Novel Silent Mutations in the HIRA Gene Associated with Litter Size in Sonid Sheep
Simple Summary
Abstract
1. Introduction
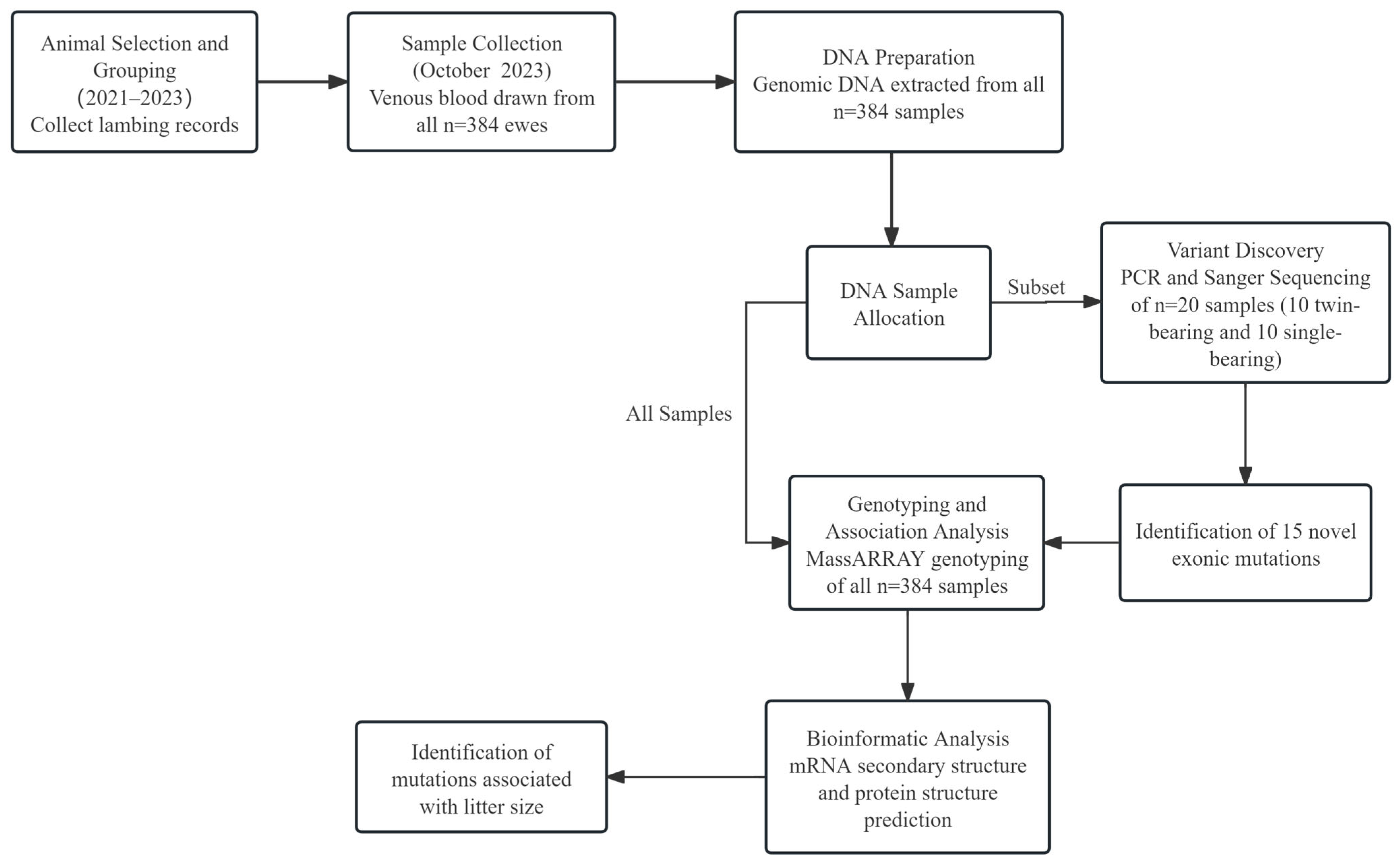
2. Materials and Methods
2.1. Samples and Date
2.2. DNA Extraction and Sequencing
2.3. SNP Genotyping Using iPLEX MassARRAY
2.4. Bioinformatic Analyses
2.5. Statistical Analyses
3. Results
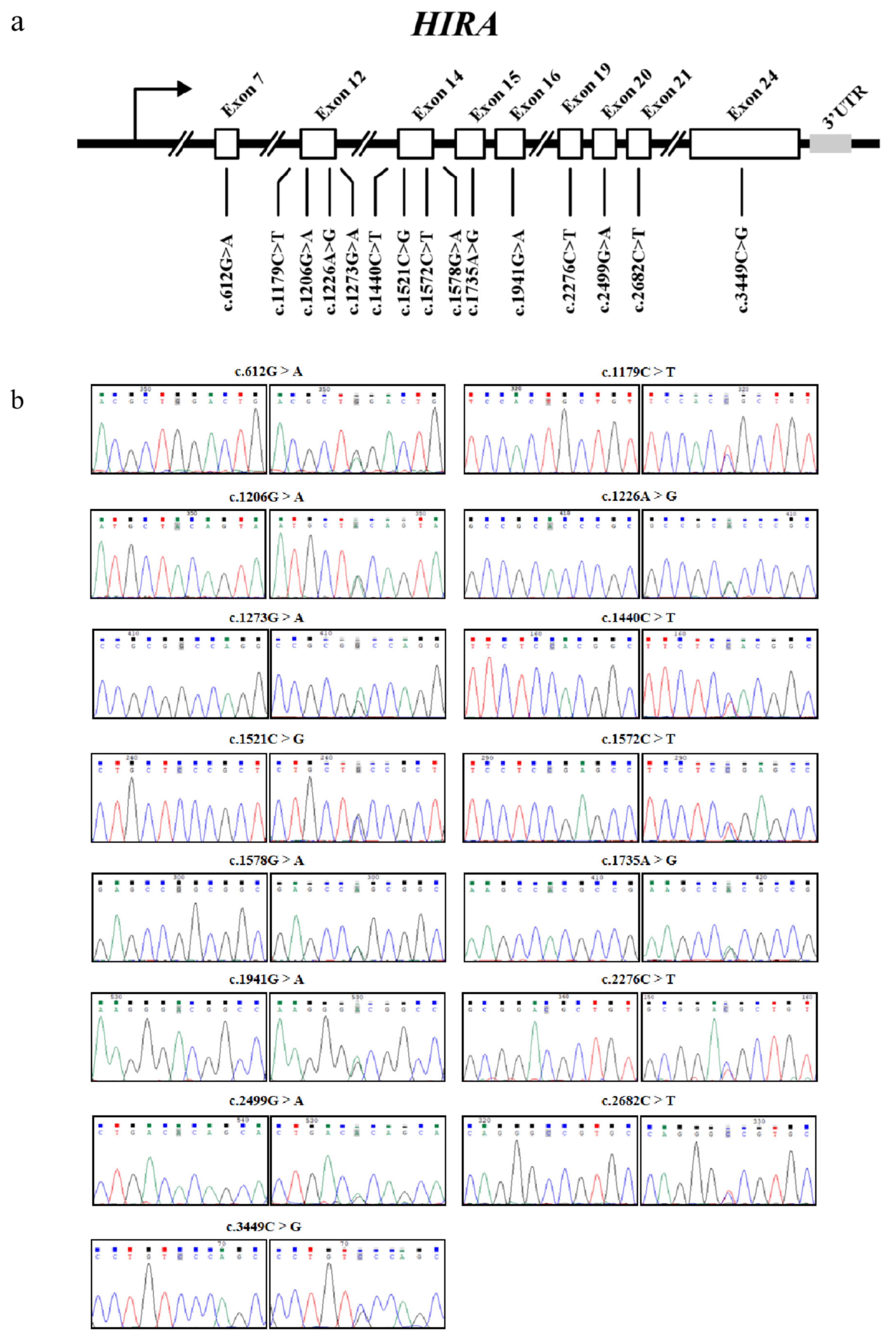
3.1. Variant Discovery in HIRA
3.2. Genetic Diversity Analysis
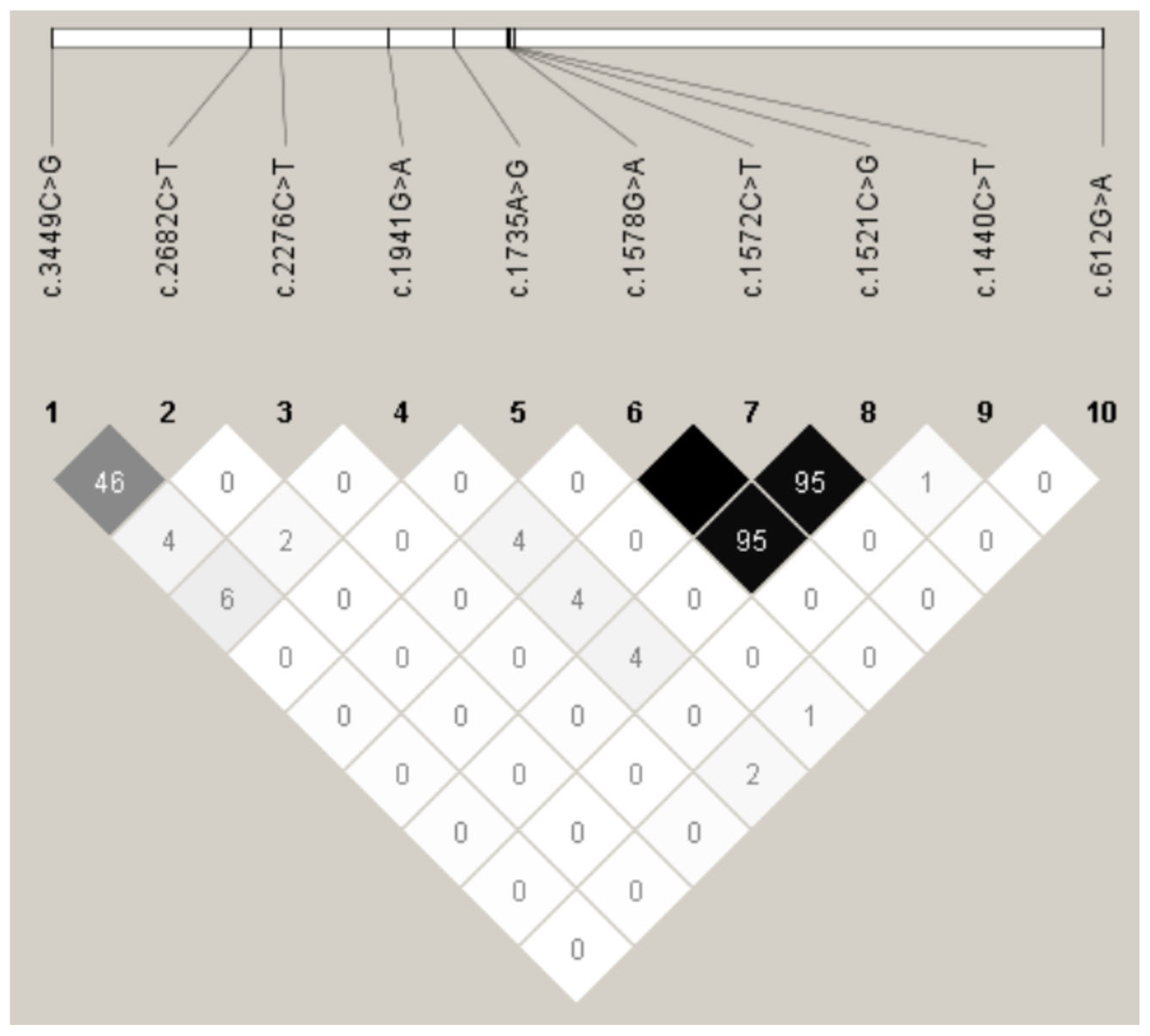
3.3. Linkage Disequilibrium Analysis of Novel Variants in HIRA
3.4. Associations Between Genetic Variants and Litter Size
3.5. Bioinformatic Analysis of Ovine HIRA
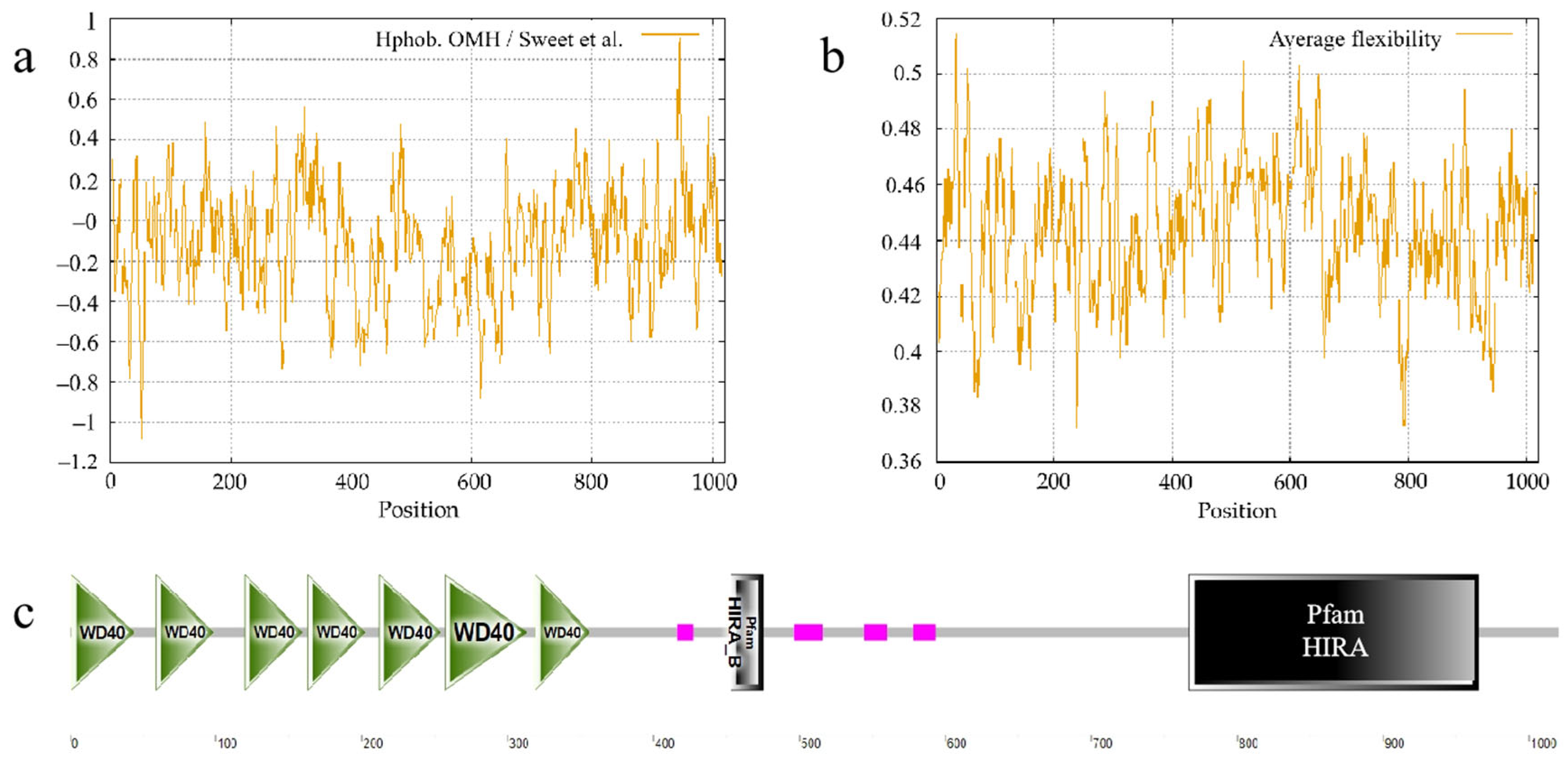
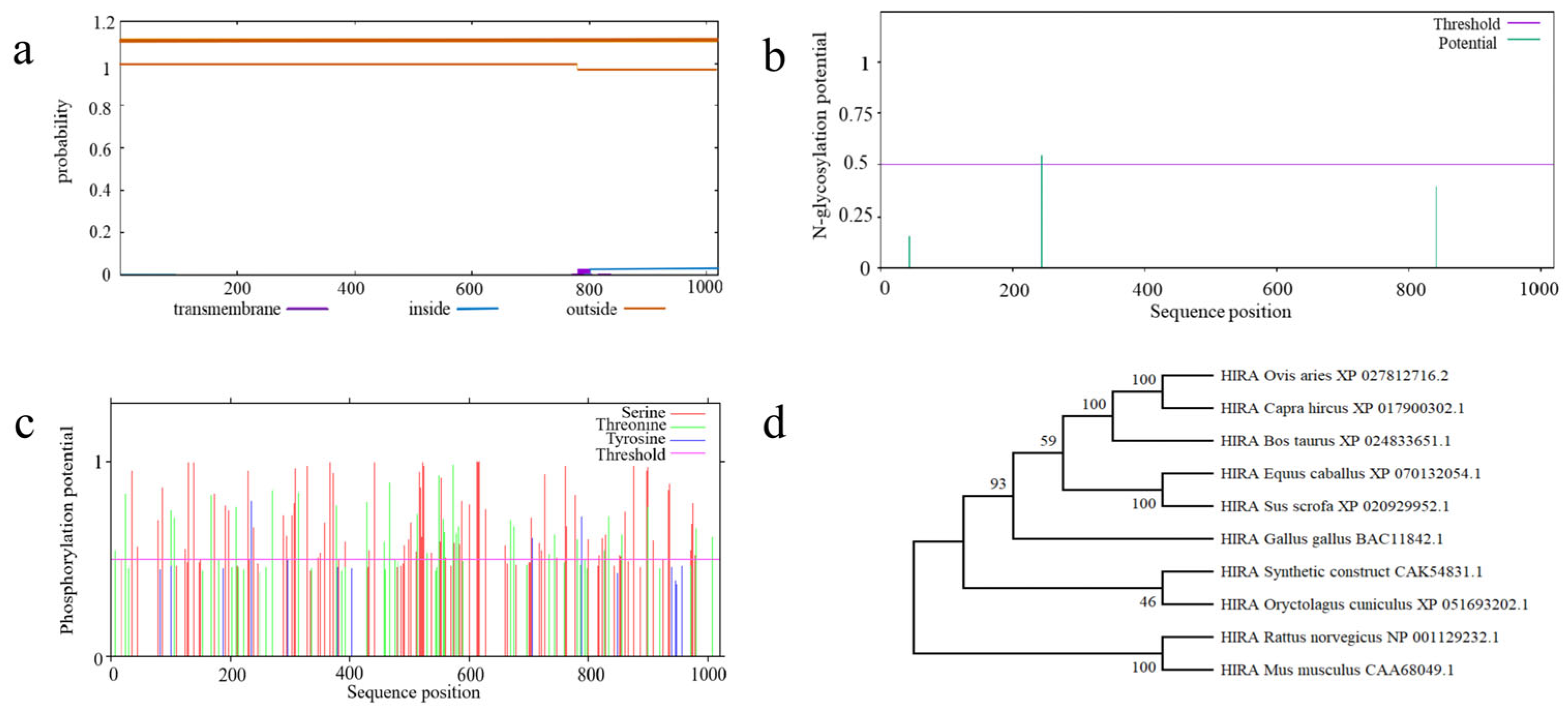
3.5.1. Structural Characterization of Ovine HIRA Protein
3.5.2. Physicochemical Properties of Ovine HIRA
3.5.3. Phylogenetic Relationships
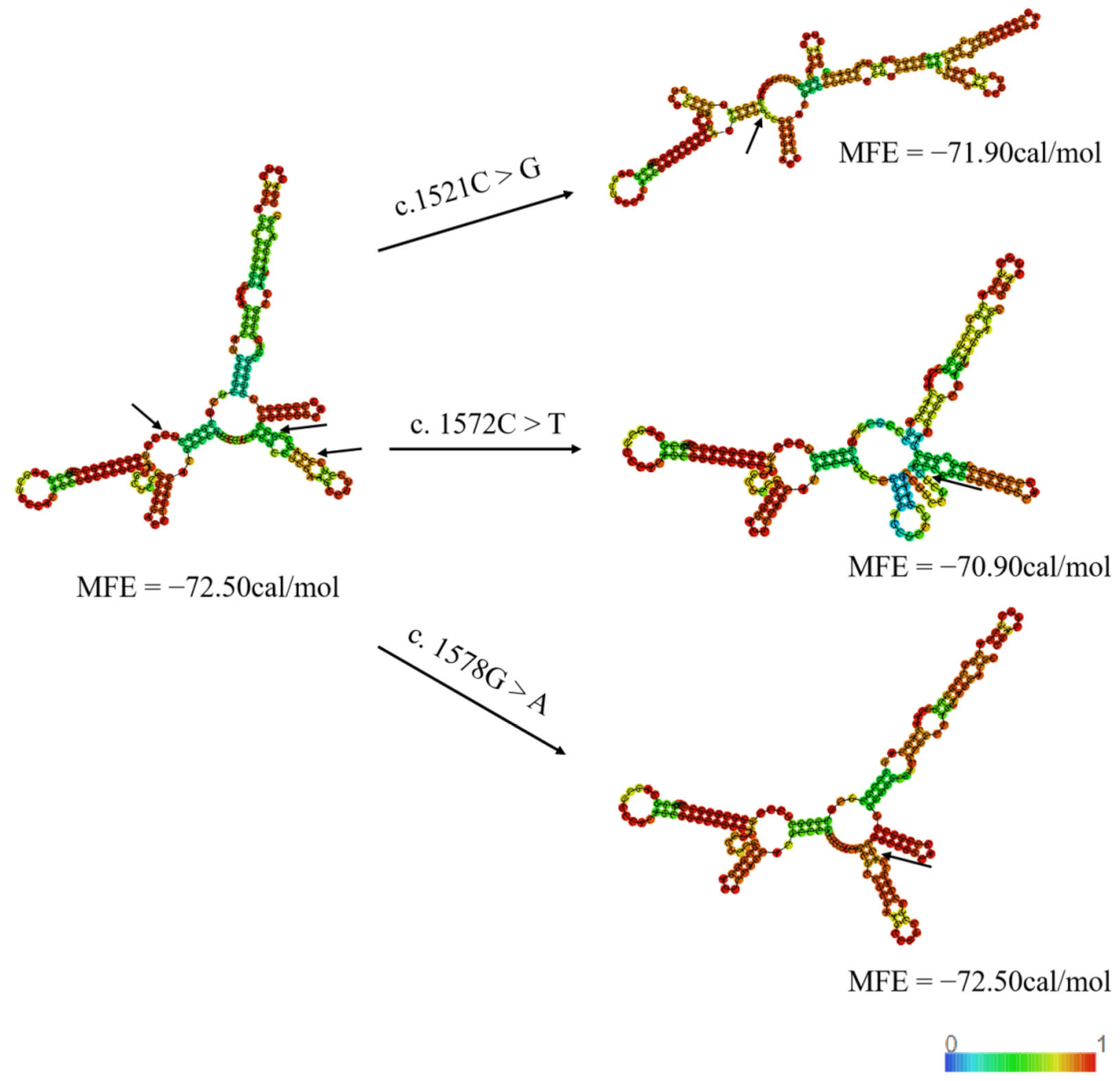
3.5.4. RNA Secondary Structure Modulation via Variants
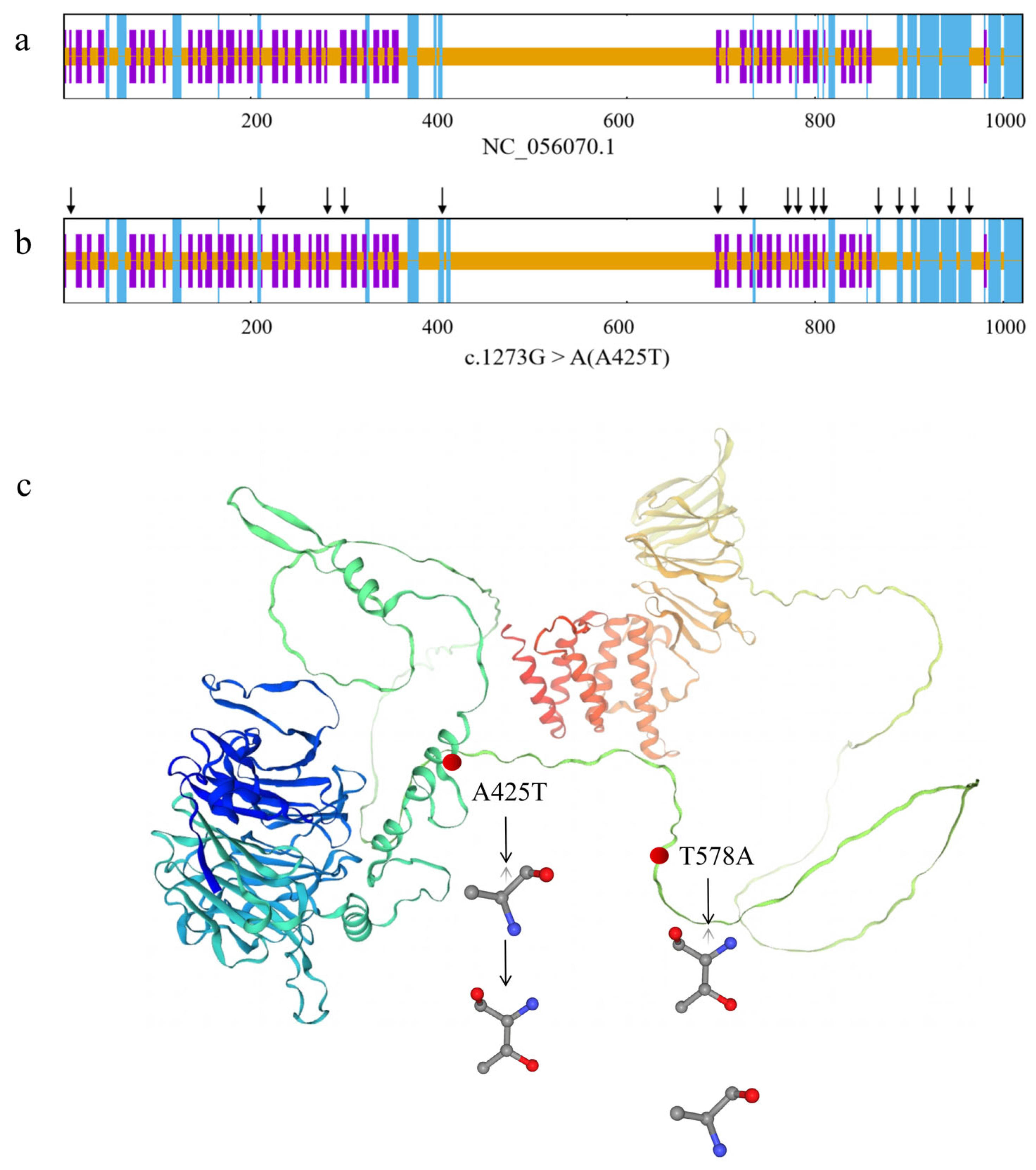
3.5.5. Structural Consequences of Missense Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- China National Commission of Animal Genetic Resources (CNCAGR). Sheep and Goats, Animal Genetic Resources in China; China Agriculture Press: Beijing, China, 2011.
- Xiang, J.; Li, H.; Guo, Z.; Li, T.; Yamada, T.; Li, X.; Bao, S.; Da, L.; Borjigin, G.; Cang, M.; et al. Effect of FABP4 Gene Polymorphisms on Fatty Acid Composition, Chemical Composition, and Carcass Traits in Sonid Sheep. Animals 2025, 15, 226. [Google Scholar] [CrossRef]
- Paz, E.; Quiñones, J.; Bravo, S.; Montaldo, H.H.; Sepúlveda, N. Genotyping of BMPR1B, BMP15 and GDF9 genes in Chilean sheep breeds and association with prolificacy. Anim. Genet. 2015, 46, 98–99. [Google Scholar] [CrossRef]
- Ahlawat, S.; Sharma, R.; Roy, M.; Mandakmale, S.; Prakash, V.; Tantia, M.S. Genotyping of Novel SNPs in BMPR1B, BMP15, and GDF9 Genes for Association with Prolificacy in Seven Indian Goat Breeds. Anim. Biotechnol. 2016, 27, 199–207. [Google Scholar] [CrossRef]
