G. (Ed.) Yearbook on Breeding Work in Sheep and Goat Breeding in Farms of the Russian Federation (2005); All Russian Research Institute of Animal Breeding: Lesnye Polyany, Russia, 2006; Available online: https://www.elibrary.ru/item.asp?id=24194102 (accessed on 27 August 2025).
- Abdelmanova, A.S.; Deniskova, T.E.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Gladyr, E.A.; Gusev, I.V.; Lebedev, S.V.; Zinovieva, N.A. Assessment of the dynamics of genetic diversity of Orenburg goat breed populations by microsatellite markers. Dostizheniya Nauki i Tekhniki APK 2024, 38, 50–56. [Google Scholar] [CrossRef]
- Al-Swailem, A.M.; Shehata, M.M.; Abu-Duhier, F.M.; Al-Yamani, E.J.; Al-Busadah, K.A.; Al-Arawi, M.S.; Al-Khider, A.Y.; Al-Muhaimeed, A.N.; Al-Qahtani, F.H.; Manee, M.M.; et al. Sequencing, analysis, and annotation of expressed sequence tags for Camelus dromedarius. PLoS ONE 2010, 5, e10720. [Google Scholar] [CrossRef]
- L.K. Ernst Federal Research Center for Animal Husbandry. Development of a Network of Bioresource Collections of Farm Animals Project No. 075-15-2025-469 (Ministry of Science Higher Education of the Russian Federation); L.K. Ernst Federal Research Center for Animal Husbandry: Dubrovitsy, Russia, 2025; Available online: https://www.vij.ru/goszadanie-i-proekty/proekty/proekty-minobrnauki/brc-2025 (accessed on 27 August 2025).
- International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature 2004, 432, 695–716. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Chang, J.L.; Gnerre, S.; Clamp, M.; Lander, E.S.; The Genome Sequencing Platform; The Genome Assembly Team. GenBank: Oryctolagus Cuniculus Breed Thorbecke Inbred, Whole Genome Shotgun Sequencing Project; GenBank: AAGW00000000.1. Annotating the Human Genome Using Low Coverage Mammalian Genomes; Direct Submission (03-May-2005); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2005. Available online: https://www.ncbi.nlm.nih.gov/nuccore/AAGW00000000.1 (accessed on 27 August 2025).
- Honeybee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature 2006, 443, 931–949. [Google Scholar] [CrossRef]
- Bovine Genome Sequencing and Analysis Consortium; Elsik, C.G.; Tellam, R.L.; Worley, K.C.; Gibbs, R.A.; Muzny, D.M.; Weinstock, G.M.; Adelson, D.L.; Eichler, E.E.; Elnitski, L.; et al. The genome sequence of taurine cattle: A window to ruminant biology and evolution. Science 2009, 324, 522–528. [Google Scholar] [CrossRef]
- Zimin, A.V.; Delcher, A.L.; Florea, L.; Kelley, D.R.; Schatz, M.C.; Puiu, D.; Hanrahan, F.; Pertea, G.; Van Tassell, C.P.; Sonstegard, T.S.; et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009, 10, R42. [Google Scholar] [CrossRef] [PubMed]
- Wade, C.M.; Giulotto, E.; Sigurdsson, S.; Zoli, M.; Gnerre, S.; Imsland, F.; Lear, T.L.; Adelson, D.L.; Bailey, E.; Bellone, R.R.; et al. Genome sequence, comparative analysis, and population genetics of the domestic horse. Science 2009, 326, 865–867. [Google Scholar] [CrossRef] [PubMed]
- Archibald, A.L.; Bolund, L.; Churcher, C.; Fredholm, M.; Groenen, M.A.; Harlizius, B.; Lee, K.T.; Milan, D.; Rogers, J.; Rothschild, M.F.; et al. Pig genome sequence–analysis and publication strategy. BMC Genom. 2010, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Xie, M.; Chen, W.; Talbot, R.; Maddox, J.F.; Faraut, T.; Wu, C.; Muzny, D.M.; Li, Y.; Zhang, W.; et al. The sheep genome illuminates biology of the rumen and lipid metabolism. Science 2014, 344, 1168–1173. [Google Scholar] [CrossRef]
- Dalloul, R.A.; Long, J.A.; Zimin, A.V.; Aslam, L.; Beal, K.; Ann Blomberg, L.; Bouffard, P.; Burt, D.W.; Crasta, O.; Crooijmans, R.P.M.A.; et al. Multi-platform next-generation sequencing of the domestic turkey (Meleagris gallopavo): Genome assembly and analysis. PLoS Biol. 2010, 8, e1000475. [Google Scholar] [CrossRef]
- Qiu, Q.; Zhang, G.; Ma, T.; Qian, W.; Wang, J.; Ye, Z.; Cao, C.; Hu, Q.; Kim, J.; Larkin, D.M.; et al. The yak genome and adaptation to life at high altitude. Nat. Genet. 2012, 44, 946–949. [Google Scholar] [CrossRef]
- Huang, Y.; Li, Y.; Burt, D.W.; Chen, H.; Zhang, Y.; Qian, W.; Kim, H.; Gan, S.; Zhao, Y.; Li, J.; et al. The duck genome and transcriptome provide insight into an avian influenza virus reservoir species. Nat. Genet. 2013, 45, 776–783. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Xie, M.; Jiang, Y.U.; Xiao, N.; Du, X.; Zhang, W.; Tosser-Klopp, G.; Wang, J.; Yang, S.; Liang, J.; et al. Sequencing and automated whole-genome optical mapping of the genome of a domestic goat (Capra hircus). Nat. Biotechnol. 2013, 31, 135–141. [Google Scholar] [CrossRef]
- Dobson, L.K.; Zimin, A.; Bayles, D.; Fritz-Waters, E.; Alt, D.; Olsen, S.; Blanchong, J.; Reecy, J.; Smith, T.P.L.; Derr, J.N. De novo assembly and annotation of the North American bison (Bison bison) reference genome and subsequent variant identification. Anim. Genet. 2021, 52, 263–274. [Google Scholar] [CrossRef]
- Lu, L.; Chen, Y.; Wang, Z.; Li, X.; Chen, W.; Tao, Z.; Shen, J.; Tian, Y.; Wang, D.; Li, G.; et al. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biol. 2015, 16, 89. [Google Scholar] [CrossRef]
- Warren, W.; Burt, D.W.; Antin, P.B.; Lanford, R.; Gros, J.; Wilson, R.K.; International Coturnix japonica Genome Analysis Consortium. GenBank: Coturnix Japonica Isolate 7356, Whole Genome Shotgun Sequencing Project; GenBank: LSZS00000000.1. Direct Submission (24-Nov-2015); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2015. Available online: https://www.ncbi.nlm.nih.gov/nuccore/LSZS00000000.1 (accessed on 27 August 2025).
- Vignal, A.; Warren, W. GenBank: Numida Meleagris Breed g44 Domestic Line Isolate 19003, Whole Genome Shotgun Sequencing Project; GenBank: MTSP00000000.1. Guinea Fowl Genome Sequence and Diversity; Direct Submission (06-Jan-2017); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2017. Available online: https://www.ncbi.nlm.nih.gov/nuccore/MTSP00000000.1 (accessed on 27 August 2025).
