Simple Summary
This comprehensive review demonstrates how metagenomic technologies have revolutionized understanding of herbivore digestive systems through culture-independent analysis of gut microbial communities. The research reveals fundamental differences between ruminant foregut fermentation strategies and non-ruminant hindgut approaches, with distinct microbial compositions dominated by Firmicutes and Bacteroidetes. Key findings show that specific microbial taxa directly correlate with feed efficiency, growth performance, and animal health outcomes. The authors identify critical research gaps in sampling methodologies and functional validation while highlighting promising applications of probiotics, prebiotics, and targeted interventions for optimizing livestock production and reducing environmental impacts through improved microbial management strategies.
Abstract
Herbivorous animals rely on complex gastrointestinal systems and microbial communities to efficiently digest plant-based diets, extract nutrients, and maintain health. Recent advances in metagenomic technologies have enabled high-resolution, culture-independent analysis of gut microbiota composition, functional potential, and host–microbe interactions, providing insights into microbial diversity across the herbivore digestive tract. This review summarizes key findings on the gastrointestinal microbiota of herbivores, focusing on ruminant foregut and non-ruminant hindgut fermentation. Ruminants like cattle, sheep, and goats host microbiota enriched with fibrolytic and methanogenic microbes that facilitate fiber degradation and volatile fatty acid production, contributing significantly to energy balance. In contrast, non-ruminants such as horses and rabbits rely on hindgut fermentation, with distinct microbial taxa contributing to carbohydrate and protein breakdown. The review further explores how specific microbial taxa, including Prevotella, Fibrobacter, and Ruminococcus, correlate with improved feed efficiency and growth performance, particularly in ruminants. Additionally, the roles of probiotics, prebiotics, and symbiotics in modulating gut microbial composition and enhancing productivity are discussed. Despite significant advances, challenges remain in microbial sampling, functional annotation, and understanding the integration of microbiota with host physiology. The review emphasizes the potential of metagenomic insights in optimizing herbivore gut microbiota to improve feed efficiency, health, and sustainable livestock production.
1. Introduction
Herbivores are animals that mainly rely on plants as their food source to obtain energy and nutrients necessary for their life activities. Herbivores are an important component of the energy and material circulation system [1]. Herbivores can be classified into ruminants, such as cattle, sheep, and goats, which rely on foregut fermentation, and non-ruminants, such as horses and donkeys, which primarily rely on hindgut fermentation [2]. The main difference between herbivores and non-herbivores lies in the structure of the digestive tract system and its interaction with the microbial flora [3]. The digestive systems of herbivores have evolved to accommodate the breakdown of complex plant materials like cellulose, which they cannot digest on their own [4]. Their gut microbiota, consisting of a diverse community of microorganisms, play a pivotal role in fermenting these materials and producing energy sources such as short-chain fatty acids [5]. However, they have evolved specialized digestive systems, like the ruminant rumen and the equine cecum, to ferment complex plant carbohydrates into volatile fatty acids that support growth and energy balance [6,7]. Unlike ruminants with their complex multi-chambered stomachs, donkeys and horses rely on hindgut fermentation, making their cecal and colonic microbiota critical for nutrient digestion and overall health. Metagenomic technology has revolutionized our understanding of microbial communities in non-ruminant herbivores, particularly in equines, which serve as important working animals and alternative livestock species since their domestication. Recent advances in metagenomic sequencing, including high-throughput 16S rRNA gene sequencing and whole-genome shotgun metagenomics, have enabled comprehensive characterization of the donkey microbiome across different anatomical sites and physiological states. Studies employing metagenome-assembled genomes (MAGs) have provided unprecedented insights into the functional capabilities of donkey hindgut microbiota, revealing complex metabolic pathways involved in fiber degradation and volatile fatty acid production [8]. Their gut microbiota is diverse, contributing to nutrient absorption and immune regulation, but its composition is influenced by diet, environment, developmental stage, host health, and genetics [9]. The gastrointestinal microbiota plays a pivotal role in the digestion and metabolism of herbivorous animals [10,11].
Recent advancements in metagenomic sequencing have revolutionized the study of microbial communities, enabling comprehensive analysis of their composition and functional potential [12,13]. Metagenomics overcomes the limitations of traditional culture methods by bypassing the need for isolated cultures, allowing direct analysis of the genetic material present in environmental samples. This approach captures the genetic information from a diverse range of microorganisms, including those that cannot be cultured by conventional techniques [14]. Through modern sequencing technology, genomic information in samples can be obtained rapidly and on a large scale [15]. Metagenomics has expanded microbial research beyond culturable strains, allowing the study of unculturable microorganisms and their interactions with hosts. It enables identification of microbial composition, genetic and functional profiles, evolutionary relationships, and environmental interactions, with advances like binning technology improving genome classification [16,17]. The exploration of gastrointestinal microbiota in herbivores has revealed complex interactions between microbial communities and host physiology [18]. Furthermore, metagenomic approaches have illuminated spatial variations in microbial composition across different intestinal segments, demonstrating distinct community structures between foregut and hindgut regions, as well as between liquid and adherent phases within the caeco-colic ecosystem [19,20]. Beyond the digestive tract, metagenomic surveys have characterized microbial communities in other body sites, including oral, skin, and rectal microbiota, showing dynamic changes during critical life stages such as weaning and gestation [21,22,23]. Microbes such as bacteria, archaea, protozoa, and fungi contribute not only to the breakdown of complex plant polysaccharides but also to the synthesis of essential nutrients, vitamins, and bioactive compounds [24]. Moreover, the microbial ecosystem influences host immune function, intestinal development, and resistance to pathogens, highlighting its integral role in maintaining overall animal health [11].
The development of sequencing technology has significantly enhanced the integrity and accuracy of genome assembly, while the development of bioinformatics tools has improved the ability to annotate microbial functional genes [25]. In recent years, the development of metagenomics has promoted the research on microorganisms at the macro level, providing a new perspective for understanding the role of microorganisms in host nutrition, health and environmental adaptation [13,26]. This review focuses on the application of metagenomics in studying herbivore gastrointestinal microbiomes, using techniques like shotgun sequencing, metatranscriptomics, and metaproteomics to analyze microbial diversity, metabolic pathways, and host–microbe interactions, highlighting their role in regulating microbial efficiency in the host [27,28,29].
Despite extensive microbiome research in individual herbivore species, previous reviews have been neither sufficiently in-depth nor species-specific in their analyses, with non-ruminants—particularly equines—receiving considerably less comprehensive coverage. This gap underscores a critical need for systematic comparative analyses that integrate metagenomic technologies with production outcomes across diverse digestive system architectures. This review addresses several key knowledge gaps that distinguish it from the existing literature. First, while previous reviews have focused primarily on either ruminants or non-ruminants separately, this work provides a systematic comparative framework examining both foregut and hindgut fermentation strategies and their distinct microbial ecosystems. Second, we critically evaluate the methodological challenges in gut microbiota sampling—from invasive rumen cannulation to emerging non-invasive approaches—an aspect often overlooked in existing reviews but essential for standardizing future research protocols. Third, this review uniquely integrates antimicrobial resistance (AMR) patterns within herbivore microbiomes, addressing an emerging concern for both animal health and public safety. Fourth, we synthesize recent advances in multi-omics approaches (metagenomics, metatranscriptomics, metaproteomics, and metabolomics) that enable functional validation beyond taxonomic profiling. Finally, by explicitly linking specific microbial taxa to production performance metrics (feed efficiency, growth rates, and health outcomes), this review provides actionable insights for precision livestock management. These integrated perspectives aim to guide future research toward (1) developing standardized, non-invasive sampling methodologies; (2) establishing functional databases for improved gene annotation; (3) elucidating mechanistic host–microbe interactions under varying environmental and dietary conditions; and (4) implementing microbiota-based interventions for enhanced productivity and sustainability in herbivore agriculture.
2. Gastrointestinal Characteristics of Herbivores
During the process of biological development and evolution, due to climate change and the influence of low-availability nutrients in the diet, a large number of herbivores became extinct. However, the interaction between animals and plants enabled some animals to evolve special digestive systems, allowing them to obtain higher nutritional value and provide energy for their bodies, thereby enabling them to survive and develop [30]. Herbivores have evolved well-developed cellulose-breaking functions due to their herbivorous habits, such as ruminants and large single-stomach herbivores. This adaptive feature can help them digest plant-based feeds well and provide energy for their bodies. Herbivores can be classified into ruminants that mainly rely on the rumen for fermentation and non-ruminants that mainly rely on the intestine for fermentation (such as the cecum) based on the site of their decomposition of plant-based feeds. The main difference between ruminants and non-ruminants lies in a series of septal pouch structures in front of their true stomachs [31]. Ruminants have a unique digestive system. Compared with other herbivores, they can make more efficient use of the energy form fibrous plants [32].
According to the anatomical structure of the gastrointestinal tract of ruminants, their digestive system is composed of a multi-compartment stomach structure (rumen, reticulum, omasum, abomasum), small intestine (duodenum, jejunum, ileum), and large intestine (colon, cecum, rectum) [33]. However, not all ruminants have the characteristic four-chambered stomach structure. For instance, camels lack an omasum. The anterior stomach includes the rumen, reticulum, and omasum [34]. It is a special digestive structure that distinguishes ruminants from other animals. It can physically process feed, undergo microbial fermentation, and absorb nutrients, and is a key structure for digesting cellulose. The rumen is the first and most important part of the stomach in ruminants and is rich in bacteria that can digest cellulose, such as Fibrobacter [35], Ruminococcus [36], Butyrivibrio [37], and Prevotella [38]. During the digestive process, ruminants, in contrast to non-ruminants, undergo rumination, where they regurgitate and re-chew their feed to break it down into smaller, more digestible particles [39]. This mainly relies on the density sorting mechanism of the anterior stomach [40]; this mechanism can increase the intake of ruminants without affecting digestive efficiency. The abomasum, known as the true stomach, is structurally and functionally similar to other monogastric animals. It can secrete gastric acid and various digestive enzymes to break down proteins and fats, facilitating their absorption and utilization in the intestinal tract.
The digestive tract of non-ruminants is composed of the same parts as that of other animals, including the stomach, duodenum, jejunum, ileum, cecum, colon, and rectum. They do not have a well-developed rumen. The stomach of non-ruminants has similar functions to those of other monogastric animals, but it has a well-developed cecum and colon, which have a fermentation function similar to that of the rumen in ruminants. The bacteria and ciliates inside can break down roughage [32]. Compared to ruminants, non-ruminants have relatively small stomachs that cannot store large quantities of feed. As a result, they require frequent feeding and are adapted to the continuous intake of pasture. The schematic diagram of the gastrointestinal tracts of representative ruminants and non-ruminants is presented in Figure 1.
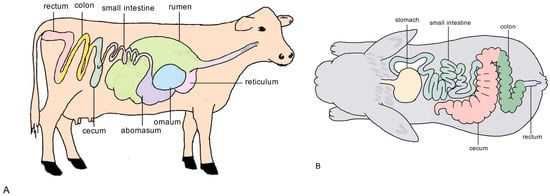
Figure 1.
Schematic diagram of the gastrointestinal tracts of representative ruminants and non-ruminants. (A) Left side: Ruminant gastrointestinal tract showing the complex, multi-chambered stomach (rumen, reticulum, omasum, and abomasum). (B) Right side: Non-ruminant gastrointestinal tract with a simpler stomach structure and reliance on the cecum and large intestine for fermentation.
Ruminants are pre-intestinal fermenters. The rumen can store a large amount of feed and is regarded as a fermentation tank, providing a perfect living environment for the proliferation, development, and metabolic activities of microorganisms. Microorganisms can digest fibrous substances into volatile fatty acids, which are then absorbed by the body, serving as the primary energy source for ruminants. Non-ruminant herbivores rely on hindgut fermentation [41]. With a well-developed cecum, their digestive capacity is limited by the volume of the intestine and the rate at which digesta passes through. The cecum and large intestine act as fermentation chambers, serving as the main sites where microorganisms degrade and ferment cellulose [42]. The volatile fatty acids produced through fermentation are then absorbed and metabolized to provide energy for the host [43]. The gastrointestinal characteristics of ruminants and non-ruminants are summarized in Table 1.

Table 1.
Gastrointestinal characteristics of ruminants and non-ruminants.
3. Research Strategies of Metagenomic Technology in Herbivores
3.1. Metagenomic Technologies
The term “metagenome” was first proposed by Handelsman et al. in 1998, referring to the collective genomes of all microorganisms in a specific environment, analyzed as a single genomic unit [71]. Metagenomics technology does not require the microbial isolation and culture by directly extracting total microbial DNA from environmental samples, constructing genomic libraries, and screening for novel functional genes and metabolites. Common approaches include whole-genome shotgun sequencing and amplicon sequencing [72,73]. DNA is fragmented by physical or enzymatic methods, sequenced, assembled, and annotated for species classification and functional potential. Early studies relied on 16S rRNA sequencing to profile microbial diversity, though this method cannot resolve subspecies, strains, or non-bacterial symbionts such as fungi, viruses, and protists [71,74]. High-throughput metagenomic sequencing combined with bioinformatics enables strain-level analysis, functional prediction, host–microbe interaction studies, and exploration of uncultured microbial “dark matter” [75].
Advances in sequencing technologies and bioinformatics have enabled reconstruction of whole microbial genomes from complex communities [76,77]. Short- and long-read sequencing, along with improved algorithms, allows for binning sequences into taxonomic clusters, improving genome completeness [75,78,79]. Long-read sequencing enhances assembly quality by increasing the continuity of metagenomic assembly, which in turn allows for more accurate reconstruction of microbial genomes, making it a crucial tool for analyzing microbial metabolic pathways [80,81,82,83,84]. Integration with multi-omics approaches, including metatranscriptomics and metabolomics, further elucidates the relationship between gene expression and ecological function [85]. Therefore, metagenomics is widely applied in various fields of microbial research, such as agriculture, biology, pollution control, energy, environment and other areas [86]. Advances in sequencing and multi-omics have made metagenomics a powerful tool for reconstructing microbial genomes and understanding their functional roles in complex ecosystems.
Despite these advances, metagenomics faces challenges. Host DNA contamination can compromise microbial resolution, necessitating selective lysis, enzymatic degradation, and enrichment of microbial DNA prior to sequencing [87,88,89,90,91,92,93,94]. Functional annotation is also limited by fragmented assemblies and incomplete reference databases, highlighting the need for improved computational tools and curated genomic resources [95]. The typical metagenomic workflow includes genome enrichment, DNA extraction, library construction, sequencing, gene prediction, and functional expression analysis.
3.2. Metagenomic Research Strategies
Metagenomic research strategies are broadly categorized into two principal approaches: (1) sequence-based metagenomics and (2) function-based metagenomics [96,97].
Sequence-based metagenomics employs shotgun sequencing of total environmental DNA to comprehensively characterize the genetic composition and functional potential of microbial communities. This approach involves the random fragmentation of genomic DNA from all microorganisms within a sample, followed by high-throughput sequencing and subsequent bioinformatic analysis to identify taxonomic composition and predict functional gene content.
Function-based metagenomics utilizes activity-driven screening methods to identify specific functional genes or biomolecules of interest. This approach typically involves constructing metagenomic expression libraries in suitable host organisms and screening clones for desired phenotypic traits or enzymatic activities, enabling the direct detection of functional variants independent of sequence homology.