- Liu, Q.Y.; Pan, Z.Y.; Wang, X.Y.; Hu, W.P.; Di, R.; Yao, Y.X. Progress on major genes for high fecundity in ewes. Front. Agric. Sci. Eng. 2014, 1, 282. [Google Scholar] [CrossRef]
- Rai, T.S.; Puri, A.; McBryan, T.; Hoffman, J.; Tang, Y.; Pchelintsev, N.A.; van Tuyn, J.; Marmorstein, R.; Schultz, D.C.; Adams, P.D. Human CABIN1 is a functional member of the human HIRA/UBN1/ASF1a histone H3.3 chaperone complex. Mol. Cell Biol. 2011, 19, 4107–4118. [Google Scholar] [CrossRef]
- Yang, J.H.; Song, T.Y.; Jo, C.; Park, J.; Lee, H.Y.; Song, I. Differential regulation of the histone chaperone HIRA during muscle cell differentiation by a phosphorylation switch. Exp. Mol. Med. 2016, 48, e252. [Google Scholar] [CrossRef]
- Dilg, D.; Saleh, R.N.; Phelps, S.E.; Rose, Y.; Dupays, L.; Murphy, C. HIRA Is Required for Heart Development and Directly Regulates Tnni2 and Tnnt3. PLoS ONE 2016, 11, e0161096. [Google Scholar] [CrossRef]
- Ray-Gallet, D.; Quivy, J.P.; Scamps, C.; Martini, E.M.; Lipinski, M.; Almouzni, G. HIRA is critical for a nucleosome assembly pathway independent of DNA synthesis. Mol. Cell 2002, 9, 1091–1100. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, G.W.; Dieker, J.W.; Derijck, A.A.; Muller, S.; Berden, J.H.; Braat, D.D. Asymmetry in histone H3 variants and lysine methylation between paternal and maternal chromatin of the early mouse zygote. Mech. Dev. 2005, 122, 1008–1022. [Google Scholar] [CrossRef]
- Horard, B.; Sapey-Triomphe, L.; Bonnefoy, E.; Loppin, B. ASF1 is required to load histones on the HIRA complex in preparation of paternal chromatin assembly at fertilization. Epigenetics Chromatin 2018, 11, 11–19. [Google Scholar] [CrossRef]
- Roberts, C.; Sutherland, H.F.; Farmer, H.; Kimber, W.; Halford, S.; Carey, A. Targeted mutagenesis of the Hira gene results in gastrulation defects and patterning abnormalities of mesoendodermal derivatives prior to early embryonic lethality. Mol. Cell Biol. 2002, 22, 2318–2328. [Google Scholar] [CrossRef]