- Li, Z.; Lin, Z.; Ba, H.; Chen, L.; Yang, Y.; Wang, K.; Qiu, Q.; Wang, W.; Li, G. Draft genome of the reindeer (Rangifer tarandus). GigaScience 2017, 6, gix102. [Google Scholar] [CrossRef]
- Kawamoto, M.; Jouraku, A.; Toyoda, A.; Yokoi, K.; Minakuchi, Y.; Katsuma, S.; Fujiyama, A.; Kiuchi, T.; Yamamoto, K.; Shimada, T. GenBank: Bombyx Mori Strain p50T, Whole Genome Shotgun Sequencing Project; GenBank: BHWX00000000.1. High-Quality Genome Assembly of the Silkworm; Bombyx Mori. Direct Submission (16-Oct-2018); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2018. Available online: https://www.ncbi.nlm.nih.gov/nuccore/BHWX00000000.1 (accessed on 27 August 2025).
- Helix pomatia (Roman Snail) RefSeq Genome. In Helix pomatia Genome Reference Project; Accession: PRJNA528055; ID: 528055. BioProject; National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2019. Available online: https://www.ncbi.nlm.nih.gov/bioproject/528055 (accessed on 27 August 2025).
- Wang, C.; Li, H.; Guo, Y.; Huang, J.; Sun, Y.; Min, J.; Wang, J.; Fang, X.; Zhao, Z.; Wang, S.; et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat. Commun. 2020, 11, 6014. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Li, H.; Liu, Z.; Zhang, C.; Li, K.; Gong, Y.; Geng, L.; Su, J.; Guan, X.; Liu, L.; et al. Chromosome-level genome assembly of the Arctic fox (Vulpes lagopus) using PacBio sequencing and Hi-C technology. Mol. Ecol. Resour. 2021, 21, 2093–2108. [Google Scholar] [CrossRef]
- Jiang, F.; Jiang, Y.; Wang, W.; Xiao, C.; Lin, R.; Xie, T.; Sung, W.-K.; Li, S.; Jakovlić, I.; Chen, J.; et al. A chromosome-level genome assembly of Cairina moschata and comparative genomic analyses. BMC Genom. 2021, 22, 581. [Google Scholar] [CrossRef]
- Khan, A.; Singh, K.; Jaiswal, S.; Raza, M.; Jasrotia, R.S.; Kumar, A.; Gurjar, A.K.S.; Kumari, J.; Nayan, V.; Iquebal, M.A.; et al. Whole-genome-based web genomic resource for water buffalo (Bubalus bubalis). Front. Genet. 2022, 13, 809741. [Google Scholar] [CrossRef] [PubMed]
- Karimi, K.; Do, D.N.; Wang, J.; Easley, J.; Borzouie, S.; Sargolzaei, M.; Plastow, G.; Wang, Z.; Miar, Y. A chromosome-level genome assembly reveals genomic characteristics of the American mink (Neogale vison). Commun. Biol. 2022, 5, 1381. [Google Scholar] [CrossRef]
- Genome, National Center for Biotechnology Information (NCBI), National Library of Medicine. Genome Assembly NIAB-ARS_B.indTharparkar_mat_pri_1.0; NCBI RefSeq Assembly GCF_029378745.1. Taxon: Bos Indicus (Zebu cattle); Genome, National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2023. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_029378745.1/ (accessed on 27 August 2025).
- Wang, Q.; Han, R.; Xing, H.; Li, H. A consensus genome of sika deer (Cervus nippon) and transcriptome analysis provided novel insights on the regulation mechanism of transcript factor in antler development. BMC Genom. 2024, 25, 617. [Google Scholar] [CrossRef]
- Biegler, M.; Keyte, A.; Tilley, T.; Traore, P.; Abueg, L.; Mathers, T.; Balacco, J.; Formenti, G.; Jarvis, E.D. GenBank: Struthio Camelus Isolate bStrCam1, Whole Genome Shotgun Sequencing Project; GenBank: JBFMIB000000000.1. Struthio camelus (North African ostrich) Genome, bStrCam1, Haplotype 1; Direct Submission (08-Jul-2024); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/nuccore/JBFMIB000000000.1 (accessed on 27 August 2025).
- Wellcome Sanger Tree of Life Programme. GenBank: Cornu aspersum, Whole Genome Shotgun Sequencing Project; GenBank: CAXLNJ000000000.1. Direct Submission (16-Jun-2024); National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2024. Available online: https://www.ncbi.nlm.nih.gov/nuccore/CAXLNJ000000000.1 (accessed on 27 August 2025).
- Lopez Colom, R.; O’Brien, M.; Natural History Museum Genome Acquisition Lab; Darwin Tree of Life Barcoding Collective; Wellcome Sanger Institute Tree of Life Management, Samples and Laboratory Team; Wellcome Sanger Institute Scientific Operations: Sequencing Operations; Wellcome Sanger Institute Tree of Life Core Informatics Team; Tree of Life Core Informatics collective; Darwin Tree of Life Consortium. The genome sequence of the red fox, Vulpes vulpes (Linnaeus, 1758). Wellcome Open Res. 2025, 10, 13. [Google Scholar] [CrossRef]
- Tomarovsky, A.; Khan, R.; Dudchenko, O.; Totikov, A.; Serdyukova, N.A.; Weisz, D.; Vorobieva, N.V.; Bulyonkova, T.; Abramov, A.V.; Nie, W.; et al. Chromosome-length genome assembly of the stone marten (Martes foina, Mustelidae): A new view on one of the cornerstones in carnivore cytogenetics. J. Hered. 2025, 116, 548–557. [Google Scholar] [CrossRef]
- Genome, National Center for Biotechnology Information (NCBI), National Library of Medicine. Genome Assembly ASM4877302v1; NCBI RefSeq Assembly GCF_048773025.1. Taxon: Camelus bactrianus (Bactrian camel); Genome, National Center for Biotechnology Information (NCBI), National Library of Medicine: Bethesda, MD, USA, 2025. Available online: https://www.ncbi.nlm.nih.gov/datasets/genome/GCF_048773025.1/ (accessed on 27 August 2025).