In intestinal microbiota studies utilizing sequence-based approaches, biological samples—including gastric contents, intestinal digesta, or fecal material—are collected for metagenomic DNA extraction. Following DNA fragmentation and sequencing library preparation, raw sequence data undergo quality control procedures to remove low-quality reads and filter host-derived contamination. High-quality reads are then assembled into contiguous sequences and subjected to binning algorithms to reconstruct metagenome-assembled genomes (MAGs). Taxonomic classification of MAGs is performed using reference databases such as the Genome Taxonomy Database (GTDB) and the National Center for Biotechnology Information (NCBI) database, while functional annotation is conducted through mapping against the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO) databases.
However, in addition to KEGG and GO, several important functional annotation databases play crucial roles in gene functional analysis. CAZy and dbCAN are essential for analyzing carbohydrate-active enzymes (CAZymes) [98], while eggNOG and COG are vital for identifying orthologous gene groups, aiding in functional annotation across species [99]. UniRef enhances protein function identification by clustering similar protein sequences [100,101,102]. Tools like eggNOG-mapper v2 assign genes to orthologous groups and integrate annotations from GO, KEGG, and CAZy, facilitating functional inference across diverse organisms [99]. Similarly, dbCAN utilizes the CAZy database for automated CAZyme annotation, highlighting its significance in annotating carbohydrate-active enzymes in microbial genomes [98].
Finally, species richness, gene enrichment, and metabolic pathway analyses are performed to characterize microbial community structure and functional potential [76,103,104,105,106,107,108]. Metagenomic research in herbivores employs a systematic analytical workflow that enables comprehensive characterization of microbial communities through sequential processes spanning DNA extraction, bioinformatics analysis, functional annotation, and customized data interpretation. This integrated approach, as demonstrated in Figure 2, facilitates the elucidation of complex microbiome structures and their ecological roles within herbivorous systems. The implementation of both function-based and sequence-based metagenomic methodologies provides complementary analytical frameworks for the comprehensive assessment of microbial diversity, functional capacity, and metabolic potential within these intricate biological communities.
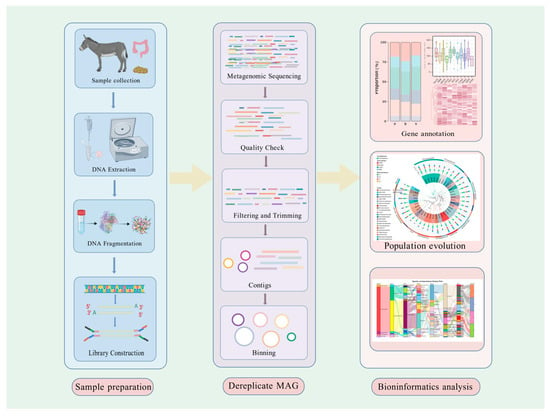
Figure 2.
Workflow of metagenomic analysis in herbivores. The diagram outlines the stepwise process starting from sample collection, DNA extraction, and fragmentation, through library construction, sequencing, and quality control, to metagenomic assembly, binning, dereplication, functional and taxonomic annotation, pathway enrichment, and personalized analysis. This workflow illustrates the integration of sequencing technologies and bioinformatics tools in characterizing microbial communities. Created with BioGDP.com [109].
4. Differences Between the Gastrointestinal Microbiota of Ruminants and Non-Ruminants
The herbivore digestive system is uniquely adapted to plant-based diets, a characteristic distinctly reflected in the composition and function of its gastrointestinal microbiota. Ruminants, including cattle, sheep, and goats, are foregut fermenters possessing a multi-chambered stomach system (rumen, reticulum, omasum, and abomasum) where microbial communities extensively degrade fibrous plant material before it encounters host digestive enzymes [110,111,112,113,114,115,116]. Beyond bacteria, methanogenic archaea (e.g., Methanobrevibacter, Methanosarcina), anaerobic fungi, and protozoa contribute synergistically to cellulose and hemicellulose breakdown, producing volatile fatty acids (VFAs) that supply up to 70% of the host’s energy requirements [10,117,118,119]. This specialized microbial ecosystem enables ruminants to efficiently utilize low-quality, high-fiber diets that remain largely indigestible to non-ruminant species. These fundamental compositional differences in microbiota architecture are comprehensively illustrated in Figure 3.
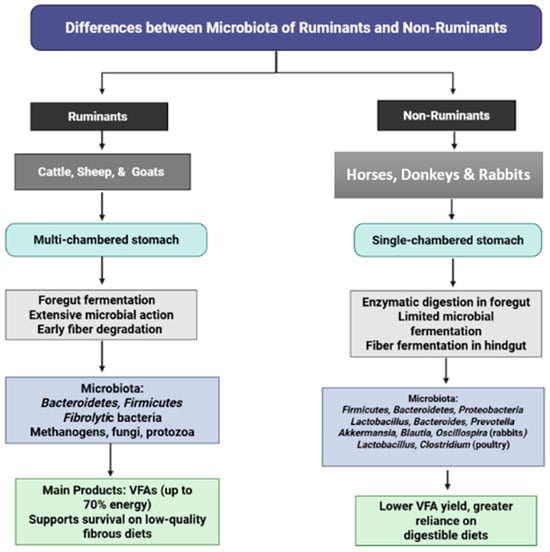
Figure 3.
Comparison of gastrointestinal microbiota between ruminants and non-ruminants.
In contrast, non-ruminant herbivores such as horses, donkeys, rabbits, and deer possess single-chambered stomachs where digestion depends primarily on host-derived enzymes, with microbial fermentation occurring predominantly in the hindgut (cecum and colon) [48,120]. Rabbits demonstrate heavy reliance on cecal fermentation and employ cecotrophy to maximize nutrient absorption, exhibiting enrichment of Akkermansia, Blautia, and Oscillospira in soft feces [68,121]. These structural and functional differences necessitate more digestible dietary inputs for non-ruminants and result in reduced efficiency in fibrous feed utilization compared to ruminants [116].
The fundamental distinction lies in the foregut fermentation strategy employed by ruminants versus the hindgut fermentation approach of non-ruminants, which profoundly influences microbial diversity patterns, energy extraction mechanisms, and host–microbe interactions [122,123]. Ruminant microbiota demonstrate enrichment with fibrolytic and methanogenic communities essential for efficient fiber degradation, whereas non-ruminant microbiota are characterized by predominant saccharolytic and lactate-utilizing taxa that complement enzymatic digestion processes [68,124,125,126]. While these distinctions are well-documented, significant research gaps persist regarding the comparative functional dynamics of these microbial ecosystems under varying dietary regimens and environmental pressures. Advanced metagenomic and multi-omics approaches could further elucidate the co-evolutionary relationships between microbial communities and host physiology, potentially providing insights for targeted dietary interventions and microbiota-based strategies aimed at improving feed efficiency and reducing methane emissions in both ruminant and non-ruminant livestock systems. Table 2 summarizes the predominant microbial groups found in the gastrointestinal tracts of both ruminant and non-ruminant herbivores.

Table 2.
Predominant microbial groups in the gastrointestinal tract of ruminants and non-ruminants.
5. Metagenomic Technology in Herbivore Gastrointestinal Microbes
Metagenomic technologies have significantly advanced our understanding of microbial communities in herbivores, particularly within their gastrointestinal (GI) tracts. These technologies allow for comprehensive analyses of microbial diversity, functional capabilities, and host–microbe interactions without the need for culturing individual species [145,146,147,148]. Unlike traditional culture-based techniques, which rely on isolating and growing individual microorganisms, metagenomics enables the direct sequencing of all microbial DNA present in environmental samples, such as feces, rumen contents, or intestinal digesta [149,150,151]. This approach allows researchers to capture the genetic material of both culturable and unculturable microorganisms, offering a more holistic view of microbial diversity [152,153]. High-throughput sequencing technologies such as Illumina and Nanopore sequencing generate large volumes of DNA data rapidly, providing insights into the full microbial community composition, including bacteria, archaea, fungi, and even viruses [154,155]. These sequencing methods, combined with advanced bioinformatics tools, facilitate taxonomic classification and functional annotation of microbial genes, enabling researchers to identify key metabolic pathways, microbial interactions, and their potential roles in herbivore nutrition and health.
Furthermore, metagenomic technology is crucial for studying herbivore microbiota’s ability to break down complex plant polysaccharides like cellulose and lignin, enabling the discovery of microbes, enzymes, and metabolic pathways vital for digestion and energy extraction [156,157,158]. The microbial communities in the gastrointestinal tract of herbivores play a pivotal role in fermenting complex substrates, producing short-chain fatty acids (SCFAs) such as acetate, propionate, and butyrate [159,160]. These SCFAs serve as primary energy sources for the host and are crucial for maintaining gut health and metabolic function.
The emergence of high-throughput omics technologies has revolutionized microbiome studies, enabling extensive and large-scale analysis of microbial communities’ structure and function. These cutting-edge techniques have made it possible to explore microbial ecosystems with unprecedented detail, greatly improving our understanding of microbial interactions and their contributions across different environments [161]. Technologies such as metagenomics, metatranscriptomics, metaproteomics, and metabolomics offer a detailed analysis of the genetic, transcriptional, protein, and metabolic characteristics of microbial communities. Unlike conventional techniques, meta-omics enable researchers to examine microbial communities in their native environments, eliminating the need for cultivation and providing a more precise and comprehensive understanding of microbial ecology and function [162].
Metagenomics has emerged as an essential method for examining the microbial communities in ruminants like cattle and sheep, allowing researchers to analyze microbial DNA directly from these animals and explore how changes in these communities correlate with animal traits [163,164,165]. Studies have revealed the intricate interactions between bacteria and archaea in the rumen, especially under conditions where methane mitigation is effective [163,164,165]. Metagenomics has revolutionized the study of microbial ecosystems, particularly in the rumen, by enabling direct DNA sequencing from environmental samples, thereby enhancing our understanding of microbial functions and providing opportunities for targeted mitigation strategies, such as reducing methane emissions in ruminants [166].
Advanced metagenomic techniques, including shotgun sequencing and metatranscriptomics, have enabled the identification of specific enzymes like cellulases and hemicellulases involved in the breakdown of plant polysaccharides. For example, the gut microbiota of Plateau pika revealed that upregulated expression of these enzymes facilitates energy extraction from grass-based diets, particularly at high altitudes [27]. Similarly, metagenomics and metatranscriptomics have revealed the role of Gangba sheep’s gut microbiota in plant biomass degradation, identifying key enzymes involved in the breakdown of plant polysaccharides. These studies highlight how diet and environment influence microbial functions and energy extraction [167]. Using metagenomic and metatranscriptomic sequencing, the rumen microbiota of Gir cattle under different dietary regimes was examined. This approach allowed for the identification of differential microbial populations and their functional dynamics, revealing key transcriptionally active genera like Caldicellulosiruptor and Paludibacter involved in fiber degradation [168]. Metatranscriptomics is a powerful meta-omics technique that provides insights into the functional dynamics of the rumen microbiome by analyzing RNA transcripts expressed by the microbial community at a given time [169]. Unlike metagenomics, which reveals the genetic potential of the microbiome, metatranscriptomics focuses on identifying the genes that are actively transcribed, offering a real-time view of microbial activity and their functional roles within the rumen ecosystem [170].
Metatranscriptomics serves as a robust tool for analyzing microbial composition and functions related to methane production within individual ruminant species, as well as comparing methane generation differences across various breeds. For example, metatranscriptomic analysis of Holstein cows (low-CH4 emitters) showed increased expression of genes associated with alternative hydrogen disposal pathways, particularly those linked to amino acid synthesis and propionate production [171]. Microbial research faces significant challenges due to the complexity of microbiota and the inherent difficulties in isolating and culturing microorganisms. The advent of metagenomic technology has provided a powerful analytical tool for investigating these complex ecosystems by enabling the sequencing and characterization of all microbial DNA within a sample without requiring individual cultivation [172]. Metagenomics allows researchers to determine microbial composition, identify functional genes, and elucidate metabolic pathways and their products through sophisticated bioinformatics approaches. Herbivore intestinal microbiota not only facilitate feed digestion but also enhance nutrient absorption, synthesize essential vitamins and amino acids, and contribute to immune regulation. These microorganisms degrade complex plant polysaccharides such as cellulose and lignin into SCFAs, which serve as primary energy sources for the host. The gastrointestinal microbiota encompasses bacteria, archaea, fungi, and protozoa, with dominant bacterial phyla including Bacteroidetes, Firmicutes, Actinobacteria, Proteobacteria, Clostridia, and Verrucomicrobia [173,174,175,176,177]. The composition and diversity of these microbial communities are influenced by multiple factors including host age, genetics, diet, environment, and physiological state.
6. Microbial Diversity in Herbivore Gastrointestinal Tract
Microbial composition exhibits distinct variation along different sections of the gastrointestinal tract. In ruminant digestive systems, the forestomach harbors the greatest microbial density, followed by the large intestine, with significantly reduced microbial populations in the small intestine [178,179]. Metagenomic studies have revealed distinct microbial patterns along the herbivore gastrointestinal tract. In ruminants, the rumen and stomach are dominated by Prevotella, Fibrobacter, and unclassified Bacteroidales and Clostridiales [180], while the small intestine exhibits lower microbial diversity but higher functional activity, with enrichment of Actinobacteria and Patescibacteria [181]. The large intestine is primarily colonized by Ruminococcaceae, Rikenellaceae, and Bacteroidaceae, facilitating fermentation and energy extraction [182]. Sequencing studies confirm that Oscillospiraceae, Lachnospiraceae, and Bacteroidaceae are dominant bacterial families in the cow rumen, where they drive the essential processes of cellulose degradation and VFA synthesis [183]
In monogastric herbivores, the hindgut, including the cecum and colon, serves as the primary fermentation site. Horses and donkeys demonstrate high abundances of Firmicutes and Bacteroidetes, with genera such as Clostridiales, Bacteroidales, and Ruminococcus dominating [7,59,184], with Bacillota and Bacteroidota representing the most abundant phyla. In the newborn goat rumen, UBA636, Bacteroides, Rothia, and Porphyromonas species constitute dominant members, although their abundance declines sharply by the tenth day. Consistently Mi et al. [185] demonstrated that most archaeal genomes belong to Methanobacteriaceae and Methanomethylophilaceae. Furthermore, a study investigated eight different gastrointestinal segments from Bactrian camels, identifying Firmicutes, Verrucomicrobia, and Bacteroidetes as dominant phyla [186]. The diversity index in the rumen was significantly higher than in other segments, while jejunum samples exhibited the lowest richness and diversity. Rabbits are dominated by Firmicutes, Bacteroidetes, and Tenericutes, with Ruminococcaceae and Lachnospiraceae prevalent at the family level [69].
Environmental and dietary adaptations significantly shape microbial communities in herbivores. Camels harbor Bacteroidetes and Firmicutes, with genera Prevotella, RC9_gut_group, and Butyrivibrio dominating [53,54]. Drought- or altitude-adapted goats and sheep exhibit increased rumen fungi and shifts in bacterial composition to optimize nutrient utilization under stress conditions [49,186,187,188,189]. Feed interventions such as solid-state fermentation, probiotics, or prebiotics can modulate microbial communities, enhancing beneficial bacteria while suppressing pathogenic species [186,188,189,190,191,192].