- Nashun, B.; Hill, P.W.; Smallwood, S.A.; Dharmalingam, G.; Amouroux, R.; Clark, S.J. Continuous Histone Replacement by Hira Is Essential for Normal Transcriptional Regulation and De Novo DNA Methylation during Mouse Oogenesis. Mol. Cell 2015, 60, 611–625. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Pan, Z.; Cao, X.; Guo, X.; He, X.; Sun, Q. Single Nucleotide Polymorphisms in the HIRA Gene Affect Litter Size in Small Tail Han Sheep. Animals 2018, 8, 71. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Sharma, V.; Bhat, A.; Singh, H.; Sharma, I.; Verma, S.; Bhat, G.R.; Sharma, B.; Bakshi, D.; Kumar, R.; et al. MassARRAY analysis of twelve cancer related SNPs in esophageal squamous cell carcinoma in J&K, India. BMC Cancer 2020, 20, 497. [Google Scholar]
- Masatoshi, N.; Roychoudhury, A.K. Sampling variances of heterozygosity and genetic distance. Genetics 1974, 76, 379–390. [Google Scholar] [CrossRef]
- Barrett, J.C.; Fry, B.; Maller, J.; Daly, M.J. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 2005, 21, 263–265. [Google Scholar] [CrossRef]
- Tong, B.; Wang, J.P.; Cheng, Z.X.; Liu, J.S.; Wu, Y.R.; Li, Y.H. Novel variants in GDF9 gene affect promoter activity and litter size in Mongolia sheep. Genes 2020, 11, 375. [Google Scholar] [CrossRef]
- Sweet, R.M.; Eisenberg, D. Correlation of sequence hydrophobicities measures similarity in three-dimensional protein structure. J. Mol. Biol. 1983, 171, 479–488. [Google Scholar] [CrossRef]
- Mathews, D.H.; Disney, M.D.; Childs, J.L.; Schroeder, S.J.; Michael, Z.; Turner, D.H. Incorporating chemical modification con-straints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. USA 2004, 101, 7287–7292. [Google Scholar] [CrossRef]
- Pchelintsev, N.A.; McBryan, T.; Rai, T.S.; van Tuyn, J.; Ray-Gallet, D.; Almouzni, G. Placing the HIRA histone chaperone com-plex in the chromatin landscape. Cell Rep. 2013, 3, 1012–1019. [Google Scholar] [CrossRef]
- Wang, Y.F.; Xu, Y.J. Effects of Hira gene overexpression on early embryonic development in Drosophila melanogaster. J. Cent. China Norm. Univ. (Nat. Sci. Ed.) 2005, 44, 530–533. [Google Scholar]