- Eggen, A. The development and application of genomic selection as a new breeding paradigm. Anim. Front. 2012, 2, 10–15. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Koshkina, O.A.; Solovieva, A.D.; Churbakova, N.A.; Petrov, S.N.; Frolov, A.N.; Platonov, S.A.; Abdelmanova, A.S.; Vladimirov, M.A.; et al. Examination of runs of homozygosity distribution patterns and relevant candidate genes of potential economic interest in Russian goat breeds using whole-genome sequencing. Genes 2025, 16, 631. [Google Scholar] [CrossRef]
- Deniskova, Т.Е.; Shakhin, A.V.; Esmailizadeh, A.; Dotsev, A.V.; Zinovieva, N.A. Analysis of polymorphism in the major genes for reproductive traits in sheep (Ovis spp.). Sel’skokhozyaistvennaya Biol. 2023, 58, 1046–1056. [Google Scholar] [CrossRef]
- Romanov, M.N.; Abdelmanova, A.S.; Fisinin, V.I.; Gladyr, E.A.; Volkova, N.A.; Koshkina, O.A.; Rodionov, A.N.; Vetokh, A.N.; Gusev, I.V.; Anshakov, D.V.; et al. Selective footprints and genes relevant to cold adaptation and other phenotypic traits are unscrambled in the genomes of divergently selected chicken breeds. J. Anim. Sci. Biotechnol. 2023, 14, 35. [Google Scholar] [CrossRef] [PubMed]
- Romanov, M.N.; Shakhin, A.V.; Abdelmanova, A.S.; Volkova, N.A.; Efimov, D.N.; Fisinin, V.I.; Korshunova, L.G.; Anshakov, D.V.; Dotsev, A.V.; Griffin, D.K.; et al. Dissecting selective signatures and candidate genes in grandparent lines subject to high selection pressure for broiler production and in a local Russian chicken breed of Ushanka. Genes 2024, 15, 524. [Google Scholar] [CrossRef] [PubMed]
- Moiseyeva, I.G.; Sevastyanova, A.A.; Aleksandrov, A.V.; Vakhrameev, A.B.; Romanov, M.N.; Dmitriev, Y.I.; Semenova, S.K.; Sulimova, G.E. Orloff chicken breed: History, current status and studies. Izv. Timiryazev. S-Kh. Akad. 2016, 1, 78–96. Available online: https://www.elibrary.ru/item.asp?id=25664565 (accessed on 27 August 2025).
- Ramos, A.M.; Crooijmans, R.P.M.A.; Affara, N.A.; Amaral, A.J.; Archibald, A.L.; Beever, J.E.; Bendixen, C.; Churcher, C.; Clark, R.; Dehais, P.; et al. Design of a high density SNP genotyping assay in the pig using SNPs identified and characterized by next generation sequencing technology. PLoS ONE 2009, 4, e6524. [Google Scholar] [CrossRef]
- Anderson, R.; McEwan, J.; Brauning, R.; Kijas, J.; Dalrymple, B.; Worley, K.; Daetwyler, H.; Heaton, M.; Van Stijn, T.; Clarke, S.; et al. Development of a High Density (600K) Illumina Ovine SNP Chip and Its Use to Fine Map the Yellow Fat Locus. In Proceedings of the International Plant and Animal Genome XII Conference, San Diego, CA, USA, 12–17 January 2014; Scherago International: San Diego, CA, USA, 2014. Abstract P611. Available online: https://pag.confex.com/pag/xxii/webprogram/Paper10725.html (accessed on 27 August 2025).
- Tosser-Klopp, G.; Bardou, P.; Bouchez, O.; Cabau, C.; Crooijmans, R.; Dong, Y.; Donnadieu-Tonon, C.; Eggen, A.; Heuven, H.C.M.; Jamli, S.; et al. Design and characterization of a 52K SNP chip for goats. PLoS ONE 2014, 9, e86227. [Google Scholar] [CrossRef]
- Groenen, M.A.; Megens, H.-J.; Zare, Y.; Warren, W.C.; Hillier, L.W.; Crooijmans, R.P.; Vereijken, A.; Okimoto, R.; Muir, W.M.; Cheng, H.H. The development and characterization of a 60K SNP chip for chicken. BMC Genom. 2011, 12, 274. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Dotsev, A.V.; Sermyagin, A.A.; Wimmers, K.; Reyer, H.; Sölkner, J.; Deniskova, T.E.; Brem, G. Study of genetic diversity and population structure of five Russian cattle breeds using whole-genome SNP analysis. Sel’skokhozyaistvennaya Biol. 2016, 51, 788–800. [Google Scholar] [CrossRef][Green Version]
- Sermyagin, A.A.; Dotsev, A.V.; Gladyr, E.A.; Traspov, A.A.; Deniskova, T.E.; Kostyunina, O.V.; Reyer, H.; Wimmers, K.; Barbato, M.; Paronyan, I.A.; et al. Whole-genome SNP analysis elucidates the genetic structure of Russian cattle and its relationship with Eurasian taurine breeds. Genet. Sel. Evol. 2018, 50, 37. [Google Scholar] [CrossRef]
- Zinovieva, N.A.; Dotsev, A.V.; Sermyagin, A.A.; Deniskova, T.E.; Abdelmanova, A.S.; Kharzinova, V.R.; Sölkner, J.; Reyer, H.; Wimmers, K.; Brem, G. Selection signatures in two oldest Russian native cattle breeds revealed using high-density single nucleotide polymorphism analysis. PLoS ONE 2020, 15, e0242200. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Barbato, M.; Traspov, A.A.; Brem, G.; et al. Population structure and genetic diversity of 25 Russian sheep breeds based on whole-genome genotyping. Genet. Sel. Evol. 2018, 50, 29. [Google Scholar] [CrossRef]
- Deniskova, T.; Dotsev, A.; Lushihina, E.; Shakhin, A.; Kunz, E.; Medugorac, I.; Reyer, H.; Wimmers, K.; Khayatzadeh, N.; Sölkner, J.; et al. Population structure and genetic diversity of sheep breeds in the Kyrgyzstan. Front. Genet. 2019, 10, 1311. [Google Scholar] [CrossRef] [PubMed]
- Ceccobelli, S.; Landi, V.; Senczuk, G.; Mastrangelo, S.; Sardina, M.T.; Ben-Jemaa, S.; Persichilli, C.; Karsli, T.; Bâlteanu, V.-A.; Raschia, M.A.; et al. A comprehensive analysis of the genetic diversity and environmental adaptability in worldwide Merino and Merino-derived sheep breeds. Genet. Sel. Evol. 2023, 55, 24. [Google Scholar] [CrossRef]
- Ghoreishifar, S.M.; Rochus, C.M.; Moghaddaszadeh-Ahrabi, S.; Davoudi, P.; Salek Ardestani, S.; Zinovieva, N.A.; Deniskova, T.E.; Johansson, A.M. Shared ancestry and signatures of recent selection in Gotland sheep. Genes 2021, 12, 433. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-density genotyping reveals signatures of selection related to acclimation and economically important traits in 15 local sheep breeds from Russia. BMC Genom. 2019, 20 (Suppl. S3), 294. [Google Scholar] [CrossRef]
- Igoshin, A.V.; Deniskova, T.E.; Yurchenko, A.A.; Yudin, N.S.; Dotsev, A.V.; Selionova, M.I.; Zinovieva, N.A.; Larkin, D.M. Copy number variants in genomes of local sheep breeds from Russia. Anim. Genet. 2022, 53, 119–132. [Google Scholar] [CrossRef]
- Deniskova, Т.Е.; Petrov, S.N.; Sermyagin, A.A.; Dosev, A.V.; Fornara, M.S.; Reyer, H.; Wimmers, K.; Bagirov, V.A.; Brem, G.; Zinovieva, N.A. A search for genomic variants associated with body weight in sheep based on high density SNP genotypes analysis. Sel’skokhozyaistvennaya Biol. 2021, 56, 279–291. [Google Scholar] [CrossRef]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Reyer, H.; Sölkner, J.; Fornara, M.S.; Aybazov, A.-M.M.; Wimmers, K.; Brem, G.; Zinovieva, N.A. SNP-based genotyping provides insight into the West Asian origin of Russian local goats. Front. Genet. 2021, 12, 708740. [Google Scholar] [CrossRef] [PubMed]