Despite diversity and compositional differences in gastrointestinal microbiota among different animal species, Firmicutes and Bacteroidetes consistently maintain dominant positions and serve as important participants in feed digestion and fermentation. Proteobacteria also occupy prominent positions in host microbiomes. This pattern has been confirmed across various animals species, including sheep [193], yaks [194], deer [188], camels [186,189], horses [190,191,192], and rabbits [56], facilitating host adaptation to complex internal environments. Across species, Firmicutes and Bacteroidetes consistently dominate and play key roles in fiber fermentation, energy production, and host adaptation. Other phyla, including Proteobacteria and Actinobacteria, contribute to host metabolism and immune regulation. Archaeal genome construction using metagenomes from 10 different ruminants revealed that most archaeal genomes belong to Methanobacteriaceae and Methanomethylophilaceae. In digestive tract, Methanobacteriaceae and Methanomethylophilaceae predominate. Methanobacteriaceae demonstrated greater abundance in the small intestine compared to the stomach and large intestine, while Methanobacteriaceae and Methanocorpusculaceae were more prevalent in the large intestine and feces. Overall relative abundance and composition of archaeal genomes vary with species composition.
He et al. [186] investigated eight different gastrointestinal segments from Bactrian camels, identifying Firmicutes, Verrucomicrobia, and Bacteroidetes as dominant phyla. Rumen diversity indices significantly exceeded those of other segments, while jejunum samples exhibited the lowest richness and diversity. Firmicutes dominated the entire intestinal microbial community, with Bacteroidetes following in the forestomach. Prevotella, Fibrobacter, unclassified Bacteroidales, unclassified BS11, and unclassified Clostridiales were significantly enriched in forestomach sites. Firmicutes and Verrucomicrobia represented the most abundant taxa in the ileum and large intestine, with unclassified Ruminococcaceae more enriched in the large intestine and ileum than in other gastrointestinal samples. Proteobacteria constituted the second most abundant microorganisms in the duodenum and jejunum.
These compositional differences may result from varying feeding conditions. Amplicon sequencing performed on rabbit cecum and fecal samples revealed Firmicutes, Tenericutes, and Bacteroidetes as the primary contributors to microbial diversity. At the family level, Ruminococcaceae and Lachnospiraceae were predominant [69]. Metabolic processes related to amino acid biosynthesis, energy production, enzyme families, vitamins, and other amino acids demonstrate greater activity in the hindgut compared to the foregut, while carbohydrate metabolism and other amino acid processes show higher activity in the foregut than hindgut, consistent with hindgut fermentation characteristics [184].
Batinah goats coping with drought and water scarcity exhibit increased rumen fungal concentrations [46]. Hu sheep introduced to arid, high-altitude regions demonstrated significant microbiota changes to facilitate environmental adaptation [195]. Solid-state fermented feeds can induce gastrointestinal tract acidification, providing appropriate conditions for beneficial bacterial establishment while reducing Enterobacteriaceae and Salmonella levels [196,197,198]. Lactobacillus supplementation increases relative abundance of the Firmicutes phylum while simultaneously decreasing Bacteroidetes phylum abundance [197]. Xylitol can promote phosphate acetyl transferase transcription and increase propionate production, thereby reducing pH values to inhibit Escherichia coli and Staphylococcus growth [198].
Uncultivated Microbial Lineages in Herbivore Guts
Recent metagenomic studies have significantly advanced our understanding of microbial diversity in herbivore gastrointestinal tracts, revealing novel, uncultivated microbial lineages [199]. Among these are UBA1242 (Firmicutes), Rs-D84 (AlphaProteobacteria), and UBA9783 (Verrucomicrobiota), identified in fecal samples from various farm animals, including cows, yaks, and sheep [200].These lineages are characterized by reduced genomes (<1 Mbp) and the absence of essential biosynthetic pathways, suggesting they rely on metabolites from their hosts, adapting to either a symbiotic or parasitic lifestyle [200]. Notably, UBA9783 possesses a nearly complete glycolytic pathway, indicating its ability to process carbohydrates, while UBA1242 and Rs-D84 exhibit more limited metabolic capabilities, emphasizing the metabolic specialization of these uncultivated taxa [200]. These microbes, particularly in regions such as the four-chambered stomach, contribute to specialized functions like polysaccharide degradation and hydrogen production [199]. The identification of these uncultivated microbial taxa and their associated metabolic pathways highlights the critical role of metagenomics in uncovering previously uncharacterized microbes within the herbivore gut microbiome [199]. These discoveries suggest potential pathways for improving feed efficiency and animal production [200].
7. Functional Roles of Gastrointestinal Microbiota in Herbivores and Antimicrobial Resistance (AMR)
The gastrointestinal microbiota of herbivores plays a critical role in nutrient digestion, energy metabolism, and overall host physiology. In ruminants, foregut microbial communities are enriched with genes responsible for degrading complex plant carbohydrates, proteins, and lipids. The small intestinal microbiota primarily focuses on nucleic acid and xenobiotic metabolism, while the large intestine hosts microbial communities dedicated to fermentation and protein synthesis pathways (Table 3) [48,180,181,201,202]. In monogastric herbivores, such as horses and donkeys, hindgut fermentation predominates, with the cecum and colon serving as primary sites of microbial carbohydrate metabolism and SCFA production (Table 4) [203,204,205].
Metagenomic studies have consistently revealed higher abundances of carbohydrate-active enzymes (CAZymes) in the large intestine compared to the small intestine, with GH13 family enzymes playing a significant role in starch degradation [48]. These microbial functions are crucial for breaking down ingested plant material into simple sugars and SCFAs, which serve as major energy sources for growth, development, and maintenance of physiological homeostasis [48,201,203]. These comprehensive metagenomic studies not only enhance our understanding of donkey biology but also establish foundational knowledge applicable to other non-ruminant equines, including horses, while contributing to improved animal health management and disease surveillance strategies [206].
Carbohydrate degradation and SCFA production represent fundamental microbial functions that sustain host energy balance. Bacteroidetes species contribute by encoding extensive CAZyme repertoires that degrade complex polysaccharides and host-unutilized glycans, producing acetate and propionate, whereas Firmicutes, particularly Ruminococcaceae and Lachnospiraceae, specialize in cellulose and hemicellulose depolymerization and butyrate synthesis [48,201,203,204,205,207,208]. These SCFAs not only supply energy but also support gluconeogenesis, regulate lipid metabolism, and modulate intestinal epithelial function. Comparative metagenomic analyses across sheep, goats, and donkeys demonstrate that carbohydrate, amino acid, lipid, and energy metabolism pathways are broadly represented in both ruminants and monogastric herbivores [48,201,203]. Despite these advances, quantitative mapping of enzyme activity to the actual, final yield of SCFA under varying dietary conditions and host genotypes remains limited, highlighting the need for integrated fluxomics and longitudinal studies [180,181]. There is a notable lack of standardized in vivo assays that link microbial enzyme families to actual SCFA production under different dietary and host genetic conditions.
Beyond nutrient metabolism, gastrointestinal microbiota exert critical immunomodulatory and barrier-protective functions. Microbial metabolites, especially butyrate, regulate tight junction protein expression, mucin secretion, and antimicrobial peptide production, enhancing mucosal integrity and resistance to pathogen colonization [209,210]. Studies in ruminants, such as sheep and goats, have shown positive correlations between the abundance of Bifidobacterium, Ruminococcus, and Enterococcus with mucin gene expression, with metagenomic analyses revealing mucin-degrading enzyme potential in host-adapted taxa [211,212]. Furthermore, probiotic interventions using Bifidobacterium, Lactobacillus, and Bacillus have been shown to stabilize microbial communities and reduce enteric disease risk, although strain-specific efficacy and dose–response relationships still require further controlled trials [213,214].
Microbial community composition also significantly influences disease susceptibility and metabolic resilience. Expansions of Proteobacteria, which include many pathobionts, are associated with dysbiosis and inflammatory disorders such as colitis, while symbiotic Enterobacteriaceae can occupy inflammatory niches without pathogenic consequences [215,216]. Conversely, Actinobacteria, including Bifidobacterium and other taxa (Eggerthellaceae, Nocardiaceae), contribute essential vitamins, amino acids, antioxidants, and bioactive metabolites with antimicrobial and immunomodulatory effects [217]. In both ruminants and non-ruminants, balanced microbial communities dominated by Firmicutes and Bacteroidetes optimize digestion, SCFA production, and nutrient synthesis, whereas microbial imbalances can disrupt metabolic homeostasis and impair host health [48,201,203,204,205,207,208]. These findings underscore the multifaceted roles of gut microbiota in supporting herbivore nutrition, immunity, and resilience across different digestive system architectures.
The occurrence of AMR genes in herbivore microbiota, particularly in yak, beef, and dairy cattle, significantly affects animal health and production. Studies show that yaks, raised in low-density, antibiotic-free environments, exhibit fewer AMR genes compared to beef and dairy cattle, which are raised in high-density conditions with frequent antibiotic use. This indicates that antibiotics in intensive farming systems contribute to the rise in AMR [218]. Furthermore, mobile genetic elements (MGEs), like integrons, play a vital role in the horizontal transfer of AMR genes. Interestingly, integron abundance was higher in yaks than in beef and dairy cattle, highlighting the role of MGEs even in low-antibiotic environments [218].
In swine, a study by Rahman et al. found that 85.3% of bacterial isolates from the gut microbiota harbored AMR genes, including those for tetracycline, macrolides, and aminoglycosides. The use of whole-genome sequencing (WGS) to analyze 129 isolates helped establish a biobank, aiding in the understanding of MGE involvement in AMR gene transmission identified 246 AMR genes across 38 families, with key resistance genes linked to tetracycline and lincosamide resistance, emphasizing the crucial role of metagenomic tools in AMR monitoring and surveillance [219]. Additionally, research revealed that animals raised with feed additives in barns had a significantly higher AMR profile compared to pasture-raised animals. The resistome in barn-raised cattle was dominated by β-lactamases and tetracycline resistance genes, underscoring the impact of antibiotic use in livestock production. This highlights the importance of integrating metagenomic techniques in AMR surveillance to protect both animal health and public safety [220].

Table 3.
Major microbial groups, metabolites, and host functions in the gut of ruminant herbivores.
Table 3.
Major microbial groups, metabolites, and host functions in the gut of ruminant herbivores.
| Microbial Group/ Exemplar Taxa | Principal Digestive/ Immune Functions | Predominant GIT Region(s) | Host Effects/ Phenotypes | Representative Hosts | References |
|---|---|---|---|---|---|
| Bacteroidetes (e.g., Bacteroides, Prevotella) | Degrade complex plant polysaccharides; utilize host-unabsorbed glycans; contribute to protein/lipid breakdown | Rumen/forestomach; large intestine | Energy harvest; suppression of pathogens; support barrier and immune tone | Cattle, sheep, goats, camels, | [180,181,182,201,203,204,205] |
| Firmicutes (Ruminococcaceae) | Degrade resistant polysaccharides, cellulose, and starch; produce degradative enzyme systems | Butyrate | Promote epithelial proliferation, energy harvest, regulate mucosal immunity | Cattle, goats, sheep | [48,201,221] |
| Firmicutes (Lachnospiraceae) | Fiber decomposition, protein hydrolysis, butyrate production | Butyrate, secondary metabolites | Support intestinal barrier, promote fat accumulation, gluconeogenesis | Sheep, goats | [207,208] |
| Phyla: Bacteroidetes, Firmicutes (>80%); Proteobacteria, Verrucomicrobia, Fibrobacteres, Spirochaetes, Tenericutes | Polysaccharide breakdown, fiber fermentation, SCFA production, immune modulation | Rumen, caecum, colon | Provide energy via VFAs; shifts with age, diet, and environmental factors; dysbiosis linked with inflammation | Goats | [222,223,224,225] |
| Families: Prevotellaceae, Veillonellaceae, Lachnospiraceae, Rikenellaceae, Ruminococcaceae | Fiber degradation, starch fermentation, butyrate production | Rumen, hindgut | Support efficient digestion, gut homeostasis, metabolic flexibility | Goats | [225,226] |
| Bacteroidetes (Prevotella, Bacteroides) | Fiber degradation, VFA production, carbohydrate | Rumen, hindgut | Improved feed efficiency, energy harvest | Sheep, Tibetan sheep, Mongolian | [227,228,229] |
| Firmicutes (Ruminococcus, Lachnospiraceae, Oscillospira, Clostridia, Lactobacillales) | Cellulose degradation, butyrate production, gut health maintenance | Rumen, intestine | Correlated with feed efficiency; role in gut homeostasis | Sheep, Qinghai | [230,231,232] |
| Bacteroidetes (Prevotella, Bacteroides) | Fiber and carbohydrate breakdown, VFA production, carbohydrate metabolism | Rumen, hindgut | Enhanced feed efficiency, energy harvest, gut homeostasis | Cattle (dairy, beef) | [233,234,235] |
| Proteobacteria (Succinivibrio, Acinetobacter) | Carbohydrate fermentation, starch metabolism | Rumen | Influenced by high-grain diets; amylolytic activity | Cattle | [178,236,237] |

Table 4.
Major microbial groups, metabolites, and host functions in the gut of non-ruminant herbivores.
Table 4.
Major microbial groups, metabolites, and host functions in the gut of non-ruminant herbivores.
| Microbial Group/Exemplar Taxa | Main Functions | Main Metabolites | Metabolite/Host Functions | Representative Animals | References |
|---|---|---|---|---|---|
| Bacteroidetes (Bacteroides spp.) | Carbohydrate and protein breakdown; enriched in arachidonic acid metabolism, pentose/glucuronate pathways | Acetate, propionate | Provide host energy, enhance barrier, reduce pro-inflammatory cytokines | horse, | [238] |
| Firmicutes (Ruminococcaceae) | Cellulose and hemicellulose degradation; resistant polysaccharide breakdown | Butyrate | Promote fat accumulation, energy harvest, barrier support | Donkey | [203,239] |
| Firmicutes (Lachnospiraceae) | Fiber decomposition, protein hydrolysis | Butyrate, secondary metabolites | Energy metabolism, gut homeostasis | Donkey, rabbit | [239,240] |
| Phyla: Firmicutes, Verrucomicrobiota | Fiber degradation, SCFA production, immune modulatio | Caecum (main fermentation site) | Seasonal abundance variations linked to productivity, physiology, and immune responses | Rabbits | [70] |
| Genera: Akkermansia, Blautia, Oscillospira | Mucus degradation (Akkermansia), fermentation, SCFA production | Soft feces (caecotropes) | Enhanced nutrient recycling via caecotrophy; improved metabolic health | Rabbits | [68] |
| Bacteroidetes + Firmicutes interplay | Co-metabolism of polysaccharides; Firmicutes specialize in cellulose fermentation, Bacteroidetes in glycan breakdown | Mixed SCFAs (acetate, propionate, butyrate) | Ensure efficient fiber digestion, provide major VFAs for host energy | Donkey, horse | [172,179,241] |
8. Role of Gastrointestinal Microbiota in Production Performance of Herbivorous Animals
Herbivorous animals depend on complex gastrointestinal systems and symbiotic microbial communities to efficiently convert fibrous feedstuffs into high-quality animal protein, thereby supporting optimal growth and production performance. The intestinal microbiota plays a central role in nutrient metabolism, immune modulation, and maintenance of intestinal barrier integrity, collectively enhancing feed utilization, daily weight gain, and disease resistance [124,242,243]. Factors such as age, diet, host genetics, and living environment significantly influence the composition and functional diversity of gut microorganisms, as illustrated in Figure 4. In ruminants, the balance between Firmicutes and Bacteroidetes in the rumen is closely associated with milk fat yield, energy storage, and average daily gain (ADG), while the presence of fibrolytic bacteria such as Fibrobacter and Eubacterium ruminantium positively correlates with milk and protein production [244,245,246,247]. Conversely, decreased populations of beneficial microbes, including Bifidobacterium and Lactobacillus, have been linked to reduced production efficiency and compromised animal health [248,249].