- Lin, C.J.; Koh, F.M.; Wong, P.; Conti, M.; Ramalho-Santos, M. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev. Cell 2014, 30, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Franco, P.; Michela, R.; Baralle, F.E. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc. Natl. Acad. Sci. USA 2005, 102, 6368–6372. [Google Scholar]
- Duan, J.; Wainwright, M.S.; Comeron, J.M.; Saitou, N.; Sanders, A.R.; Gelernter, J. Synonymous mutations in the human do-pamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum. Mol. Genet. 2003, 12, 205–216. [Google Scholar] [CrossRef]
- Chava, K.S.; Jung, M.O.; Kim, I.W.; Sauna, Z.E.; Anna, M.C.; Ambudkar, S.V. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007, 315, 525–528. [Google Scholar]
- Mohan, G.A. A-Adducin nsSNPs affect mRNA secondary structure, protein modification and stability. Meta Gene 2018, 17, 153–162. [Google Scholar]
- Rachel, S.; Cygan, K.J.; Rhine, C.L.; Glidden, D.T.; Taggart, A.J.; Lin, C.L. The effects of structure on pre-mRNA processing and stability. Methods 2017, 125, 36–44. [Google Scholar]
- Sauna, Z.E.; Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011, 12, 683–691. [Google Scholar] [CrossRef]
- Imran, F.S.; Al-Thuwaini, T.M. The novel C268A variant of BMP2 is linked to the reproductive performance of Awassi and Hamdani sheep. Mol. Biol. Rep. 2024, 51, 267. [Google Scholar] [CrossRef]
- Rose, J.; Kraft, T.; Brenner, B.; Montag, J. Hypertrophic cardiomyopathy MYH7 mutation R723G alters mRNA secondary structure. Physiol. Genom. 2020, 52, 15–19. [Google Scholar] [CrossRef]
- Kong, L.; Gong, Y.; Wang, Y.; Yuan, M.; Liu, W.; Zhou, H. Multi-omics revealed that DCP1A and SPDL1 determine embryo-genesis defects in postovulatory ageing oocytes. Cell Prolif. 2025, 58, e13766. [Google Scholar] [CrossRef] [PubMed]
- Sha, Q.Q.; Zheng, W.; Wu, Y.W.; Li, S.; Guo, L.; Zhang, S. Dynamics and clinical relevance of maternal mRNA clearance during the oocyte-to-embryo transition in humans. Nat. Commun. 2020, 11, 4917. [Google Scholar] [CrossRef]