- Deniskova, T.E.; Dotsev, A.V.; Selionova, M.I.; Aibazov, A.-M.M.; Zinovieva, N.A. Search for signatures of selection in the genomes of domestic goats (Capra hircus L.) raised in Russia using detection of ROH islands. Sel’skokhozyaistvennaya Biol. 2024, 59, 620–632. [Google Scholar] [CrossRef]
- Manunza, A.; Diaz, J.R.; Sayre, B.L.; Cozzi, P.; Bobbo, T.; Deniskova, T.; Dotsev, A.; Zinovieva, N.; Stella, A. Discovering novel clues of natural selection on four worldwide goat breeds. Sci. Rep. 2023, 13, 2110. [Google Scholar] [CrossRef] [PubMed]
- Sermyagin, A.A.; Deniskova, T.E.; Gusev, I.V.; Petrov, S.N.; Rodionov, A.N.; Dotsev, A.V.; Zinovieva, N.A. Identification of SNPs associated with growth and development traits of goats (Capra hircus Linnaeus, 1758) from the resource population in age dynamics. Sel’skokhozyaistvennaya Biol. 2024, 59, 633–648. [Google Scholar] [CrossRef]
- Traspov, A.; Deng, W.; Kostyunina, O.; Ji, J.; Shatokhin, K.; Lugovoy, S.; Zinovieva, N.; Yang, B.; Huang, L. Population structure and genome characterization of local pig breeds in Russia, Belorussia, Kazakhstan and Ukraine. Genet. Sel. Evol. 2016, 48, 16. [Google Scholar] [CrossRef]
- Kharzinova, V.R.; Dotsev, A.V.; Deniskova, T.E.; Solovieva, A.D.; Fedorov, V.I.; Layshev, K.A.; Romanenko, T.M.; Okhlopkov, I.M.; Wimmers, K.; Reyer, H.; et al. Genetic diversity and population structure of domestic and wild reindeer (Rangifer tarandus L. 1758): A novel approach using BovineHD BeadChip. PLoS ONE 2018, 13, e0207944. [Google Scholar] [CrossRef]
- Kharzinova, V.R.; Zinovieva, N.A. The pattern of genetic diversity of different breeds of pigs based on microsatellite analysis. Vavilovskii Zh. Genet. Sel. 2020, 24, 747–754. [Google Scholar] [CrossRef]
- Dementieva, N.V.; Kudinov, A.A.; Larkina, T.A.; Mitrofanova, O.V.; Dysin, A.P.; Terletsky, V.P.; Tyshchenko, V.I.; Griffin, D.K.; Romanov, M.N. Genetic variability in local and imported germplasm chicken populations as revealed by analyzing runs of homozygosity. Animals 2020, 10, 1887. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Romanov, M.N.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Terletsky, V.P.; Fedorova, E.S.; Nikitkina, E.V.; Plemyashov, K.V. Studying the structure of a gene pool population of the Russian White chicken breed by genome-wide SNP scan. Sel’skokhozyaistvennaya Biol. 2017, 52, 1166–1174. [Google Scholar] [CrossRef]
- Dementeva, N.V.; Kudinov, A.A.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Romanov, M.N. Genome-wide association study of reproductive traits in a gene pool breed of the Russian White chickens. Reprod. Domest. Anim. 2018, 53 (Suppl. S2), 123–124. [Google Scholar] [CrossRef]
- Kudinov, A.A.; Dementieva, N.V.; Mitrofanova, O.V.; Stanishevskaya, O.I.; Fedorova, E.S.; Larkina, T.A.; Mishina, A.I.; Plemyashov, K.V.; Griffin, D.K.; Romanov, M.N. Genome-wide association studies targeting the yield of extraembryonic fluid and production traits in Russian White chickens. BMC Genom. 2019, 20, 270. [Google Scholar] [CrossRef]
- Abdelmanova, A.S.; Dotsev, A.V.; Romanov, M.N.; Stanishevskaya, O.I.; Gladyr, E.A.; Rodionov, A.N.; Vetokh, A.N.; Volkova, N.A.; Fedorova, E.S.; Gusev, I.V.; et al. Unveiling comparative genomic trajectories of selection and key candidate genes in egg-type Russian White and meat-type White Cornish chickens. Biology 2021, 10, 876. [Google Scholar] [CrossRef]
- Fedorova, E.S.; Dementieva, N.V.; Shcherbakov, Y.S.; Stanishevskaya, O.I. Identification of key candidate genes in runs of homozygosity of the genome of two chicken breeds, associated with cold adaptation. Biology 2022, 11, 547. [Google Scholar] [CrossRef]
- Putman, A.I.; Carbone, I. Challenges in analysis and interpretation of microsatellite data for population genetic studies. Ecol. Evol. 2014, 4, 4399–4428. [Google Scholar] [CrossRef]
- Romanov, M.N.; Weigend, S. Genetic diversity in chicken populations based on microsatellite markers. In Proceedings of the Conference “From Jay Lush to Genomics: Visions for Animal Breeding and Genetics”, Ames, IA, USA, 16–18 May 1999; Iowa State University, Department of Animal Science: Ames, IA, USA, 1999; p. 174. [Google Scholar]
- Kulibaba, R.O.; Srikulnath, K.; Singchat, W.; Liashenko, Y.V.; Griffin, D.K.; Romanov, M.N. The application of microsatellite markers as molecular tools for studying genomic variability in vertebrate populations. Curr. Issues Mol. Biol. 2025, 47, 447. [Google Scholar] [CrossRef] [PubMed]
- Laoun, A.; Harkat, S.; Lafri, M.; Gaouar, S.B.S.; Belabdi, I.; Ciani, E.; De Groot, M.; Blanquet, V.; Leroy, G.; Rognon, X.; et al. Inference of breed structure in farm animals: Empirical comparison between SNP and microsatellite performance. Genes 2020, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Brenig, B.; Schütz, E. Recent development of allele frequencies and exclusion probabilities of microsatellites used for parentage control in the German Holstein Friesian cattle population. BMC Genet. 2016, 17, 18. [Google Scholar] [CrossRef]
- Fox, G.; Preziosi, R.F.; Antwis, R.E.; Benavides-Serrato, M.; Combe, F.J.; Harris, W.E.; Hartley, I.R.; Kitchener, A.C.; de Kort, S.R.; Nekaris, A.I.; et al. Multi-individual microsatellite identification: A multiple genome approach to microsatellite design (MiMi). Mol. Ecol. Resour. 2019, 19, 1672–1680. [Google Scholar] [CrossRef]
- Li, M.-H.; Kantanen, J. Genetic structure of Eurasian cattle (Bos taurus) based on microsatellites: Clarification for their breed classification. Anim. Genet. 2010, 41, 150–158. [Google Scholar] [CrossRef]
- Cañón, J.; Alexandrino, P.; Bessa, I.; Carleos, C.; Carretero, Y.; Dunner, S.; Ferran, N.; Garcia, D.; Jordana, J.; Laloë, D.; et al. Genetic diversity measures of local European beef cattle breeds for conservation purposes. Genet. Sel. Evol. 2001, 33, 311. [Google Scholar] [CrossRef][Green Version]
- Hauser, S.S.; Athrey, G.; Leberg, P.L. Waste not, want not: Microsatellites remain an economical and informative technology for conservation genetics. Ecol. Evol. 2021, 11, 15800–15814. [Google Scholar] [CrossRef]
- FAO. Measurement of Domestic Animal Diversity (MoDAD): New Recommended Microsatellite Markers; New Microsatellite Marker Sets—Recommendations of Joint ISAG/FAO Standing Committee; Food and Agriculture Organization of the United Nations: Rome, Italy, 2004; Available online: http://www.fao.org/3/a-aq569e.pdf (accessed on 27 August 2025).