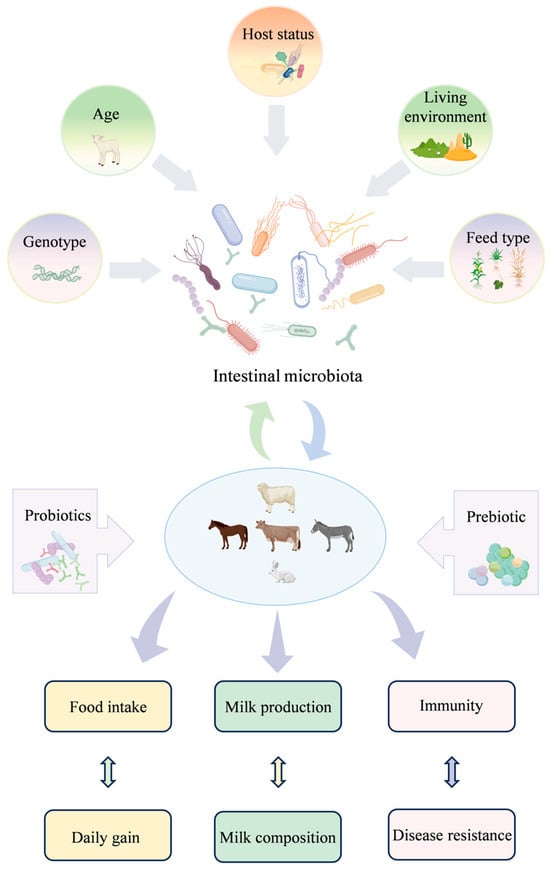
Figure 4.
Integrated model of the gastrointestinal microbiota as the metabolic engine of production performance in herbivorous animals.
Feed efficiency (FE) and residual feed intake (RFI) represent critical indicators of production performance, reflecting how effectively animals convert feed into body mass or milk production. Metagenomic investigations have also revealed that concentrate feeding sequences influence volatile fatty acid production and microbial community composition in both weaned and adult donkeys [250]. The application of metagenomic technology has been particularly valuable in understanding how dietary interventions and physiological conditions modulate the donkey microbiome and subsequent health outcomes. Research utilizing metagenomic analyses has demonstrated that dietary energy levels significantly impact cecal microbial diversity and metabolome profiles, with low-energy diets causing oxidative stress and growth reduction through alterations in microbial metabolite production [251].Recent studies have demonstrated that specific microbial species or strains, rather than entire microbial communities, exert stronger influences on feed efficiency in both ruminants and monogastric animals [252,253,254,255]. For example, Ruminococcus gauvreauii abundance is positively associated with dry matter intake (DMI) in dairy cows, whereas Howardella correlates with reduced DMI [244,256]. Similarly, microbial populations in the jejunum, cecum, and colon—including Prevotella, Clostridium, Oscillospira, and Faecalibacterium prausnitzii—have been shown to influence feed utilization and growth performance in beef cattle [257]. In monogastric herbivores such as donkeys, dietary interventions including corn silage supplementation can modulate the abundance of carbohydrate-metabolizing microbes such as Prevotella-1 and Alloprevotella, thereby improving ADG and overall growth performance [105,258,259,260]. Despite these significant findings, the complex interactions between feed characteristics, energy requirements, and microbial activity remain incompletely understood, highlighting the critical need for additional mechanistic studies.
The strategic application of probiotics, prebiotics, and synbiotics represents a promising approach to enhance gastrointestinal microbial balance and promote animal health and productivity. Prebiotics, including fructooligosaccharides (FOS) and galactooligosaccharides (GOS), selectively stimulate beneficial microbial populations, preventing pathogen colonization and improving nutrient absorption [257,258,259,260,261]. Probiotic supplementation, including Bacillus subtilis natto and Faecalibacterium prausnitzii, has been demonstrated to enhance ADG, feed efficiency, and immune function while reducing the incidence of gastrointestinal diseases [261,262]. Synbiotic supplementation further supports growth performance by combining the synergistic effects of probiotics and prebiotics on gut microbial composition, immune function, and nutrient metabolism [263,264]. Additionally, metagenomic technology has facilitated the assessment of dietary supplements, such as yeast polysaccharides, multienzymes, and methionine, on gut microbial composition and host immune function [265,266,267].
Collectively, these findings underscore the inseparable relationship between gastrointestinal microbiota and production performance, suggesting that strategic microbial modulation can optimize growth, lactation, and overall health in herbivorous animals. This approach ultimately promotes sustainable animal husbandry practices by maximizing production efficiency while maintaining animal welfare and reducing environmental impacts through improved feed conversion ratios.
9. Challenges and Advances in Sampling Gut Microbiota
Studying gut microbiota in large herbivores and monogastric livestock presents significant challenges due to anatomical constraints and limited accessibility. While fecal sampling remains the most commonly employed non-invasive method, it primarily reflects luminal microbial communities and may not accurately represent mucosa-associated bacteria, which are critical for host–microbe interactions [268]. Fecal microbial composition can also be influenced by factors such as transit time, dietary composition, and environmental exposure, potentially introducing biases in microbiota analysis [233,269]. In ruminants, rumen cannulation (fistulation) allows repeated, direct sampling of ruminal contents and is considered the gold standard for rumen microbiota studies [270]. However, this invasive procedure requires surgical intervention and is largely restricted to controlled research settings. Less invasive alternatives, such as oral stomach tubing, often underrepresent particle-associated microbes essential for fiber digestion [271,272]. Post-mortem sampling provides comprehensive access to gut contents and mucosal scrapings but is constrained by rapid microbial shifts following tissue death and the inability to conduct longitudinal studies [273,274]. Endoscopic and biopsy-based approaches enable investigation of mucosa-associated microbiota, yet these techniques are technically challenging in large animals and frequently necessitate anesthesia or surgical procedures, limiting their application in large-scale studies [268,275].
Recent advances in non-invasive sampling techniques have highlighted buccal swabs as a promising alternative for rumen microbiota assessment. Kittelmann et al. [276] demonstrated that buccal swabs can effectively capture bacterial, archaeal, and eukaryotic community structures, while Tapio et al. [277] reported that regurgitated bolus samples exhibit higher similarity to rumen contents compared to buccal swabs, likely due to distinct gingival microbiota composition. Time-course sampling combined with machine learning approaches further indicated that buccal swabs can detect key microbial taxa, although their accuracy depends on collection timing and environmental conditions [278]. More recently, MinION amplicon sequencing has enhanced the resolution and throughput of buccal swab microbiome profiling, enabling improved characterization of rumen microbiomes while maintaining a non-invasive approach [279]. Despite these promising developments, further research is needed to optimize sampling protocols, address site-specific microbial variability, and validate buccal swabs as reliable proxies for direct rumen sampling. This represents a critical research gap in large-animal gut microbiota studies, emphasizing the urgent need for standardized, non-invasive methodologies that balance animal welfare considerations, sampling accuracy, and practical feasibility.
10. Conclusions
This review highlights the pivotal role of metagenomics in advancing our understanding of herbivore gastrointestinal microbiota and its profound impact on animal health, feed efficiency, and production performance. By comparing the microbial ecosystems of ruminants and non-ruminants, it is evident that the specialized fermentation processes in the foregut and hindgut result in distinct microbial compositions that directly influence nutrient metabolism and energy extraction. Key microbial taxa, such as Prevotella, Fibrobacter, and Ruminococcus, play critical roles in fiber degradation, while targeted interventions like probiotics and prebiotics offer promising strategies for optimizing microbial balance and improving productivity.
Despite these advances, challenges such as microbial sampling accuracy, functional gene annotation, and understanding the complex interactions between microbiota and host physiology remain. Moving forward, integrating multi-omics approaches and improving sampling methodologies will be essential for a more comprehensive understanding of microbial dynamics. The continued application of metagenomics in herbivore microbiota research holds great potential for enhancing livestock management practices, improving feed efficiency, reducing environmental impacts, and supporting sustainable agricultural systems.
Author Contributions
J.W., M.Z.K., C.W. (Chunming Wang) and Z.Z.: Conceptualization, writing—original draft; J.W., M.Z.K., C.W. (Changfa Wang), C.W. (Chunming Wang), Z.Z., L.W., A.U., M.G. and X.Z.: writing—review and editing, literature search, proofreading, data curation, software, M.Z.K., C.W. (Chunming Wang) and Z.Z.: supervision, validation, resources and funding. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the National Key R&D Program of China (grant number 2023YFD1302004; 2022YFD1600103), Shandong Province Modern Agricultural Technology System Donkey Industrial Innovation Team (grant number SDAIT-27), Livestock and Poultry Breeding Industry Project of the Ministry of Agriculture and Rural Affairs (grant number 19211162), Shandong Province Agricultural Major Technology Collaborative Promotion Plan (SDNYXTTG-2024-13); The Open Project of Liaocheng University Animal Husbandry Discipline (grant number 319312101–14), The Open Project of Shandong Collaborative Innovation Center for Donkey Industry Technology (grant number 3193308), Research on Donkey Pregnancy Improvement (grant number K20LC0901), Key R&D Program Project of Shandong Province (2021TZXD012), Liaocheng University Scientific Research Fund (grant number 318052025) and Liaocheng Municipal Bureau of Science and Technology, High-talented Foreign Expert Introduction Program (GDWZ202401).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
No new data were created or analyzed in this study.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Pringle, R.M.; Abraham, J.O.; Anderson, T.M.; Coverdale, T.C.; Davies, A.B.; Dutton, C.L.; Gaylard, A.; Goheen, J.R.; Holdo, R.M.; Hutchinson, M.C.; et al. Impacts of large herbivores on terrestrial ecosystems. Curr. Biol. 2023, 33, R584–R610. [Google Scholar]
- Mottet, A.; Teillard, F.; Boettcher, P.; De, G.; Besbes, B. Review: Domestic herbivores and food security: Current contribution, trends and challenges for a sustainable development. Animal 2018, 12, s188–s198. [Google Scholar]
- Mackie, R.I. Mutualistic fermentative digestion in the gastrointestinal tract: Diversity and evolution. Integr. Comp. Biol. 2002, 42, 319–326. [Google Scholar] [CrossRef] [PubMed]
- Kurniasih, T.; Zulkarnain, R.; Wijaya, R.; Lesmana, D.; Mumpuni, F.S.; Panigoro, N.; Sutisna, E.; Heptarina, D.; Fitria, Y. Digestibility of plant-based feeds in omnivorous, carnivorous, and herbivorous fish: A review of Nile tilapia (Oreochromis niloticus (Linnaeus, 1758)), North African catfish (Clarias gariepinus (Burchell, 1822)), and grass carp (Ctenopharyngodon idella (Valenciennes, 1844)). Aquac. Aquar. Conserv. Legis. 2024, 17, 2994–3014. [Google Scholar]
- Singh, B.; Mal, G.; Kalra, R.S.; Marotta, F. Microbial-Rich Niches in Herbivores. In Probiotics as Live Biotherapeutics for Veterinary and Human Health, Volume 1: Functional Feed and Industrial Applications; Springer: Cham, Switzerland, 2024; pp. 235–256. [Google Scholar]
- Mizrahi, I.; Wallace, R.J.; Moraïs, S. The rumen microbiome: Balancing food security and environmental impacts. Nat. Rev. Microbiol. 2021, 19, 553–566. [Google Scholar] [CrossRef]
- Li, C.; Li, X.; Guo, R.; Ni, W.; Liu, K.; Liu, Z.; Dai, J.; Xu, Y.; Abduriyim, S.; Wu, Z.; et al. Expanded catalogue of metagenome-assembled genomes reveals resistome characteristics and athletic performance-associated microbes in horse. Microbiome 2023, 11, 7. [Google Scholar] [CrossRef]
- Kou, X.; Liu, Y.; Xiang, F.; Zhang, X.; Khan, M.Z.; Wu, B.; Wang, H.; Gong, Y.; Wang, C.; Ma, Q.; et al. Insights into the Donkey Hindgut Microbiome Using Metagenome-Assembled Genomes. Animals 2024, 14, 3625. [Google Scholar] [CrossRef]
- Cui, X.; Zhang, Q.; Zhang, Q.; Zhang, Y.; Chen, H.; Liu, G.; Zhu, L. Research Progress of the Gut Microbiome in Hybrid Fish. Microorganisms 2022, 10, 891. [Google Scholar] [CrossRef]
- Xu, Q.; Qiao, Q.; Gao, Y.; Hou, J.; Hu, M.; Du, Y.; Zhao, K.; Li, X. Gut Microbiota and Their Role in Health and Metabolic Disease of Dairy Cow. Front. Nutr. 2021, 8, 701511. [Google Scholar] [CrossRef] [PubMed]
- Negash, A. Gut microbiota ecology role in animal nutrition and health performance. J. Clin. Microbiol. Antimicrob. 2022, 6, 1–14. [Google Scholar]
- Roumpeka, D.D.; Wallace, R.J.; Escalettes, F.; Fotheringham, I.; Watson, M. A Review of Bioinformatics Tools for Bio-Prospecting from Metagenomic Sequence Data. Front. Genet. 2017, 8, 23. [Google Scholar] [CrossRef]
- Nam, N.N.; Do, H.D.K.; Trinh, K.T.L.; Lee, N.Y. Metagenomics: An Effective Approach for Exploring Microbial Diversity and Functions. Foods 2023, 12, 2140. [Google Scholar] [CrossRef]