- Peng, F.; Nordgren, C.E.; Murray, J.I. A spatiotemporally resolved atlas of mRNA decay in the C. elegans embryo reveals differential regulation of mRNA stability across stages and cell types. Genome Res. 2024, 34, 1235–1252. [Google Scholar] [CrossRef]
- Niu, X.; Huang, Y.; Lu, H.; Li, S.; Huang, S.; Ran, X. CircRNAs in Xiang pig ovaries among diestrus and estrus stages. Porc. Health Manag. 2022, 8, 29. [Google Scholar] [CrossRef]
- Shim, J.S.; Park, S.H.; Lee, D.K.; Kim, Y.S.; Park, S.C.; Redillas, M.C.F.R. The Rice GLYCINE-RICH PROTEIN 3 Confers Drought Tolerance by Regulating mRNA Stability of ROS Scavenging-Related Genes. Rice 2021, 14, 31. [Google Scholar] [CrossRef]
- Geisberg, J.V.; Moqtaderi, Z.; Struhl, K. Condition-specific 3′ mRNA isoform half-lives and stability elements in yeast. Proc. Natl. Acad. Sci. USA 2023, 120, e2301117120. [Google Scholar] [CrossRef]
- Froebel, B.R.; Trujillo, A.J.; Sullivan, J.M. Effects of Pathogenic Variations in the Human Rhodopsin Gene (hRHO) on the Predicted Accessibility for a Lead Candidate Ribozyme. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3576–3591. [Google Scholar]
- Zeng, X.; Wei, Z.; Du, Q.; Li, J.; Xie, Z.; Wang, X. Unveil cis-acting combinatorial mRNA motifs by interpreting deep neural network. Bioinformatics 2024, 40, i381–i389. [Google Scholar] [CrossRef] [PubMed]
- Beltran, A.; Jiang, X.; Shen, Y.; Lehner, B. Site-saturation mutagenesis of 500 human protein domains. Nature 2025, 637, 885–894. [Google Scholar] [CrossRef]
- Zhu, J.; Chen, K.; Sun, Y.H.; Ye, W.; Liu, J.; Zhang, D. LSM1-mediated Major Satellite RNA decay is required for nonequilibrium histone H3.3 incorporation into parental pronuclei. Nat. Commun. 2023, 14, 957. [Google Scholar] [CrossRef]
- Samie, K.A.; Kowalewski, M.P.; Schuler, G.; Gastal, G.D.A.; Bollwein, H.; Scarlet, D. Roles of GDF9 and BMP15 in equine fol-licular development: In vivo content and in vitro effects of IGF1 and cortisol on granulosa cells. BMC Vet. Res. 2025, 21, 292. [Google Scholar] [CrossRef]