- FAO. Molecular Genetic Characterization of Animal Genetic Resources; FAO Animal Production and Health Guidelines No. 9; Commission on Genetic Resources for Food and Agriculture, Food and Agriculture Organization of the United Nations: Rome, Italy, 2011; Available online: https://www.fao.org/4/i2413e/i2413e00.htm (accessed on 27 August 2025).
- Volkova, V.V.; Abdelmanova, A.S.; Deniskova, T.E.; Romanenkova, O.S.; Khozhokov, A.A.; Ozdemirov, A.A.; Sermyagin, A.A.; Zinovieva, N.A. Investigation of the genetic diversity of Dagestan Mountain cattle using STR-markers. Diversity 2022, 14, 569. [Google Scholar] [CrossRef]
- Kharzinova, V.R.; Dotsev, A.V.; Solovieva, A.D.; Fedorov, V.I.; Shimit, L.D.O.; Romanenko, T.M.; Senchik, A.V.; Sergeeva, O.K.; Goncharov, V.V.; Laishev, K.A.; et al. Genetic variability of Russian domestic reindeer populations (Rangifer tarandus) by microsatellites. J. Anim. Sci. 2020, 98 (Suppl. S4), 237–238. [Google Scholar] [CrossRef]
- Kharzinova, V.; Dotsev, A.; Solovieva, A.; Sergeeva, O.; Bryzgalov, G.; Reyer, H.; Wimmers, K.; Brem, G.; Zinovieva, N. Insight into the current genetic diversity and population structure of domestic reindeer (Rangifer tarandus) in Russia. Animals 2020, 10, 1309. [Google Scholar] [CrossRef] [PubMed]
- Imperial Russian Poultry Society, A.F. Stolzenburg Printing House. Album of Commercial Breeds of Poultry. Handbook of a Poultry Farmer; Imperial Russian Poultry Society, A.F. Stolzenburg Printing House: St. Petersburg, Russia, 1905; Available online: https://exponat-online.ru/exhibit/22217870/ (accessed on 27 August 2025).
- Romanov, M.N. Geese. In Encyclopedia of Animal Science, Two-Volume Set, 2nd ed.; Taylor & Francis Inc.: London, UK; CRC Press Inc.: Boca Raton, FL, USA, 2018; Volume 1, pp. 487–489. [Google Scholar] [CrossRef]
- Poultry Laboratory. Upper Volga Federal Agricultural Scientific Center, Novy, Suzdal District, Vladimir Oblast, Russia. Available online: https://vnish.org/laboratoriya-ptitsevodstva/ (accessed on 27 August 2025).
- Grishina, D.S.; Zharkova, I.P. Comparative assessment of geese of gene pool herd by their exterior. Vladimir. Zemled. 2020, 3, 64–68. Available online: https://elibrary.ru/item.asp?id=44069770 (accessed on 27 August 2025).
- Kharzinova, V.R.; Volkova, V.V.; Dotsev, A.V.; Nikipelov, V.I.; Rodionov, A.V.; Nikipelova, A.K.; Churbakova, N.A.; Bakoev, N.F.; Grishina, D.S.; Shchukin, I.M.; et al. Conservation and Genetic Structure of Geese Breeds in the Russian Federation: Insights from Microsatellite Analysis. In Proceedings of the 10th International Conference on Sustainable Animal Agriculture for Developing Countries Conference (SAADC2025), Can Tho City, Vietnam, 1–4 October 2025; Can Tho University Publishing House: Can Tho City, Vietnam, 2025; p. 68. [Google Scholar]
- Keenan, K.; McGinnity, P.; Cross, T.F.; Crozier, W.W.; Prodohl, P.A. diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol. Evol. 2013, 4, 782–788. [Google Scholar] [CrossRef]
- Jombart, T. adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics 2008, 24, 1403–1405. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2009. [Google Scholar] [CrossRef]
- Francis, R.M. pophelper: An R package and web app to analyze and visualize population structure. Mol. Ecol. Resour. 2017, 17, 27–32. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- FGBNU Rosinformagrotech. State Register of Breeding Achievements Approved for Use. Vol. 2. Animal Breeds (Official Publication); FGBNU Rosinformagrotech: Moscow, Russia, 2022; Available online: https://gossortrf.ru/wp-content/uploads/2022/09/%D0%A0%D0%B5%D0%B5%D1%81%D1%82%D1%80_%D0%B6%D0%B8%D0%B2%D0%BE%D1%82%D0%BD%D1%8B%D1%85_2022.pdf (accessed on 27 August 2025).
- Fisinin, V.I.; Zlochevskaya, K.V. Geese. In Animal Genetic Resources of the USSR; FAO Animal Production and Health Paper 65; FAO: Rome, Italy, 1989; pp. 469–506. Available online: https://www.fao.org/4/ah759e/AH759E23.htm (accessed on 27 August 2025).
- Martsev, A. Origin and exterior features analysis of geese breeds genealogical groups in Russian gene pool herd. E3S Web Conf. 2020, 203, 01025. [Google Scholar] [CrossRef]
- Romanov, M.N. Using Phenetic Approaches for Studying Poultry Populations under Preservation and Breeding. In Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, ON, Canada, 7–12 August 1994; International Committee for World Congresses on Genetics Applied to Livestock Production, University of Guelph: Guelph, ON, Canada, 1994; Volume 21, pp. 556–559. Available online: https://kar.kent.ac.uk/46403/ (accessed on 27 August 2025).
- Romanov, M.N. Genetic Studies on Geese of Ukraine and Russia. 3. Phenetic and Phylogenetic Approaches. In Proceedings of the 10th International Symposium on Waterfowl, Halle (Saale), Germany, 26–31 March 1995; World’s Poultry Science Association: Halle (Saale), Germany, 1995; pp. 429–432. Available online: https://kar.kent.ac.uk/46301/ (accessed on 27 August 2025).