- Chettri, D.; Verma, A.K.; Chirania, M.; Verma, A.K. Metagenomic approaches in bioremediation of environmental pollutants. Environ. Pollut. 2024, 363, 125297. [Google Scholar] [CrossRef]
- Degois, J.; Clerc, F.; Simon, X.; Bontemps, C.; Leblond, P.; Duquenne, P. First Metagenomic Survey of the Microbial Diversity in Bioaerosols Emitted in Waste Sorting Plants. Ann. Work. Expo. Health 2017, 61, 1076–1086. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Z.; Lu, Y.Y.; Sun, F.; Zhu, S. SolidBin: Improving metagenome binning with semi-supervised normalized cut. Bioinformatics 2019, 35, 4229–4238. [Google Scholar] [CrossRef]
- Du, Y.; Sun, F. HiCBin: Binning metagenomic contigs and recovering metagenome-assembled genomes using Hi-C contact maps. Genome Biol. 2022, 23, 63. [Google Scholar] [CrossRef]
- Milani, C.; Alessandri, G.; Mancabelli, L.; Mangifesta, M.; Lugli, G.A.; Viappiani, A.; Longhi, G.; Anzalone, R.; Duranti, S.; Turroni, F.; et al. Multi-omics Approaches To Decipher the Impact of Diet and Host Physiology on the Mammalian Gut Microbiome. Appl. Environ. Microbiol. 2020, 86, e01864-20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hu, T.; Liang, K.; Li, S.; Zhang, Q.; Li, W.; Qu, H.; Dong, B.; Zhang, H.; Ma, Q.; et al. Spatial variations in the microbiota: Comparative analysis of microbial composition and predicted functions across different intestinal segments and feces in donkeys. Front. Microbiol. 2024, 15, 1494926. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, B.; Wang, Y.; Zhu, M.; Wang, C. Could Weaning Remodel the Oral Microbiota Composition in Donkeys? An Exploratory Study. Animals 2022, 12, 2024. [Google Scholar] [PubMed]
- Zhang, Z.; Huang, B.; Shi, X.; Wang, T.; Wang, Y.; Zhu, M.; Wang, C. Comparative Analysis of Bacterial Diversity between the Liquid Phase and Adherent Fraction within the Donkey Caeco-Colic Ecosystem. Animals 2022, 12, 1116. [Google Scholar] [CrossRef]
- Ma, Q.; Yue, Y.; Kou, X.; Hou, W.; Wang, M.; Yang, X.; Liu, G.; Li, Y.; Wang, C. Dynamic Distribution of Skin Microorganisms in Donkeys at Different Ages and Various Sites of the Body. Animals 2023, 13, 1566. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ma, Q.; Liu, G.; Zhang, Z.; Zhan, Y.; Zhu, M.; Wang, C. Metabolic Alternations During Gestation in Dezhou Donkeys and the Link to the Gut Microbiota. Front. Microbiol. 2022, 13, 801976. [Google Scholar] [CrossRef] [PubMed]
- Rani, A.; Saini, K.C.; Bast, F.; Mehariya, S.; Bhatia, S.K.; Lavecchia, R.; Zuorro, A. Microorganisms: A Potential Source of Bioactive Molecules for Antioxidant Applications. Molecules 2021, 26, 1142. [Google Scholar] [CrossRef]
- Lema, N.K.; Gemeda, M.T.; Woldesemayat, A.A. Recent Advances in Metagenomic Approaches, Applications, and Challenge. Curr. Microbiol. 2023, 80, 347. [Google Scholar] [CrossRef]
- Alves, L.F.; Westmann, C.A.; Lovate, G.L.; de Siqueira, G.M.V.; Borelli, T.C.; Guazzaroni, M.E. Metagenomic Approaches for Understanding New Concepts in Microbial Science. Int. J. Genom. 2018, 2018, 2312987. [Google Scholar] [CrossRef]
- Gan, X.; Yu, Q.; Hu, X.; Qian, Y.; Mu, X.; Li, H. Metagenomic and metatranscriptomic analysis reveals the enzymatic mechanism of plant polysaccharide degradation through gut microbiome in plateau model animal (Ochotona curzoniae). FEMS Microbiol. Lett. 2025, 372, fnaf045. [Google Scholar] [CrossRef] [PubMed]
- Hu, F.; Cheng, Y.; Fan, B.; Li, W.; Ye, B.; Wu, Z.; Tan, Z.; He, Z. Ruminal microbial metagenomes and host transcriptomes shed light on individual variability in the growth rate of lambs before weaning: The regulated mechanism and potential long-term effect on the host. mSystems 2024, 9, e0087324. [Google Scholar] [CrossRef]
- Xia, X.; Gurr, G.M.; Vasseur, L.; Zheng, D.; Zhong, H.; Qin, B.; Lin, J.; Wang, Y.; Song, F.; Li, Y.; et al. Metagenomic Sequencing of Diamondback Moth Gut Microbiome Unveils Key Holobiont Adaptations for Herbivory. Front. Microbiol. 2017, 8, 663. [Google Scholar] [CrossRef]
- De Tarso, S.; Oliveira, D.; Afonso, J. Ruminants as part of the global food system: How evolutionary adaptation sand diversity of the digestive system brought them to the future. J. Dairy Vet. Anim. Res. 2016, 3, 171–176. [Google Scholar] [CrossRef]
- Harfoot, C.G. Anatomy, physiology and microbiology of the ruminant digestive tract. Prog. Lipid Res. 1978, 17, 1–19. [Google Scholar] [CrossRef]
- Harris, P.A.; Ellis, A.D.; Fradinho, M.J.; Jansson, A.; Julliand, V.; Luthersson, N.; Santos, A.S.; Vervuert, I. Review: Feeding conserved forage to horses: Recent advances and recommendations. Animal 2017, 11, 958–967. [Google Scholar] [CrossRef] [PubMed]
- Przybyło, M.; Flaga, J.; Clauss, M.; Szczepanik, K.; Miltko, R.; Bełżecki, G.; Kowalski, Z.M.; Górka, P. Increased intake of mono- and disaccharides by Reeves’s muntjac (Muntiacus reevesi). Effect on gastrointestinal tract structure and function and blood parameters. J. Anim. Physiol. Anim. Nutr. 2022, 106, 922–938. [Google Scholar]
- Górka, P.; Sliwinski, B.; Flaga, J.; Olszewski, J.; Wojciechowski, M.; Krupa, K.; Godlewski, M.M.; Zabielski, R.; Kowalski, Z.M. Effect of exogenous butyrate on the gastrointestinal tract of sheep. I. Structure and function of the rumen, omasum, and abomasum. J. Anim. Sci. 2018, 96, 5311–5324. [Google Scholar]
- Raut, M.; Couto, N.; Karunakaran, E.; Biggs, C.; Wright, P. Deciphering the unique cellulose degradation mechanism of the ruminal bacterium Fibrobacter succinogenes S85. Sci. Rep. 2019, 9, 16542. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hou, C.; Li, N.; Zhang, X.; Zhang, G.; Yang, F.; Zeng, X.; Liu, Z.; Qiao, S. Microbial and metabolic alterations in gut microbiota of sows during pregnancy and lactation. FASEB J. 2019, 33, 4490–4501. [Google Scholar] [CrossRef] [PubMed]
- Derakhshani, H.; Tun, H.M.; Cardoso, F.C.; Plaizier, J.C.; Khafipour, E.; Loor, J.J. Linking Peripartal Dynamics of Ruminal Microbiota to Dietary Changes and Production Parameters. Front. Microbiol. 2016, 7, 2143. [Google Scholar] [CrossRef]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef]
- Fritz, J.; Hummel, J.; Kienzle, E.; Arnold, C.; Nunn, C.; Clauss, M. Comparative chewing efficiency in mammalian herbivores. Oikos 2009, 118, 1623–1632. [Google Scholar] [CrossRef]
- Clauss, M.; Hummel, J. Physiological adaptations of ruminants and their potential relevance for production systems. Rev. Bras. Zootec. 2017, 46, 606–613. [Google Scholar] [CrossRef]
- Dougal, K.; de la Fuente, G.; Harris, P.A.; Girdwood, S.E.; Pinloche, E.; Geor, R.J.; Nielsen, B.D.; Schott, H.C., II; Elzinga, S.; Newbold, C.J. Characterisation of the faecal bacterial community in adult and elderly horses fed a high fibre, high oil or high starch diet using 454 pyrosequencing. PLoS ONE 2014, 9, e87424. [Google Scholar] [CrossRef]
- Al Jassim, R.A.; Scott, P.T.; Trebbin, A.L.; Trott, D.; Pollitt, C.C. The genetic diversity of lactic acid producing bacteria in the equine gastrointestinal tract. FEMS Microbiol. Let. 2005, 248, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Biddle, A.S.; Tomb, J.F.; Fan, Z. Microbiome and Blood Analyte Differences Point to Community and Metabolic Signatures in Lean and Obese Horses. Front. Vet. Sci. 2018, 5, 225. [Google Scholar] [CrossRef]
- Souza, J.G.; Ribeiro, C.; Harvatine, K.J. Meta-analysis of rumination behavior and its relationship with milk and milk fat production, rumen pH, and total-tract digestibility in lactating dairy cows. J. Dairy Sci. 2022, 105, 188–200. [Google Scholar] [CrossRef]
- Zhou, J.W.; Zhong, C.L.; Liu, H.; Degen, A.A.; Titgemeyer, E.C.; Ding, L.M.; Shang, Z.H.; Guo, X.S.; Qiu, Q.; Li, Z.P.; et al. Comparison of nitrogen utilization and urea kinetics between yaks (Bos grunniens) and indigenous cattle (Bos taurus). J. Anim. Sci. 2017, 95, 4600–4612. [Google Scholar] [CrossRef]
- Ramadhan, M.R.; Schlecht, E.; Dickhoefer, U.; Mahgoub, O.; Joergensen, R.G. Feed digestibility, digesta passage and faecal microbial biomass in desert-adapted goats exposed to mild water restriction. J. Anim. Physiol. Anim. Nutr. 2022, 106, 721–732. [Google Scholar] [CrossRef]
- Chai, J.; Zhuang, Y.; Cui, K.; Bi, Y.; Zhang, N. Metagenomics reveals the temporal dynamics of the rumen resistome and microbiome in goat kids. Microbiome 2024, 12, 14. [Google Scholar] [CrossRef]
- Li, B.; Zhang, K.; Li, C.; Wang, X.; Chen, Y.; Yang, Y. Characterization and Comparison of Microbiota in the Gastrointestinal Tracts of the Goat (Capra hircus) During Preweaning Development. Front. Microbiol. 2019, 10, 2125. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Jin, L.; Xue, B.; Wang, Z.; Peng, Q. Characterizing the bacterial community across the gastrointestinal tract of goats: Composition and potential function. Microbiologyopen 2019, 8, e00820. [Google Scholar] [CrossRef]
- Zhang, B.; Shi, M.; Xu, S.; Zhang, H.; Li, Y.; Hu, D. Analysis on Changes and Influencing Factors of the Intestinal Microbiota of Alpine Musk Deer between the Place of Origin and Migration. Animals 2023, 13, 3791. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Gao, H.; Qin, W.; Song, P.; Wang, H.; Zhang, J.; Liu, D.; Wang, D.; Zhang, T. Marked Seasonal Variation in Structure and Function of Gut Microbiota in Forest and Alpine Musk Deer. Front. Microbiol. 2021, 12, 699797. [Google Scholar] [CrossRef]
- Hu, X.; Liu, G.; Li, Y.; Wei, Y.; Lin, S.; Liu, S.; Zheng, Y.; Hu, D. High-Throughput Analysis Reveals Seasonal Variation of the Gut Microbiota Composition Within Forest Musk Deer (Moschus berezovskii). Front. Microbiol. 2018, 9, 1674. [Google Scholar] [CrossRef] [PubMed]
- Rabee, A.E.; El Rahman, T.A.; Lamara, M. Changes in the bacterial community colonizing extracted and non-extracted tannin-rich plants in the rumen of dromedary camels. PLoS ONE 2023, 18, e0282889. [Google Scholar] [CrossRef] [PubMed]
- Hinsu, A.T.; Tulsani, N.J.; Panchal, K.J.; Pandit, R.J.; Jyotsana, B.; Dafale, N.A.; Patil, N.V.; Purohit, H.J.; Joshi, C.G.; Jakhesara, S.J. Characterizing rumen microbiota and CAZyme profile of Indian dromedary camel (Camelus dromedarius) in response to different roughages. Sci. Rep. 2021, 11, 9400. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ren, X.; Wu, H.; Hu, H.; Cheng, C.; Du, M.; Huang, Y.; Zhao, X.; Wang, L.; Yi, L.; et al. Diversity and functional prediction of fungal communities in different segments of mongolian horse gastrointestinal tracts. BMC Microbiol. 2023, 23, 253. [Google Scholar] [CrossRef]
- O’Donnell, M.M.; Harris, H.M.; Ross, R.P.; O’Toole, P.W. Core fecal microbiota of domesticated herbivorous ruminant, hindgut fermenters, and monogastric animals. Microbiologyopen 2017, 6, e00509. [Google Scholar] [CrossRef]
- Janis, C. The evolutionary strategy of the equidae and the origins of rumen and cecal digestion. Evolution 1976, 30, 757–774. [Google Scholar] [CrossRef]
- Sha, Y.; Yu, J.; Xia, D.; Zhang, Y.; Liu, J.; Wang, H. Remodeling of intestinal bacterial community and metabolome of Dezhou donkey induced by corn silage. Sci. Rep. 2024, 14, 17032. [Google Scholar] [CrossRef]
- Guo, R.; Zhang, W.; Shen, W.; Zhang, G.; Xie, T.; Li, L.; Jinmei, J.; Liu, Y.; Kong, F.; Guo, B.; et al. Analysis of gut microbiota in chinese donkey in different regions using metagenomic sequencing. BMC Genom. 2023, 24, 524. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, C.; Wang, Y.; Du, M.; Zhang, G.; Lee, Y. Dietary Energy Level Impacts the Performance of Donkeys by Manipulating the Gut Microbiome and Metabolome. Front. Vet. Sci. 2021, 8, 694357. [Google Scholar] [CrossRef]
- Naumova, E.I.; Zharova, G.K.; Chistova, T.Y.; Kuznetsova, T.A. The Effect of Coprophagy on the Size of Plant Fibers in the Digestive Tract of Hares Lepus europaeus and L. timidus (Lagomorpha, Leporidae). Biol. Bull. 2015, 42, 426–431. [Google Scholar] [CrossRef]
- Abecia, L.; Rodríguez-Romero, N.; Yañez-Ruiz, D.R.; Fondevila, M. Biodiversity and fermentative activity of caecal microbial communities in wild and farm rabbits from Spain. Anaerobe 2012, 18, 344–349. [Google Scholar] [CrossRef]
- Amiry, A.F.; Kigata, T.; Shibata, H. Wall thickness and mucous cell distribution in the rabbit large intestine. J. Vet. Med. Sci. 2019, 81, 990–999. [Google Scholar] [CrossRef]
- Combes, S.; Gidenne, T.; Cauquil, L.; Bouchez, O.; Fortun-Lamothe, L. Coprophagous behavior of rabbit pups affects implantation of cecal microbiota and health status. J. Anim. Sci. 2014, 92, 652–665. [Google Scholar] [CrossRef]
- Cauquil, L.; Beaumont, M.; Schmaltz-Panneau, B.; Liaubet, L.; Lippi, Y.; Naylies, C.; Bluy, L.; Poli, M.; Gress, L.; Lencina, C.; et al. Coprophagia in early life tunes expression of immune genes after weaning in rabbit ileum. Sci. Rep. 2024, 14, 8898. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; He, H.; Chen, M.; Ni, M.; Guo, C.; Wan, Z.; Zhou, J.; Wang, Z.; Wang, Y.; Cai, H.; et al. Novel mechanism of Clostridium butyricum alleviated coprophagy prevention-induced intestinal inflammation in rabbit. Int. Immunopharmacol. 2024, 130, 111773. [Google Scholar] [CrossRef]