- Abedal-Majed, M.A.; Abuajamieh, M.; Al-Qaisi, M.; Sargent, K.M.; Titi, H.H.; Alnimer, M.A. Sheep with ovarian androgen excess have fibrosis and follicular arrest with increased mRNA abundance for steroidogenic enzymes and gonadotropin receptors. J. Anim. Sci. 2023, 101, skad082. [Google Scholar] [CrossRef]
- Morozov, V.M.; Riva, A.; Sarwar, S.; Kim, W.J.; Li, J.; Zhou, L. HIRA-mediated loading of histone variant H3.3 controls andro-gen-induced transcription by regulation of AR/BRD4 complex assembly at enhancers. Nucleic Acids Res. 2023, 51, 10194–10217. [Google Scholar] [CrossRef]
- Ma, S.J.; Ji, X.W.; Cang, M.; Wang, J.G.; Yu, H.Q.; Liu, Y.B. Association analysis between novel variants in LEPR gene and litter size in Mongolia and Ujimqin sheep breeds. Theriogenology 2022, 183, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.Y.; Hao, Q.; Cang, M.; Wang, J.G.; Yu, H.Q.; Liu, Y.B. Association between novel variants in BMPR1B gene and litter size in Mongolia and Ujimqin sheep breeds. Reprod. Domest. Anim. 2021, 56, 1562–1571. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chi, Z.; Jia, S.; Zhao, S.; Cao, G.; Purev, C. Effects of novel variants in BMP15 gene on litter size in Mongolia and Ujimqin sheep breeds. Theriogenology 2023, 198, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mura, M.C.; Cosso, G.; Ouadday, M.; Hosri, C.; Starič, J.; Nehme, M. Identifying Key Genetic Factors Influencing Reproductive Performance in Dairy and Meat Sheep Breeds of the Mediterranean Region. Reprod. Domest. Anim. 2025, 60, e70. [Google Scholar] [CrossRef]

| Primer | Primer Sequence (5′–3′) | Target Region | Annealing Temperature Tm (°C) | Product Size (bp) |
|---|---|---|---|---|
| HIRA-1 | F: CGTGACTTCCGGCGTTGC R: CAGGACCTGCCCCTTCCAG | Exon 1 | 54 | 613 (178 bp 5′flanking region + 128 bp exon 1 + 307 bp intron 1) |
| HIRA-2 | F: TCGCTTGCGGCGTTTGTT R: CGCTCGTCCTTCTCGTCATC | Exon 2 | 56 | 1071 (120 bp intron 1 + 68 bp exon 2 + 810 bp intron 2 + 73 bp exon 3) |