- Romanov, M.N. Evolution of domestic geese. Reconstruction of microphylogenesis by methods of population phenetics. In New Investigations on Palearctic Geese; Zaporizhya Branch of the Ukrainian Ornithological Society, Zaporizhya State University: Zaporizhya, Ukraine, 1995; pp. 115–120. Available online: https://kar.kent.ac.uk/46274/ (accessed on 27 August 2025).
- Baydevlyatova, O.N.; Ogurtsova, N.S.; Shomina, N.V.; Tereshchenko, A.V. Morphological indicators of egg quality in a new chicken subpopulation of the meat-egg type of productivity. Ptakhivnytstvo 2009, 64, 109–115. Available online: http://avianua.com/archiv/ptahivnictvo/64/15.pdf (accessed on 27 August 2025).
- Nikiforov, A.A.; Moiseeva, I.G.; Zakharov, I.A. Position of Russian chicken breeds in the diversity of Eurasian breeds. Genetika 1998, 34, 850–851. Available online: https://pubmed.ncbi.nlm.nih.gov/9719931/ (accessed on 27 August 2025).
- Moiseyeva, I.G.; Kovalenko, A.T.; Mosyakina, T.V.; Romanov, M.N.; Bondarenko, Y.V.; Kutnyuk, P.I.; Podstreshny, A.P.; Nikiforov, A.A.; Tkachik, T.E. Origin, history, genetics and economic traits of the Poltava chicken breed. Elektron. Zh. 2006, 4. Available online: https://web.archive.org/web/20120205195904/http://www.lab-cga.ru/articles/Jornal04/Statia2.htm (accessed on 5 February 2012).
- Bondarenko, Y.V.; Khvostik, V.P. Improving the productivity of meat and egg chickens of domestic selection. Vìsn. Sumsʹkogo nac. agrar. unìv. Ser. Tvarynnytstvo 2020, 2, 29–32. [Google Scholar] [CrossRef]
- Semyenova, S.K.; Moiseev, I.G.; Vasil’ev, V.A.; Filenko, A.L.; Nikiforov, A.A.; Sevast’yanova, A.A.; Ryskov, A.P. Genetic polymorphism of Russian, European, and Asian chicken breeds as revealed with DNA and protein markers. Russ. J. Genet. 2002, 38, 1109–1112. [Google Scholar] [CrossRef]
- Romanov, M.N.; Sazanov, A.A.; Moiseyeva, I.G.; Smirnov, A.F. Poultry. In Genome Mapping and Genomics in Animals, Volume 3: Genome Mapping and Genomics in Domestic Animals; Cockett, N.E., Kole, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 75–141. [Google Scholar] [CrossRef]
- Moiseyeva, I.G.; Bannikova, L.V.; Altukhov, Y.u.P. State of poultry breeding in Russia: Genetic monitoring. Mezhdunar. S.-Kh. Zh. 1993, 5–6, 66–69. Available online: https://scholar.google.com/citations?view_op=view_citation&citation_for_view=tFr9MGIAAAAJ:olpn-zPbct0C (accessed on 27 August 2025).
- Larkina, T.A.; Barkova, O.Y.; Peglivanyan, G.K.; Mitrofanova, O.V.; Dementieva, N.V.; Stanishevskaya, O.I.; Vakhrameev, A.B.; Makarova, A.V.; Shcherbakov, Y.S.; Pozovnikova, M.V.; et al. Evolutionary subdivision of domestic chickens: Implications for local breeds as assessed by phenotype and genotype in comparison to commercial and fancy breeds. Agriculture 2021, 11, 914. [Google Scholar] [CrossRef]






| # | Animal Species | Number | |||
|---|---|---|---|---|---|
| Breeds * | Types | Crosses | |||
| 1. | Altai wapiti | Cervus canadensis sibiricus | 2 (2 **) | 2 | |
| 2. | American bison | Bison bison | 1 | ||
| 3. | American mink | Neogale vison | 17 | 10 | |
| 4. | Arctic fox | Vulpes lagopus | 2 (2) | 3 | |
| 5. | Bactrian camel | Camelus bactrianus | 2 (1) | 1 | |
| 6. | Cattle | Bos taurus | 80 (25) | 55 | |
| 7. | Chicken | Gallus gallus | 79 (32) | 100 | |
| 8. | Common ostrich | Struthio camelus | 1 | ||
| 9. | Common raccoon dog | Nyctereutes procyonoides | 1 (1 **) | ||
| 10. | Domestic duck | Anas platyrhynchos | 3 (1) | 10 | |
| 11. | Domestic guinea fowl | Numida meleagris | 5 (5) | ||
| 12. | Donkey | Equus asinus | 1 (1 **) | ||
| 13. | European rabbit | Oryctolagus cuniculus | 13 (9) | 4 | |
| 14. | Ferret | Mustela furo | 3 (3) | ||
| 15. | Garden snail | Cornu aspersum | 2 | ||
| 16. | Goat | Capra hircus | 35 (12) | 3 | |
| 17. | Graylag goose | Anser anser | 28 (17) | 1 | |
| 18. | Horse | Equus caballus | 70 (33) | 7 | |
| 19. | Japanese quail | Coturnix japonica | 13 (3) | 1 | |
| 20. | Long-tailed chinchilla | Chinchilla lanigera | 1 (1) | ||
| 21. | Muscovy duck | Cairina moschata | 1 (1 **) | 3 | |
| 22. | Nutria | Myocastor coypus | 1 | 2 | |
| 23. | Pig | Sus domesticus | 27 (10) | 15 | |
| 24. | Red fox | Vulpes vulpes | 4 (3) | 8 | |
| 25. | Reindeer | Rangifer tarandus | 4 (4) | 1 | |
| 26. | Roman snail | Helix pomatia | 2 (2 **) | ||
| 27. | Sable | Martes zibellina | 2 (2) | 1 | |
| 28. | Sheep | Ovis aries | 77 (41) | 24 | |
| 29. | Sika deer | Cervus nippon | 2 (2 **) | ||
| 30. | Silkworm | Bombyx mori | 9 (9) | 12 | |
| 31. | Turkey | Meleagris gallopavo | 7 (6) | 10 | |
| 32. | Water buffalo | Bubalus arnee (bubalis) | 1 (1) | ||
| 33. | Western honeybee | Apis mellifera | 7 (6) | 7 | |
| 34. | Yak | Bos grunniens | 2 (1) | 1 | |
| 35. | Zebu | Bos indicus | 2 | ||
| Total | 507 (236) | 140 | 141 | ||
| # | Breed | Population Size | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Total, Thousand Animals | Cows, Thousand Animals | Bulls, Animals | ||||||||
| Year of Recording | 2022 | 2023 | 2024 | 2022 | 2023 | 2024 | 2022 | 2023 | 2024 | |
| 1. | Bestuzhev | 0.72 | – | – | 0.45 | – | – | – | – | – |