- Li, R.; Li, X.; Huang, T.; Wang, Y.; Xue, M.; Sun, S.; Yan, D.; Song, G.; Sun, G.; Li, M. Influence of cecotrophy on fat metabolism mediated by caecal microorganisms in New Zealand white rabbits. J. Anim. Physiol. Anim. Nutr. 2020, 104, 749–757. [Google Scholar] [CrossRef]
- Zeng, B.; Han, S.; Wang, P.; Wen, B.; Jian, W.; Guo, W.; Yu, Z.; Du, D.; Fu, X.; Kong, F.; et al. The bacterial communities associated with fecal types and body weight of rex rabbits. Sci. Rep. 2015, 5, 9342. [Google Scholar] [CrossRef] [PubMed]
- Velasco-Galilea, M.; Piles, M.; Viñas, M.; Rafel, O.; González-Rodríguez, O.; Guivernau, M.; Sánchez, J.P. Rabbit Microbiota Changes Throughout the Intestinal Tract. Front. Microbiol. 2018, 9, 2144. [Google Scholar] [CrossRef]
- Ye, D.; Ding, X.; Pang, S.; Gan, Y.; Li, Z.; Gan, Q.; Fang, S. Seasonal Variations in Production Performance, Health Status, and Gut Microbiota of Meat Rabbit Reared in Semi-Confined Conditions. Animals 2023, 14, 113. [Google Scholar]
- Handelsman, J.; Rondon, M.R.; Brady, S.F.; Clardy, J.; Goodman, R.M. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 1998, 5, R245–R249. [Google Scholar] [CrossRef]
- Fox, J.D.; Sims, A.; Ross, M.; Bettag, J.; Wilder, A.; Natrop, D.; Borsotti, A.; Kolli, S.; Mehta, S.; Verma, H.; et al. Bioinformatic Methodologies in Assessing Gut Microbiota. Microbiol. Res. 2024, 15, 2554–2574. [Google Scholar] [CrossRef]
- Bature, I.; Liang, Z.; Xiaohu, W.; Yang, F.; Yang, Y.; Dong, P.; Ding, X. Isolation, cloning, and characterization of a novel GH5 cellulase from yak rumen metagenome for enhanced lignocellulose hydrolysis in biofuel production and ruminant feed utilization. Enzym. Microb. Technol. 2025, 191, 110737. [Google Scholar] [CrossRef]
- Han, H.; Ji, M.; Li, Y.; Gong, X.; Song, W.; Zhou, J.; Ma, K.; Zhou, Y.; Liu, X.; Wang, M.; et al. Tracing non-fungal eukaryotic diversity via shotgun metagenomes in the complex mudflat intertidal zones. mSystems 2025, 10, e0041325. [Google Scholar] [CrossRef]
- Pavlopoulos, G.A.; Baltoumas, F.A.; Liu, S.; Selvitopi, O.; Camargo, A.P.; Nayfach, S.; Azad, A.; Roux, S.; Call, L.; Ivanova, N.N.; et al. Unraveling the functional dark matter through global metagenomics. Nature 2023, 622, 594–602. [Google Scholar] [CrossRef] [PubMed]
- Breitwieser, F.P.; Lu, J.; Salzberg, S.L. A review of methods and databases for metagenomic classification and assembly. Brief. Bioinform. 2019, 20, 1125–1136. [Google Scholar] [CrossRef]
- Frioux, C.; Singh, D.; Korcsmaros, T.; Hildebrand, F. From bag-of-genes to bag-of-genomes: Metabolic modelling of communities in the era of metagenome-assembled genomes. Comput. Struct. Biotechnol. J. 2020, 18, 1722–1734. [Google Scholar] [CrossRef]
- de Oliveira Martins, L.; Page, A.J.; Mather, A.E.; Charles, I.G. Taxonomic resolution of the ribosomal RNA operon in bacteria: Implications for its use with long-read sequencing. NAR Genom. Bioinform. 2020, 2, lqz016. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Ma, J.; Kim, W.; Kim, J.; Belenky, P.; Lee, I. Genome-resolved metagenomics: A game changer for microbiome medicine. Exp. Mol. Med. 2024, 56, 1501–1512. [Google Scholar] [CrossRef]
- Herbert, J.; Thompson, S.; Beckett, A.H.; Robson, S.C. Impact of microbiological molecular methodologies on adaptive sampling using nanopore sequencing in metagenomic studies. Environ. Microbiome 2025, 20, 47. [Google Scholar] [CrossRef]
- Orellana, L.H.; Krüger, K.; Sidhu, C.; Amann, R. Comparing genomes recovered from time-series metagenomes using long- and short-read sequencing technologies. Microbiome 2023, 11, 105. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Yang, C.; Sun, Y.; Igarashi, Y.; Jin, T.; Luo, F. PacBio Long Reads Improve Metagenomic Assemblies, Gene Catalogs, and Genome Binning. Front. Genet. 2020, 11, 516269. [Google Scholar] [CrossRef]
- Haro-Moreno, J.M.; López-Pérez, M.; Rodriguez-Valera, F. Enhanced Recovery of Microbial Genes and Genomes From a Marine Water Column Using Long-Read Metagenomics. Front. Microbiol. 2021, 12, 708782. [Google Scholar] [CrossRef] [PubMed]
- Goussarov, G.; Mysara, M.; Vandamme, P.; Van Houdt, R. Introduction to the principles and methods underlying the recovery of metagenome-assembled genomes from metagenomic data. Microbiologyopen 2022, 11, e1298. [Google Scholar] [CrossRef]
- Mallawaarachchi, V.; Wickramarachchi, A.; Xue, H.; Papudeshi, B.; Grigson, S.R.; Bouras, G.; Prahl, R.E.; Kaphle, A.; Verich, A.; Talamantes-Becerra, B.; et al. Solving genomic puzzles: Computational methods for metagenomic binning. Brief. Bioinform. 2024, 25, bbae372. [Google Scholar] [CrossRef]
- Stewart, R.D.; Auffret, M.D.; Warr, A.; Walker, A.W.; Roehe, R.; Watson, M. Compendium of 4941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat. Biotechnol. 2019, 37, 953–961. [Google Scholar] [CrossRef]
- Ghurye, J.S.; Cepeda-Espinoza, V.; Pop, M. Metagenomic Assembly: Overview, Challenges and Applications. Yale J. Biol. Med. 2016, 89, 353–362. [Google Scholar]
- Olson, N.D.; Treangen, T.J.; Hill, C.M.; Cepeda-Espinoza, V.; Ghurye, J.; Koren, S.; Pop, M. Metagenomic assembly through the lens of validation: Recent advances in assessing and improving the quality of genomes assembled from metagenomes. Brief. Bioinform. 2019, 20, 1140–1150. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Liang, J.L.; Huang, L.N.; Mengoni, A.; Shu, W.S. Metagenomic Assembly: Reconstructing Genomes from Metagenomes. Bact. Pangenomics: Methods Protoc. 2021, 2242, 139–152. [Google Scholar]
- Wu, C.; Yin, Y.; Zhu, L.; Zhang, Y.; Li, Y.Z. Metagenomic sequencing-driven multidisciplinary approaches to shed light on the untapped microbial natural products. Drug Discov. Today 2022, 27, 730–742. [Google Scholar] [CrossRef]
- Datta, S.; Rajnish, K.N.; Samuel, M.S.; Pugazlendhi, A.; Selvarajan, E. Metagenomic applications in microbial diversity, bioremediation, pollution monitoring, enzyme and drug discovery. A review. Environ. Chem. Lett. 2020, 18, 1229–1241. [Google Scholar] [CrossRef]
- Yang, S.; Gao, X.; Meng, J.; Zhang, A.; Zhou, Y.; Long, M.; Li, B.; Deng, W.; Jin, L.; Zhao, S.; et al. Metagenomic Analysis of Bacteria, Fungi, Bacteriophages, and Helminths in the Gut of Giant Pandas. Front. Microbiol. 2018, 9, 1717. [Google Scholar] [CrossRef]
- Wilson, M.R.; Sample, H.A.; Zorn, K.C.; Arevalo, S.; Yu, G.; Neuhaus, J.; Federman, S.; Stryke, D.; Briggs, B.; Langelier, C.; et al. Clinical Metagenomic Sequencing for Diagnosis of Meningitis and Encephalitis. N. Engl. J. Med. 2019, 380, 2327–2340. [Google Scholar] [CrossRef]
- Liang, G.; Bushman, F.D. The human virome: Assembly, composition and host interactions. Nat. Rev. Microbiol. 2021, 19, 514–527. [Google Scholar] [CrossRef]
- Bruggeling, C.E.; Garza, D.R.; Achouiti, S.; Mes, W.; Dutilh, B.E.; Boleij, A. Optimized bacterial DNA isolation method for microbiome analysis of human tissues. Microbiologyopen 2021, 10, e1191. [Google Scholar] [CrossRef]
- Oechslin, C.P.; Lenz, N.; Liechti, N.; Ryter, S.; Agyeman, P.; Bruggmann, R.; Leib, S.L.; Beuret, C.M. Limited Correlation of Shotgun Metagenomics Following Host Depletion and Routine Diagnostics for Viruses and Bacteria in Low Concentrated Surrogate and Clinical Samples. Front. Cell. Infect. Microbiol. 2018, 8, 375. [Google Scholar] [CrossRef]
- Nelson, M.T.; Pope, C.E.; Marsh, R.L.; Wolter, D.J.; Weiss, E.J.; Hager, K.R.; Vo, A.T.; Brittnacher, M.J.; Radey, M.C.; Hayden, H.S.; et al. Human and Extracellular DNA Depletion for Metagenomic Analysis of Complex Clinical Infection Samples Yields Optimized Viable Microbiome Profiles. Cell Rep. 2019, 26, 2227–2240.e5. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Mao, X.; Yang, J.; Chen, X.; Mao, F.; Xu, Y. dbCAN: A web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012, 40, W445–W451. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Zheng, J.; Hu, B.; Zhang, X.; Ge, Q.; Yan, Y.; Akresi, J.; Piyush, V.; Huang, L.; Yin, Y. dbCAN-seq update: CAZyme gene clusters and substrates in microbiomes. Nucleic Acids Res. 2023, 51, D557–D563. [Google Scholar] [CrossRef] [PubMed]
- Ameri, R.; García, J.L.; Derenfed, A.B.; Pradel, N.; Neifar, S.; Mhiri, S.; Mezghanni, M.; Jaouadi, N.Z.; Barriuso, J.; Bejar, S. Genome sequence and Carbohydrate Active Enzymes (CAZymes) repertoire of the thermophilic Caldicoprobacter algeriensis TH7C1T. Microb. Cell Factories 2022, 21, 91. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Ge, Q.; Yan, Y.; Zhang, X.; Huang, L.; Yin, Y. dbCAN3: Automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 2023, 51, W115–W121. [Google Scholar] [CrossRef]
- Ojo-Okunola, A.; Claassen-Weitz, S.; Mwaikono, K.S.; Gardner-Lubbe, S.; Zar, H.J.; Nicol, M.P.; Toit, E.D. The Influence of DNA Extraction and Lipid Removal on Human Milk Bacterial Profiles. Methods Protoc. 2020, 3, 39. [Google Scholar] [CrossRef]
- Amar, Y.; Lagkouvardos, I.; Silva, R.L.; Ishola, O.A.; Foesel, B.U.; Kublik, S.; Schöler, A.; Niedermeier, S.; Bleuel, R.; Zink, A.; et al. Pre-digest of unprotected DNA by Benzonase improves the representation of living skin bacteria and efficiently depletes host DNA. Microbiome 2021, 9, 123. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, G.; Lau, H.C.; Yu, J. Metagenomic Sequencing for Microbial DNA in Human Samples: Emerging Technological Advances. Int. J. Mol. Sci. 2022, 23, 2181. [Google Scholar] [CrossRef]
- Simon, C.; Daniel, R. Metagenomic analyses: Past and future trends. Appl. Environ. Microbiol. 2011, 77, 1153–1161. [Google Scholar] [CrossRef]
- Woloszynek, S.; Zhao, Z.; Ditzler, G.; Price, J.R.; Reichenberger, E.R.; Lan, Y.; Chen, J.; Earl, J.; Langroodi, S.K.; Ehrlich, G. Analysis methods for shotgun metagenomics. In Theoretical and Applied Aspects of Systems Biology; Springer: Cham, Switzerland, 2018; pp. 71–112. [Google Scholar]
- Jia, L.; Wu, Y.; Dong, Y.; Chen, J.; Chen, W.H.; Zhao, X.M. A survey on computational strategies for genome-resolved gut metagenomics. Brief. Bioinform. 2023, 24, bbad162. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Li, H.; Zhang, L.; Mu, W.; Zhang, Y.; Chen, T.; Wu, J.; Tang, H.; Zheng, S.; Liu, Y.; et al. Generic Diagramming Platform (GDP): A comprehensive database of high-quality biomedical graphics. Nucleic Acids Res. 2025, 53, D1670–D1676. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Fan, Z.; Li, J.; Wang, X.; Lan, Y.; Yue, B.; He, M.; Zhang, A.; Li, J. Assembly of novel microbial genomes from gut metagenomes of rhesus macaque (Macaca mulatta). Gut Microbes 2023, 15, 2188848. [Google Scholar] [CrossRef]
- Feng, Y.; Wang, Y.; Zhu, B.; Gao, G.F.; Guo, Y.; Hu, Y. Metagenome-assembled genomes and gene catalog from the chicken gut microbiome aid in deciphering antibiotic resistomes. Commun. Biol. 2021, 4, 1305. [Google Scholar] [CrossRef] [PubMed]
- Saheb Kashaf, S.; Almeida, A.; Segre, J.A.; Finn, R.D. Recovering prokaryotic genomes from host-associated, short-read shotgun metagenomic sequencing data. Nat. Protoc. 2021, 16, 2520–2541. [Google Scholar] [CrossRef]
- Ma, T.; Zhuang, Y.; Lu, W.; Tu, Y.; Diao, Q.; Fan, X.; Zhang, N. Seven hundred and ninety-seven metagenome-assembled genomes from the goat rumen during early life. Sci. Data 2024, 11, 897. [Google Scholar] [CrossRef]
- Conteville, L.C.; Silva, J.V.D.; Andrade, B.G.N.; Coutinho, L.L.; Palhares, J.C.P.; Regitano, L.C.A. Recovery of metagenome-assembled genomes from the rumen and fecal microbiomes of Bos indicus beef cattle. Sci. Data 2024, 11, 1385. [Google Scholar] [CrossRef]
- Denman, S.E.; Morgavi, D.P.; McSweeney, C.S. Review: The application of omics to rumen microbiota function. Animal 2018, 12, s233–s245. [Google Scholar] [CrossRef]
- Chesworth, J.; Stuchbury, T.; Scaife, J. Digestion and Absorption in Ruminants and Non-Ruminants. In An Introduction to Agricultural Biochemistry; Springer: Cham, Switzerland, 1998; pp. 395–411. [Google Scholar]
- Chen, X.; Yan, F.; Liu, T.; Zhang, Y.; Li, X.; Wang, M.; Zhang, C.; Xu, X.; Deng, L.; Yao, J.; et al. Ruminal Microbiota Determines the High-Fiber Utilization of Ruminants: Evidence from the Ruminal Microbiota Transplant. Microbiol. Spectr. 2022, 10, e0044622. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Forano, E.; Martin, C.; Newbold, C.J. Microbial ecosystem and methanogenesis in ruminants. Animal 2010, 4, 1024–1036. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial Community Structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef]