| HIRA-3 | F: GAGTGGGCTCCTGCTTTAGG R: ATCAGGCGGGAATGGCTC | Exon 3 | 62 | 1026 (670 bp intron 2 + 111 bp exon 3 + 245 bp intron 3) |
| HIRA-4 | F: CCCGCAGTAGCGCCATAGT R: GGTGCCAGGGTGGGATAAA | Exon 4 | 62 | 782 (618 bp intron 3 + 91 bp exon 4 + 73 bp intron 4) |
| HIRA-5 | F: TTGAGAAACAGAAGCAAATCACAGTCCCACC R: TCCAGACCTGAAAGAGCCCACTGAAGACA | Exon 5 | 54 | 740 (311 bp intron 4 + 95 bp exon 5 + 334 bp intron 5) |
| HIRA-6 | F: CTTCAGGGTGGTTTGGACAGT R: CGACAGGGCAGGAACAGGA | Exon 6 | 62 | 645 (166 bp intron 5 + 96 bp exon 6 + 383 bp intron 6) |
| HIRA-7 | F: CATTGAGGACGAGCAGGGAC R: CCAACGAGGCAAGGGAGA | Exon 7 | 54 | 751 (260 bp intron 6 + 161 bp exon 7 + 330 bp intron 7) |
| HIRA-8 | F: ATAGCAGCATCACCTCACCCT R: AAAGAAGAACCGGACGACAAC | Exon 8 | 60 | 790 (467 bp intron 7 + 168 bp exon 8 +155 bp intron 8) |
| HIRA-9 | F: GCAAAGCGTCACCATCTGTT R: ACCTCCAGTCCTGCAACTCC | Exon 9 | 58 | 841 (226 bp intron 8 + 114 bp exon 9 + 501 bp intron 9) |
| HIRA-10/11 | F: GTGGTGGGCTGAGCGGACTT R: CAGGCTAACCAAGGAGGGAC | Exon 10/11 | 62 | 863 (124 bp intron 9 + 71 bp exon 10 + 207 bp intron 10 + 106 bp exon 11 + 355 bp intron 11) |
| HIRA-12 | F: CCGAGGGTCTCAATTTCAGTT R: TGCAGCCACAAGCACAGG | Exon 12 | 62 | 883 (281 bp intron 11 + 231 bp exon 12 + 371 bp intron 12) |
| HIRA-13 | F: TCGGTTATGAGCCTTTGGTG R: CGCTGAGAAAGAGGGTGCTAC | Exon 13 | 62 | 710 (341 bp intron 12 + 86 bp exon 13 + 283 bp intron 13) |
| HIRA-14 | F: TCGGGTCTGACCGAATGTAG R: TGTCGCAGAGGCTGTGGTA | Exon 14 | 62 | 896 (180 bp intron 13 + 192 bp exon 14 + 524 bp intron 14) |
| HIRA-15 | F: ATGCGCTTCCGGTTAGGC R: GGAGGCGAGGATACGCTGTG | Exon 15 | 54 | 634 (321 bp intron 14 + 162 bp exon 15 + 151 bp intron 15) |
| HIRA-16/17 | F: CGTGCTGTTTCCAAAAGGTGTCATCG R: TGCGTGGGAGCAGCCAGAGTCGG | Exon 16/17 | 62 | 1008 (403 bp intron 15 + 205 bp exon 16 + 237 bp intron 16 + 105 bp exon 17 + 58 bp intron 17) |
| HIRA-18 | F: CTGACGCCAACCCGCACCAT R: AGAAAGCCTTGCTCACTTCCCT | Exon 18 | 60 | 830 (440 bp intron 17 + 149 bp exon 18 + 241 bp intron 18) |
| HIRA-19/20 | F: ACCACGGGTGCTCACTG R: CCTGGACTATGGCTAAGGGT | Exon 19/20 | 60 | 948 (143 bp intron 18 + 221 bp exon 19 +154 bp intron 19 + 106 bp exon 20 + 324 bp intron 20) |