| 2. | Brown Caucasian | 2.02 | 0.67 | 1.46 | 1.22 | 0.34 | 1.13 | – | – | 4 |
| 3. | Dagestan Mountain | 0.64 | 0.27 | 0.18 | 0.39 | 0.22 | 0.11 | 3 | 3 | 2 |
| 4. | Istobensk | 0.73 | 0.75 | 0.74 | 0.46 | 0.46 | 0.46 | – | – | – |
| 5. | Pechora type 1 | 0.30 | 0.18 | 0.19 | 0.29 | 0.14 | 0.14 | – | – | – |
| 6. | Red Gorbatov | – | 0.28 | – | – | 0.21 | – | – | – | – |
| 7. | Red Steppe | 1.76 | 0.68 | 0.77 | 1.04 | 0.42 | 0.42 | – | – | – |
| 8. | Sychyovka | – | – | 0.32 | – | – | 0.18 | – | – | – |
| 9. | Tagil | 0.14 | 0.12 | 0.11 | 0.09 | 0.08 | 0.08 | – | – | – |
| 10. | Yakut | 0.83 | 0.70 | 0.96 | 0.35 | 0.32 | 0.39 | 24 | 24 | 28 |
| Total | 7.14 | 3.74 | 4.72 | 4.29 | 2.18 | 2.90 | 27 | 27 | 34 | |
| # | Animal Species | No. of Chromosomes (2n) | Size, in Gb | Year | References | n |
|---|---|---|---|---|---|---|
| 1. | Chicken (Gallus gallus) | 78 | 1.05 | 2004 | [83] | 183 |
| 2. | European rabbit (Oryctolagus cuniculus) | 44 | 2.67 | 2005 | [84] | – |
| 3. | Western honeybee (Apis mellifera) | 16 | 0.23 | 2006 | [85] | – |
| 4. | Cattle (Bos taurus) | 60 | 2.91 | 2009 | [86,87] | 269 |
| 5. | Horse (Equus caballus) | 64 | 2.47 | 2009 | [88] | – |
| 6. | Pig (Sus scrofa) | 38 | 2.20 | 2009 | [89] | 47 |
| 7. | Sheep (Ovis aries) | 54 | 2.71 | 2009 | [90] | 204 |
| 8. | Turkey (Meleagris gallopavo) | 80 | 1.08 | 2009 | [91] | 10 |
| 9. | Yak (Bos grunniens) | 60 | 2.8 | 2012 | [92] | 12 |
| 10. | Domestic duck (Anas platyrhynchos) | 80 | 1.21 | 2013 | [93] | – |
| 11. | Goat (Capra hircus) | 60 | 2.92 | 2013 | [94] | 83 |
| 12. | American bison (Bison bison) | 60 | 2.8 (3.0) | 2014 | [95] | – |
| 13. | Swan goose (Anser cygnoides) | 80 | 1.12 | 2015 | [96] | 60 |
| 14. | Japanese quail (Coturnix japonica) | 31 | 0.93 | 2015 | [97] | – |
| 15. | Domestic guinea fowl (Numida meleagris) | 31 | 1 | 2017 | [98] | – |
| 16. | Reindeer (Rangifer tarandus) | 70 | 2.64 | 2017 | [99] | 96 |
| 17. | Silkworm (Bombyx mori) | 28 | 0.460 | 2018 | [100] | – |
| 18. | Roman snail (Helix pomatia) | 54 | – | 2019 | [101] | – |
| 19. | Donkey (Equus asinus) | 31 | 2.79 | 2020 | [102] | – |
| 20. | Arctic fox (Vulpes lagopus) | 25 | 2.35 | 2021 | [103] | – |
| 21. | Muscovy duck (Cairina moschata) | 39 | 1.1 | 2021 | [104] | – |
| 22. | Water buffalo (Bubalus arnee (bubalis)) | 50 | 2.6 | 2022 | [105] | – |
| 23. | American mink (Neogale vison) | 15 | 2.68 | 2022 | [106] | – |
| 24. | Zebu (Bos indicus) | 60 | 2.7 | 2023 | [107] | – |
| 25. | Sika deer (Cervus nippon) | 34 | 2.78 | 2024 | [108] | – |
| 26. | Common ostrich (Struthio camelus) | 41 | 1.5 | 2024 | [109] | – |
| 27. | Garden snail (Cornu aspersum) | 27 | 2.91 | 2024 | [110] | – |
| 28. | Red fox (Vulpes vulpes) | 17 | 2.4 | 2025 | [111] | – |
| 29. | Sable (Martes zibellina) | 19 | 2.39 | 2025 | [112] | – |
| 30. | Bactrian camel (Camelus bactrianus) | 37 | 2.5 | 2025 | [113] | – |
| Animal Species | Manufacturer | DNA Chip Name | No. of SNPs | n 1 |
|---|---|---|---|---|
| Cattle | Illumina | Bovine HD BeadChip | >777,000 | 329 |
| Affymetrix (Santa Clara, CA, USA) | Axiom® Genome-Wide BOS1 | >640,000 | – | |
| Illumina | Bovine GGP HD BeadChip | ~150,000 | 1078 | |
| Affymetrix | Axiom Bovine 100K Array | ~100,000 | – | |
| Affymetrix | Axiom Bovine Genotyping v3 | 63,988 | – | |
| Illumina | Bovine SNP50 v3 BeadChip | 53,714 | 1385 | |
| Illumina | Bovine LD v2 BeadChip | 7931 | – | |
| Pig | Affymetrix | Axiom® Porcine Array | 658,692 | – |
| Illumina | Porcine GGP BeadChip | ~80,000 | 1313 | |
| Illumina | Porcine SNP50 v2 BeadChip [120] | 64,232 | 4400 | |
| Affymetrix | Axiom Porcine Breeders Genotyping Array | 55,232 | – | |
| Sheep | Illumina | Ovine Infinium® HD SNP BeadChip [121] | 603,350 | 864 |
| Illumina | Ovine SNP50 BeadChip | 54,241 | 1530 | |
| Affymetrix | Axiom Ovine Genotyping Array (51K) | ~51,000 | – | |
| Goat | Affymetrix | Axiom Goat Genotyping v2 | 59,795 | – |
| Affymetrix | Axiom Goat Genotyping v1 | 58,655 | – | |
| Illumina | Goat SNP50 BeadChip [122] | 52,295 | 668 | |
| Horse | Affymetrix | Axiom® Equine Array | 670,796 | – |
| Illumina | GGP Equine BeadChip | ~70,000 | – | |
| Illumina | Equine 50K BeadChip | ~50,000 | – | |
| Buffalo | Affymetrix | Axiom® Buffalo Array | 90,000 | – |
| Reindeer | Affymetrix | OVSNP600 | 702,183 | – |
| Affymetrix | OVSNP60 | 72,723 | – | |
| Camelids | Affymetrix | Axiom Camelids Genotyping | 59,938 2 | – |
| Chicken | Affymetrix | Axiom® Genome-Wide Chicken Array | >580,000 | – |
| Illumina | Chicken SNP BeadChip [123] | 57,636 | 528 | |
| Turkey | Affymetrix | Axiom Turkey Genotyping | 643,845 | – |
| Animal Species | ISAG 1 | Custom Panels 2 | No. of Loci 3 | n 4 | |
|---|---|---|---|---|---|
| Cattle | Bos taurus | + | 12 | 117,230 | |
| Pig | Sus domesticus | + | 10 | 96,675 | |
| Sheep | Ovis aries | + | 12 | 11,068 | |
| Reindeer | Rangifer tarandus | + | 9 | 4929 | |
| Goat | Capra hircus | + | 14 | 2130 | |
| Western honeybee | Apis mellifera | + | 7 | 1921 | |
| Chicken | Gallus gallus | + | 25 | 1527 | |
| Yak | Bos grunniens | + | 11 | 850 | |