- Dal Pont, G.C.; Eyng, C.; Bortoluzzi, C.; Kogut, M.H. Enzymes and gut health in monogastric animals: Effects beyond digestibility. In Gut Microbiota, Immunity, and Health in Production Animals; Springer: Cham, Switzerland, 2022; pp. 33–55. [Google Scholar]
- Wang, Z.; He, H.; Chen, M.; Ni, M.; Yuan, D.; Cai, H.; Chen, Z.; Li, M.; Xu, H. Impact of coprophagy prevention on the growth performance, serum biochemistry, and intestinal microbiome of rabbits. BMC Microbiol. 2023, 23, 125. [Google Scholar] [CrossRef] [PubMed]
- Bagwan, W.; Durgawale, P.; Jadhav, M.; Vidyapeeth, K. Exploring the gut microbiota of ruminants and its impact on digestive efficiency and methane emissions in livestock production systems. Afr. J. Biol. Sci. 2024, 6, 2226–2236. [Google Scholar]
- Garber, A.; Hastie, P.; Murray, J.A. Factors Influencing Equine Gut Microbiota: Current Knowledge. J. Equine Vet. Sci. 2020, 88, 102943. [Google Scholar] [CrossRef]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef]
- Costa, M.C.; Silva, G.; Ramos, R.V.; Staempfli, H.R.; Arroyo, L.G.; Kim, P.; Weese, J.S. Characterization and comparison of the bacterial microbiota in different gastrointestinal tract compartments in horses. Vet. J. 2015, 205, 74–80. [Google Scholar] [CrossRef]
- Verbeek, E.; Keeling, L.; Landberg, R.; Lindberg, J.E.; Dicksved, J. The gut microbiota and microbial metabolites are associated with tail biting in pigs. Sci. Rep. 2021, 11, 20547. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.D.; Martinez-Fernandez, G.; Padmanabha, J.; Long, R.; Denman, S.E.; McSweeney, C.S. Methanogen diversity in indigenous and introduced ruminant species on the Tibetan plateau. Archaea 2016, 2016, 5916067. [Google Scholar] [CrossRef] [PubMed]
- Misiukiewicz, A.; Gao, M.; Filipiak, W.; Cieslak, A.; Patra, A.; Szumacher-Strabel, M. Methanogens and methane production in the digestive systems of nonruminant farm animals. Animal 2021, 15, 100060. [Google Scholar] [CrossRef] [PubMed]
- Lambie, S.C.; Kelly, W.J.; Leahy, S.C.; Li, D.; Reilly, K.; McAllister, T.A.; Valle, E.R.; Attwood, G.T.; Altermann, E. The complete genome sequence of the rumen methanogen Methanosarcina barkeri CM1. Stand. Genom. Sci. 2015, 10, 57. [Google Scholar] [CrossRef]
- Betancur-Murillo, C.L.; Aguilar-Marín, S.B.; Jovel, J. Prevotella: A key player in ruminal metabolism. Microorganisms 2022, 11, 1. [Google Scholar] [CrossRef]
- Niwińska, B. Digestion in ruminants. In Carbohydrates-Comprehensive Studies on Glycobiology and Glycotechnology; InTech Open: Rijeka, Croatia, 2012; Volume 10, pp. 245–258. [Google Scholar]
- Mackenzie, A.; Naas, A.; Kracun, S.; Schückel, J.; Fangel, J.; Agger, J.W.; Willats, W.; Eijsink, V.; Pope, P.B. A polysaccharide utilization locus from an uncultured Bacteroidetes phylotype suggests ecological adaptation and substrate versatility. Appl. Environ. Microbiol. 2015, 81, 187–195. [Google Scholar] [CrossRef]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Johnston, L.J.; Noll, S.; Renteria, A.; Shurson, J. Feeding by-products high in concentration of fiber to nonruminants. In Proceedings of the National Symposium on Alternative Feeds for Livestock and Poultry, Kansas City, MO, USA, 4 November 2003. [Google Scholar]
- Russell, J.B.; Muck, R.E.; Weimer, P.J. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 2009, 67, 183–197. [Google Scholar] [CrossRef]
- Niba, A.; Beal, J.; Kudi, A.; Brooks, P. Bacterial fermentation in the gastrointestinal tract of non-ruminants: Influence of fermented feeds and fermentable carbohydrates. Trop. Anim. Health Prod. 2009, 41, 1393. [Google Scholar] [CrossRef]
- Chaucheyras-Durand, F.; Chevaux, E.; Martin, C.; Forano, E. Rumen pH, Fibre Degradation, and Microbiota According to the Diet; Intech: Sao Paulo, Brazil, 2012. [Google Scholar]
- Guo, P.; Zhang, K.; Ma, X.; He, P. Clostridium species as probiotics: Potentials and challenges. J. Anim. Sci. Biotechnol. 2020, 11, 24. [Google Scholar] [CrossRef]
- Zeitz, J.O.; Amelchanka, S.L.; Michałowski, T.; Wereszka, K.; Meile, L.; Hartnack, S.; Kreuzer, M.; Soliva, C.R. Effect of the rumen ciliates Entodinium caudatum, Epidinium ecaudatum and Eudiplodinium maggii, and combinations thereof, on ruminal fermentation and total tract digestion in sheep. Arch. Anim. Nutr. 2012, 66, 180–199. [Google Scholar] [CrossRef]
- Sahu, N.; Kamra, D. Microbial eco-system of the gastro-intestinal tract of wild herbivorous animals. J. Appl. Anim. Res. 2002, 21, 207–230. [Google Scholar] [CrossRef]
- Wei, Y.-Q.; Long, R.-J.; Yang, H.; Yang, H.-J.; Shen, X.-H.; Shi, R.-F.; Wang, Z.-Y.; Du, J.-G.; Qi, X.-J.; Ye, Q.-H. Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 2016, 39, 158–164. [Google Scholar] [CrossRef]
- Cheng, Y.; Shi, Q.; Sun, R.; Liang, D.; Li, Y.; Li, Y.; Jin, W.; Zhu, W. The biotechnological potential of anaerobic fungi on fiber degradation and methane production. World J. Microbiol. Biotechnol. 2018, 34, 155. [Google Scholar] [CrossRef] [PubMed]
- Sirohi, S.K.; Choudhury, P.K.; Dagar, S.S.; Puniya, A.K.; Singh, D. Isolation, characterization and fibre degradation potential of anaerobic rumen fungi from cattle. Ann. Microbiol. 2013, 63, 1187–1194. [Google Scholar] [CrossRef]
- Teunissen, M.; Smits, A.; den Camp, H.O.; Huis in, J.; Vogels, G. Fermentation of cellulose and production of cellulolytic and xylanolytic enzymes by anaerobic fungi from ruminant and non-ruminant herbivores. Arch. Microbiol. 1991, 156, 290–296. [Google Scholar] [CrossRef]
- Cao, Y.; Feng, T.; Wu, Y.; Xu, Y.; Du, L.; Wang, T.; Luo, Y.; Wang, Y.; Li, Z.; Xuan, Z.; et al. The multi-kingdom microbiome of the goat gastrointestinal tract. Microbiome 2023, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Guo, J.; Shi, W.; Tong, W.; Li, X.; Yang, B.; Xiang, Z.; Qin, C. Metagenomic analysis reveals the community composition of the microbiome in different segments of the digestive tract in donkeys and cows: Implications for microbiome research. BMC Microbiol. 2024, 24, 530. [Google Scholar] [CrossRef]
- Hui, T.K.L.; Lo, I.C.N.; Wong, K.K.W.; Tsang, C.T.T.; Tsang, L.M. Metagenomic analysis of gut microbiome illuminates the mechanisms and evolution of lignocellulose degradation in mangrove herbivorous crabs. BMC Microbiol. 2024, 24, 57. [Google Scholar] [CrossRef]
- Hu, D.; Zhang, T.; He, S.; Pu, T.; Yin, Y.; Hu, Y. Mining metagenomic data to gain a new insight into the gut microbial biosynthetic potential in placental mammals. Microbiol. Spectr. 2024, 12, e00864-24. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Zhang, Y.; Thompson, K.N.; Branck, T.; Yan, Y.; Nguyen, L.H.; Franzosa, E.A.; Huttenhower, C. Metatranscriptomics for the human microbiome and microbial community functional profiling. Annu. Rev. Biomed. Data Sci. 2021, 4, 279–311. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Edet, U.; Antai, S.; Brooks, A.; Asitok, A.; Enya, O.; Japhet, F. An overview of cultural, molecular and metagenomic techniques in description of microbial diversity. J. Adv. Microbiol. 2017, 7, 1–19. [Google Scholar] [CrossRef]
- Garza, D.R.; Dutilh, B.E. From cultured to uncultured genome sequences: Metagenomics and modeling microbial ecosystems. Cell. Mol. Life Sci. 2015, 72, 4287–4308. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef]
- Singh, B.; Mal, G.; Gautam, S.K.; Mukesh, M. Metagenomics for utilizing herbivore gut potential. In Advances in Animal Biotechnology; Springer: Cham, Switzerland, 2019; pp. 3–15. [Google Scholar]
- Nwachukwu, B.C.; Babalola, O.O. Metagenomics: A tool for exploring key microbiome with the potentials for improving sustainable agriculture. Front. Sustain. Food Syst. 2022, 6, 886987. [Google Scholar] [CrossRef]
- Hoon, K.K. Effects of TMR and Separate Feeding System on Ruminal Methane Production, Total Digestibility, Rumen Metabolic and Microbial Profile. Ph.D. Thesis, Graduate School of Seoul National Universit, Seoul, Republic of Korea, 2017. [Google Scholar]
- Stevens, C.E.; Hume, I.D. Contributions of microbes in vertebrate gastrointestinal tract to production and conservation of nutrients. Physiol. Rev. 1998, 78, 393–427. [Google Scholar] [CrossRef]
- He, Z.; Dong, H. The roles of short-chain fatty acids derived from colonic bacteria fermentation of non-digestible carbohydrates and exogenous forms in ameliorating intestinal mucosal immunity of young ruminants. Front. Immunol. 2023, 14, 1291846. [Google Scholar] [CrossRef] [PubMed]
- Di Bella, J.M.; Bao, Y.; Gloor, G.B.; Burton, J.P.; Reid, G. High throughput sequencing methods and analysis for microbiome research. J. Microbiol. Methods 2013, 95, 401–414. [Google Scholar] [CrossRef]
- Helbling, D.E.; Ackermann, M.; Fenner, K.; Kohler, H.-P.E.; Johnson, D.R. The activity level of a microbial community function can be predicted from its metatranscriptome. ISME J. 2012, 6, 902–904. [Google Scholar] [CrossRef]
- Denman, S.E.; McSweeney, C.S. The early impact of genomics and metagenomics on ruminal microbiology. Annu. Rev. Anim. Biosci. 2015, 3, 447–465. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M. Addressing global ruminant agricultural challenges through understanding the rumen microbiome: Past, present, and future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Saborío-Montero, A.; Gutiérrez-Rivas, M.; García-Rodríguez, A.; Atxaerandio, R.; Goiri, I.; de Maturana, E.L.; Jiménez-Montero, J.A.; Alenda, R.; González-Recio, O. Structural equation models to disentangle the biological relationship between microbiota and complex traits: Methane production in dairy cattle as a case of study. J. Anim. Breed. Genet. 2020, 137, 36–48. [Google Scholar] [CrossRef]
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Ding, H.; Zhang, X.; Ta, N.; Zhao, J.; Zhang, Q.; Liu, H.; Sun, M.; Zhang, X. Dynamic changes in the gastrointestinal microbial communities of Gangba sheep and analysis of their functions in plant biomass degradation at high altitude. Microbiome 2025, 13, 17. [Google Scholar] [CrossRef]
- Pandit, R.J.; Hinsu, A.T.; Patel, S.H.; Jakhesara, S.J.; Koringa, P.G.; Bruno, F.; Psifidi, A.; Shah, S.; Joshi, C.G. Microbiota composition, gene pool and its expression in Gir cattle (Bos indicus) rumen under different forage diets using metagenomic and metatranscriptomic approaches. Syst. Appl. Microbiol. 2018, 41, 374–385. [Google Scholar] [CrossRef]
- Pitta, D.W.; Indugu, N.; Baker, L.; Vecchiarelli, B.; Attwood, G. Symposium review: Understanding diet–microbe interactions to enhance productivity of dairy cows. J. Dairy Sci. 2018, 101, 7661–7679. [Google Scholar] [CrossRef]
- Shakya, M.; Lo, C.-C.; Chain, P.S. Advances and challenges in metatranscriptomic analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef]
- Li, Q.; Ma, Z.; Huo, J.; Zhang, X.; Wang, R.; Zhang, S.; Jiao, J.; Dong, X.; Janssen, P.H.; Ungerfeld, E.M. Distinct microbial hydrogen and reductant disposal pathways explain interbreed variations in ruminant methane yield. ISME J. 2024, 18, wrad016. [Google Scholar] [CrossRef]
- Kuziel, G.A.; Rakoff-Nahoum, S. The gut microbiome. Curr. Biol. 2022, 32, R257–R264. [Google Scholar] [CrossRef]
- Ley, R.E.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Biagi, E.; Nylund, L.; Candela, M.; Ostan, R.; Bucci, L.; Pini, E.; Nikkïla, J.; Monti, D.; Satokari, R.; Franceschi, C.; et al. Through ageing, and beyond: Gut microbiota and inflammatory status in seniors and centenarians. PLoS ONE 2010, 5, e10667. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Metcalf, J.L.; Song, S.J.; Morton, J.T.; Weiss, S.; Seguin-Orlando, A.; Joly, F.; Feh, C.; Taberlet, P.; Coissac, E.; Amir, A.; et al. Evaluating the impact of domestication and captivity on the horse gut microbiome. Sci. Rep. 2017, 7, 15497. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, M.; Liu, J.; Zhu, W. Characterising the bacterial microbiota across the gastrointestinal tracts of dairy cattle: Membership and potential function. Sci. Rep. 2015, 5, 16116. [Google Scholar] [CrossRef]
- de Oliveira, M.N.; Jewell, K.A.; Freitas, F.S.; Benjamin, L.A.; Tótola, M.R.; Borges, A.C.; Moraes, C.A.; Suen, G. Characterizing the microbiota across the gastrointestinal tract of a Brazilian Nelore steer. Vet. Microbiol. 2013, 164, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Xie, F.; Jin, W.; Si, H.; Yuan, Y.; Tao, Y.; Liu, J.; Wang, X.; Yang, C.; Li, Q.; Yan, X.; et al. An integrated gene catalog and over 10,000 metagenome-assembled genomes from the gastrointestinal microbiome of ruminants. Microbiome 2021, 9, 137. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, H.; Hu, L.; Zhang, G.; Lu, H.; Luo, H.; Zhao, S.; Zhu, H.; Wang, Y. Characterization of the Microbial Communities along the Gastrointestinal Tract in Crossbred Cattle. Animals 2022, 12, 825. [Google Scholar] [CrossRef]