| HIRA-21/22 | F: CTACTGCTTCAGCCCGTCATTG R: CCCAGCCTTTCCGCTCACA | Exon 21/22 | 60 | 796 (244 bp intron 20 + 123 bp exon 21 + 178 bp intron 21 + 164 bp exon 22 + 87 bp intron 22) |
| HIRA-23 | F: TGCGTTTGTGGTAACTTGGC R: AGGAAGGCGGAAGGCTCA | Exon 23 | 62 | 719 (78 bp intron 22 + 89 bp exon 23 + 552 bp intron 23) |
| HIRA-24-1 | F: CCTTCCTTTATAGGTACGATGTGCCTAATCTTTGG R: TCCTTGGGCTGGGACAGGCTTTCCTTTTA | Exon 24 | 54 | 1434 (919 bp intron 23 + 515 bp exon 24) |
| HIRA-24-2 | F: CGGTGGATGAGCCATTGTG R: GCCTGCCGCCTAAGAAAA | Exon 24 | 62 | 669 (378 bp exon 24 + 291 bp 3′ untranslated region) |
| Variant | Genotype | Number | Litter Size |
|---|---|---|---|
| c.612G>A | GG | 356 | 1.14 ± 0.02 |
| GA | 28 | 1.07 ± 0.05 | |
| c.1521C>G in LD-Sonid | CC | 340 | 1.07 ± 0.01 A |
| CG | 42 | 1.64 ± 0.07 B | |
| c.1941G>A | GG | 142 | 1.18 ± 0.03 |
| GA | 167 | 1.14 ± 0.03 | |
| AA | 74 | 1.05 ± 0.03 | |
| c.2682C>T | CC | 349 | 1.13 ± 0.02 |
| CT | 33 | 1.33 ± 0.06 | |
| c.3449C>G | CC | 320 | 1.14 ± 0.02 |
| CG | 60 | 1.13 ± 0.04 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Naren, Z.; Bu, H.; Cang, M.; Cao, G.; Nashun, B.; Tong, B. Novel Silent Mutations in the HIRA Gene Associated with Litter Size in Sonid Sheep. Animals 2025, 15, 2936. https://doi.org/10.3390/ani15202936
Wang C, Naren Z, Bu H, Cang M, Cao G, Nashun B, Tong B. Novel Silent Mutations in the HIRA Gene Associated with Litter Size in Sonid Sheep. Animals. 2025; 15(20):2936. https://doi.org/10.3390/ani15202936
Chicago/Turabian StyleWang, Chen, Zhana Naren, He Bu, Ming Cang, Guifang Cao, Buhe Nashun, and Bin Tong. 2025. "Novel Silent Mutations in the HIRA Gene Associated with Litter Size in Sonid Sheep" Animals 15, no. 20: 2936. https://doi.org/10.3390/ani15202936
APA StyleWang, C., Naren, Z., Bu, H., Cang, M., Cao, G., Nashun, B., & Tong, B. (2025). Novel Silent Mutations in the HIRA Gene Associated with Litter Size in Sonid Sheep. Animals, 15(20), 2936. https://doi.org/10.3390/ani15202936