| Turkey | Meleagris gallopavo | + | 6 | 436 | |
| Goose | Anser anser/A. cygnoides | + | 11 | 149 | |
| Altai wapiti | Cervus canadensis sibiricus | + | 14 | 126 | |
| American bison | Bison bison | + | 15 | 94 | |
| Bactrian camel | Camelus bactrianus | + | 17 | 46 | |
| Breed | Description | References |
|---|---|---|
| Chinese Gray (CN_GR) (Figure 10c) | Originated in Manchuria, spread across China and reached Europe and Russia in the 1700s. Russian breeders crossed it with local geese to create the Kuban, Pereyaslav, Kholmogory and Linda breeds. Distinguished from standard Chinese geese solely by gray plumage, it was included in the State Register of Breeding Achievements (SRBA) [172] in 1993. | [173,174] |
| Tula Fighting (TULA_FH) (Figure 10b) | One of Russia’s oldest native breeds, developed through traditional folk selection specifically for goose fighting—a popular pastime in Russia, particularly in Tula Governorate from the 17th–18th centuries onward. Characterized by a sturdy build, remarkable endurance and an aggressive fighting spirit. Included in the SRBA [172] in 1993. | [174] |
| Shadrinsk (SHAD) (Figure 10f) | An ancient indigenous breed developed between the 17th–19th centuries in Trans-Urals (Shadrinsk, Kurgan Oblast) through traditional folk selection of domesticated wild graylag geese. Remarkable for its hardiness, adaptation to harsh climates and excellent meat qualities. Included in the SRBA [172] in 1993. | [173,174] |
| Large Gray (LR_GR) (Figure 10e) | A relatively young breed developed in the second half of the 20th century. Selective breeding began in Ukraine in the 1930s. In 1941, the breed development continued in Tambov region through crossing Romny and Toulouse geese. Classified as a dual-purpose meat-and-fat type breed. Included in the SRBA [172] in 1993. | [173,174] |
| Vladimir Clay (VLAD_CL) (Figure 10g) | Developed in Vladimir Oblast in the 1960s–1980s by crossing Kholmogory (for size and hardiness), Tula Fighting (for robust physique) and Chinese (for enhanced egg production) geese. Included in the SRBA [172] in 1993. | [173,174] |
| Kholmogory (KHLM) (Figure 10a) | One of the Russia’s oldest and most popular breeds. Developed in the 19th century through crossbreeding of Chinese and Arzamas geese near the town of Kholmogory (Arkhangelsk Governorate). The breed became widely valued across Russia due to targeted selection for cold tolerance and high meat yield. Primarily distributed in Oryol, Bryansk, Voronezh, Kursk and Belgorod Oblasts. Included in the SRBA [172] in 1993 (without color specification). | [173,174] |
| Kholmogory Gray (KHLM_GR) (Figure 10d) | A variety of the Kholmogory goose that is differentiated solely by its distinctive plumage that mimics wild goose coloration (gray-brown feathers on the back and wings with white ventral sections). All other characteristics remain identical to the standard Kholmogory breed. | [173,174] |
| Breed 1 | n 2 | AR 3 (M 7 ± SE 8) | HO 4 (M ± SE) | UHE 5 (M ± SE) | UFIS 6 (M ± SE) |
|---|---|---|---|---|---|
| CN_GR | 21 | 2.913 ± 0.352 | 0.481 ± 0.079 | 0.492 ± 0.079 | 0.007 [−0.116; 0.130] |
| TULA_FH | 8 | 2.727 ± 0.273 | 0.352 ± 0.067 | 0.403 ± 0.068 | 0.138 [−0.085; 0.361] |
| SHAD | 11 | 2.123 ± 0.209 | 0.306 ± 0.064 | 0.322 ± 0.055 | 0.039 [−0.165; 0.243] |
| LR_GR | 14 | 2.337 ± 0.214 | 0.377 ± 0.059 | 0.371 ± 0.058 | −0.032 [−0.15; 0.086] |
| VLAD_CL | 18 | 2.064 ± 0.215 | 0.278 ± 0.069 | 0.299 ± 0.066 | 0.086 [−0.084; 0.256] |
| KHLM | 14 | 3.236 ± 0.341 | 0.481 ± 0.093 | 0.492 ± 0.078 | −0.003 [−0.218; 0.212] |
| KHLM_GR | 14 | 3.215 ± 0.269 | 0.506 ± 0.08 | 0.504 ± 0.063 | −0.004 [−0.181; 0.173] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zinovieva, N.A.; Deniskova, T.E.; Kharzinova, V.R.; Bagirov, V.A.; Romanov, M.N.; Volkova, V.V.; Grishina, D.S.; Abdelmanova, A.S.; Gusev, I.V.; Shchukin, I.M.; et al. Conservation of Native Livestock Breeds in Russia: Current State and Promising Prospects. Animals 2025, 15, 3103. https://doi.org/10.3390/ani15213103
Zinovieva NA, Deniskova TE, Kharzinova VR, Bagirov VA, Romanov MN, Volkova VV, Grishina DS, Abdelmanova AS, Gusev IV, Shchukin IM, et al. Conservation of Native Livestock Breeds in Russia: Current State and Promising Prospects. Animals. 2025; 15(21):3103. https://doi.org/10.3390/ani15213103
Chicago/Turabian StyleZinovieva, Natalia A., Tatiana E. Deniskova, Veronika R. Kharzinova, Vugar A. Bagirov, Michael N. Romanov, Valeriya V. Volkova, Dinara S. Grishina, Alexandra S. Abdelmanova, Igor V. Gusev, Ivan M. Shchukin, and et al. 2025. "Conservation of Native Livestock Breeds in Russia: Current State and Promising Prospects" Animals 15, no. 21: 3103. https://doi.org/10.3390/ani15213103
APA StyleZinovieva, N. A., Deniskova, T. E., Kharzinova, V. R., Bagirov, V. A., Romanov, M. N., Volkova, V. V., Grishina, D. S., Abdelmanova, A. S., Gusev, I. V., Shchukin, I. M., Trukhachev, V. I., & Boronetskaya, O. I. (2025). Conservation of Native Livestock Breeds in Russia: Current State and Promising Prospects. Animals, 15(21), 3103. https://doi.org/10.3390/ani15213103