- Jiao, J.; Wu, J.; Zhou, C.; He, Z.; Tan, Z.; Wang, M. Ecological niches and assembly dynamics of diverse microbial consortia in the gastrointestine of goat kids. ISME J. 2024, 18, wrae002. [Google Scholar] [CrossRef]
- Akram, A.; Shahin, F.; Asif, I.; Bilal, A.; Abbas, K.J.; Younas, E. Exploring the role of gut bacteria in digestive system of cow. J. Med. Health Sci. Rev. 2025, 2. [Google Scholar] [CrossRef]
- Liu, G.; Bou, G.; Su, S.; Xing, J.; Qu, H.; Zhang, X.; Wang, X.; Zhao, Y.; Dugarjaviin, M. Microbial diversity within the digestive tract contents of Dezhou donkeys. PLoS ONE 2019, 14, e0226186. [Google Scholar] [CrossRef]
- Mi, J.; Jing, X.; Ma, C.; Shi, F.; Cao, Z.; Yang, X.; Yang, Y.; Kakade, A.; Wang, W.; Long, R. A metagenomic catalogue of the ruminant gut archaeome. Nat. Commun. 2024, 15, 9609. [Google Scholar] [CrossRef]
- He, J.; Yi, L.; Hai, L.; Ming, L.; Gao, W.; Ji, R. Characterizing the bacterial microbiota in different gastrointestinal tract segments of the Bactrian camel. Sci. Rep. 2018, 8, 654. [Google Scholar] [CrossRef] [PubMed]
- Langda, S.; Zhang, C.; Zhang, K.; Gui, B.; Ji, D.; Deji, C.; Cuoji, A.; Wang, X.; Wu, Y. Diversity and Composition of Rumen Bacteria, Fungi, and Protozoa in Goats and Sheep Living in the Same High-Altitude Pasture. Animals 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Glendinning, L.; Genç, B.; Wallace, R.J.; Watson, M. Metagenomic analysis of the cow, sheep, reindeer and red deer rumen. Sci. Rep. 2021, 11, 1990. [Google Scholar] [CrossRef] [PubMed]
- Gharechahi, J.; Sarikhan, S.; Han, J.L.; Ding, X.Z.; Salekdeh, G.H. Functional and phylogenetic analyses of camel rumen microbiota associated with different lignocellulosic substrates. NPJ Biofilms Microbiomes 2022, 8, 46. [Google Scholar] [CrossRef]
- Reed, K.J.; Kunz, I.G.Z.; Scare, J.A.; Nielsen, M.K.; Turk, P.J.; Coleman, R.J.; Coleman, S.J. The pelvic flexure separates distinct microbial communities in the equine hindgut. Sci. Rep. 2021, 11, 4332. [Google Scholar] [CrossRef]
- Husso, A.; Jalanka, J.; Alipour, M.J.; Huhti, P.; Kareskoski, M.; Pessa-Morikawa, T.; Iivanainen, A.; Niku, M. The composition of the perinatal intestinal microbiota in horse. Sci. Rep. 2020, 10, 441. [Google Scholar] [CrossRef]
- Lindenberg, F.; Krych, L.; Kot, W.; Fielden, J.; Frøkiær, H.; van Galen, G.; Nielsen, D.S.; Hansen, A.K. Development of the equine gut microbiota. Sci. Rep. 2019, 9, 14427. [Google Scholar] [CrossRef] [PubMed]
- McLoughlin, S.; Spillane, C.; Claffey, N.; Smith, P.E.; O’Rourke, T.; Diskin, M.G.; Waters, S.M. Rumen Microbiome Composition Is Altered in Sheep Divergent in Feed Efficiency. Front. Microbiol. 2020, 11, 1981. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhu, Y.; Wang, Z.; Yu, X.; Hu, R.; Wang, X.; Cao, G.; Zou, H.; Shah, A.M.; Peng, Q.; et al. Comparing the Bacterial Community in the Gastrointestinal Tracts Between Growth-Retarded and Normal Yaks on the Qinghai-Tibetan Plateau. Front. Microbiol. 2020, 11, 600516. [Google Scholar] [CrossRef]
- He, H.; Fang, C.; Liu, L.; Li, M.; Liu, W. Environmental Driving of Adaptation Mechanism on Rumen Microorganisms of Sheep Based on Metagenomics and Metabolomics Data Analysis. Int. J. Mol. Sci. 2024, 25, 10957. [Google Scholar] [CrossRef]
- Yang, L.; Zeng, X.; Qiao, S. Advances in research on solid-state fermented feed and its utilization: The pioneer of private customization for intestinal microorganisms. Anim. Nutr. 2021, 7, 905–916. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Tong, W.; Han, S.; Wu, M.; Li, P.; Li, Y.; Liang, Y. Effects of Dietary Supplementation with Lactobacillus reuteri Postbiotics on Growth Performance, Intestinal Flora Structure and Plasma Metabolome of Weaned Piglets. Animals 2025, 15, 204. [Google Scholar] [CrossRef]
- Xiang, S.; Ye, K.; Li, M.; Ying, J.; Wang, H.; Han, J.; Shi, L.; Xiao, J.; Shen, Y.; Feng, X.; et al. Xylitol enhances synthesis of propionate in the colon via cross-feeding of gut microbiota. Microbiome 2021, 9, 62. [Google Scholar] [CrossRef]
- Lin, L.; Lai, Z.; Zhang, J.; Zhu, W.; Mao, S. The gastrointestinal microbiome in dairy cattle is constrained by the deterministic driver of the region and the modified effect of diet. Microbiome 2023, 11, 10. [Google Scholar] [CrossRef]
- Begmatov, S.; Beletsky, A.V.; Mardanov, A.V.; Lukina, A.P.; Glukhova, L.B.; Karnachuk, O.V.; Ravin, N.V. Novel lineages of bacteria with reduced genomes from the gut of farm animals. mSphere 2025, 10, e00294-25. [Google Scholar] [CrossRef]
- Al-Masaudi, S.; El Kaoutari, A.; Drula, E.; Al-Mehdar, H.; Redwan, E.M.; Lombard, V.; Henrissat, B. A Metagenomics Investigation of Carbohydrate-Active Enzymes along the Gastrointestinal Tract of Saudi Sheep. Front. Microbiol. 2017, 8, 666. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, C.; Li, B.; Jiang, F.; Song, P.; Gu, H.; Gao, H.; Cai, Z.; Zhang, T. Gut microbiome and diet contribute to ecological niche differentiation between argali (Ovis ammon hodgsoni) and blue sheep (Pseudois nayaur) on the Qinghai-Tibet Plateau. Commun. Biol. 2025, 8, 930. [Google Scholar]
- Cai, C.; Xie, L.; Xing, J.; Lu, T.; Qi, X.; Li, L.; Chen, X.; Akhtar, M.F.; Jin, Y.; Liu, G. Effects of concentrate feeding sequence on VFA production, and cecal microbiota of Dezhou donkeys by metagenomic technology. Front. Vet. Sci. 2024, 11, 1401980. [Google Scholar] [CrossRef]
- Guo, R.; Zhang, S.; Chen, J.; Shen, W.; Zhang, G.; Wang, J.; Zhang, F.; Pan, Q.; Xie, T.; Ai, D.; et al. Comparison of gut microflora of donkeys in high and low altitude areas. Front. Microbiol. 2022, 13, 964799. [Google Scholar] [CrossRef]
- Nkosi, B.V.Z.; Padayachee, T.; Gront, D.; Nelson, D.R.; Syed, K. Contrasting Health Effects of Bacteroidetes and Firmicutes Lies in Their Genomes: Analysis of P450s, Ferredoxins, and Secondary Metabolite Clusters. Int. J. Mol. Sci. 2022, 23, 5057. [Google Scholar] [CrossRef]
- Khan, M.Z.; Li, Y.; Zhu, M.; Li, M.; Wang, T.; Zhang, Z.; Liu, W.; Ma, Q.; Wang, C. Advances in Donkey Disease Surveillance and Microbiome Characterization in China. Microorganisms 2025, 13, 749. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Han, Y.; Zhao, J.; Zhou, Z. Characterization of the microbial communities along the gastrointestinal tract of sheep by 454 pyrosequencing analysis. Asian-Australas. J. Anim. Sci. 2017, 30, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Xue, S.Y.; Ma, W.; Li, M.Y.; Meng, W.K.; Ding, Y.L.; Yang, B.; Lv, Y.R.; Chen, R.B.; Wu, Z.H.; Tunala, S.; et al. The Impact of Mycobacterium avium subsp. paratuberculosis on Intestinal Microbial Community Composition and Diversity in Small-Tail Han Sheep. Pathogens 2024, 13, 1118. [Google Scholar] [PubMed]
- Facchin, S.; Vitulo, N.; Calgaro, M.; Buda, A.; Romualdi, C.; Pohl, D.; Perini, B.; Lorenzon, G.; Marinelli, C.; D’Incà, R.; et al. Microbiota changes induced by microencapsulated sodium butyrate in patients with inflammatory bowel disease. Neurogastroenterol. Motil. 2020, 32, e13914. [Google Scholar] [CrossRef] [PubMed]
- Sicard, J.F.; Le Bihan, G.; Vogeleer, P.; Jacques, M.; Harel, J. Interactions of Intestinal Bacteria with Components of the Intestinal Mucus. Front. Cell. Infect. Microbiol. 2017, 7, 387. [Google Scholar] [CrossRef]
- Jia, L.; Wu, J.; Lei, Y.; Kong, F.; Zhang, R.; Sun, J.; Wang, L.; Li, Z.; Shi, J.; Wang, Y.; et al. Oregano Essential Oils Mediated Intestinal Microbiota and Metabolites and Improved Growth Performance and Intestinal Barrier Function in Sheep. Front. Immunol. 2022, 13, 908015. [Google Scholar] [CrossRef]
- Zhang, K.; He, C.; Wang, L.; Suo, L.; Guo, M.; Guo, J.; Zhang, T.; Xu, Y.; Lei, Y.; Liu, G.; et al. Compendium of 5810 genomes of sheep and goat gut microbiomes provides new insights into the glycan and mucin utilization. Microbiome 2024, 12, 104. [Google Scholar] [CrossRef]
- Zábranský, L.; Poborská, A.; Gálik, B.; Šoch, M.; Brož, P.; Kantor, M.; Kernerová, N.; Řezáč, I.; Rolinec, M.; Hanušovský, O.; et al. Influence of Probiotic Strains Bifidobacterium, Lactobacillus, and Enterococcus on the Health Status and Weight Gain of Calves, and the Utilization of Nitrogenous Compounds. Antibiotics 2022, 11, 1273. [Google Scholar] [CrossRef] [PubMed]
- Zafar, M.; Alam, S.; Sabir, M.; Saba, N.; Din, A.U.; Ahmad, R.; Khan, M.R.; Muhammad, A.; Dayisoylu, K.S. Isolation, characterization, bacteriocin production and biological potential of Bifidobacteria of ruminants. Anal. Biochem. 2022, 658, 114926. [Google Scholar] [CrossRef] [PubMed]
- Shin, N.R.; Whon, T.W.; Bae, J.W. Proteobacteria: Microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015, 33, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Mukhopadhya, I.; Hansen, R.; El-Omar, E.M.; Hold, G.L. IBD-what role do Proteobacteria play? Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 219–230. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.H.B.; Lauridsen, C.; Nielsen, B.; Jørgensen, L.; Schönherz, A.; Canibe, N. Early Inoculation of a Multi-Species Probiotic in Piglets-Impacts on the Gut Microbiome and Immune Responses. Microorganisms 2025, 13, 1292. [Google Scholar] [CrossRef]
- Wang, W.; Wei, X.; Wu, L.; Shang, X.; Cheng, F.; Li, B.; Zhou, X.; Zhang, J. The occurrence of antibiotic resistance genes in the microbiota of yak, beef and dairy cattle characterized by a metagenomic approach. J. Antibiot. 2021, 74, 508–518. [Google Scholar] [CrossRef]
- Rahman, N.; McCullough, T.; Orozco, D.F.; Walkowiak, S.; Farzan, A.; Shekarriz, S.; Surette, M.G.; Cicek, N.; Derakhshani, H. Genomic characterization of antimicrobial resistance and mobile genetic elements in swine gut bacteria isolated from a Canadian research farm. Anim. Microbiome 2025, 7, 66. [Google Scholar] [CrossRef]
- Begmatov, S.A.; Beletsky, A.; Rakitin, A.; Lukina, A.; Sokolyanskaya, L.; Rakitin, A.; Glukhova, L.; Mardanov, A.; Karnachuk, O.; Ravin, N. Antibiotic resistance genes in cattle gut microbiota: Influence of housing conditions. Mol. Biol. 2024, 58, 1101–1110. [Google Scholar] [CrossRef]
- Kim, M.; Morrison, M.; Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 2011, 76, 49–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shah, A.M.; Liu, Y.; Jin, L.; Wang, Z.; Xue, B.; Peng, Q. Relationship between true digestibility of dietary phosphorus and gastrointestinal bacteria of goats. PLoS ONE 2020, 15, e0225018. [Google Scholar] [CrossRef]
- Zou, X.; Liu, G.; Meng, F.; Hong, L.; Li, Y.; Lian, Z.; Yang, Z.; Luo, C.; Liu, D. Exploring the Rumen and Cecum Microbial Community from Fetus to Adulthood in Goat. Animals 2020, 10, 1639. [Google Scholar] [CrossRef] [PubMed]
- Fliegerova, K.O.; Podmirseg, S.M.; Vinzelj, J.; Grilli, D.J.; Kvasnová, S.; Schierová, D.; Sechovcová, H.; Mrázek, J.; Siddi, G.; Arenas, G.N.; et al. The Effect of a High-Grain Diet on the Rumen Microbiome of Goats with a Special Focus on Anaerobic Fungi. Microorganisms 2021, 9, 157. [Google Scholar] [CrossRef]
- Cremonesi, P.; Conte, G.; Severgnini, M.; Turri, F.; Monni, A.; Capra, E.; Rapetti, L.; Colombini, S.; Chessa, S.; Battelli, G.; et al. Evaluation of the effects of different diets on microbiome diversity and fatty acid composition of rumen liquor in dairy goat. Animal 2018, 12, 1856–1866. [Google Scholar] [CrossRef]
- Wang, Z.; Yin, L.; Liu, L.; Lan, X.; He, J.; Wan, F.; Shen, W.; Tang, S.; Tan, Z.; Yang, Y. Tannic acid reduced apparent protein digestibility and induced oxidative stress and inflammatory response without altering growth performance and ruminal microbiota diversity of Xiangdong black goats. Front. Vet. Sci. 2022, 9, 1004841. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Yao, X.; Zuo, C.; Qi, Y.; Chen, D.; Ma, W. The gut bacterial diversity of sheep associated with different breeds in Qinghai province. BMC Vet. Res. 2020, 16, 254. [Google Scholar] [CrossRef]
- Lv, W.; Liu, X.; Sha, Y.; Shi, H.; Wei, H.; Luo, Y.; Wang, J.; Li, S.; Hu, J.; Guo, X.; et al. Rumen Fermentation-Microbiota-Host Gene Expression Interactions to Reveal the Adaptability of Tibetan Sheep in Different Periods. Animals 2021, 11, 3529. [Google Scholar] [CrossRef]
- Cui, X.; Wang, Z.; Guo, P.; Li, F.; Chang, S.; Yan, T.; Zheng, H.; Hou, F. Shift of Feeding Strategies from Grazing to Different Forage Feeds Reshapes the Rumen Microbiota to Improve the Ability of Tibetan Sheep (Ovis aries) to Adapt to the Cold Season. Microbiol. Spectr. 2023, 11, e0281622. [Google Scholar]
- Wang, X.; Hu, L.; Liu, H.; Xu, T.; Zhao, N.; Zhang, X.; Geng, Y.; Kang, S.; Xu, S. Characterization of the bacterial microbiota across the different intestinal segments of the Qinghai semi-fine wool sheep on the Qinghai-Tibetan Plateau. Anim. Biosci. 2021, 34, 1921–1929. [Google Scholar] [CrossRef]
- Minozzi, G.; Biscarini, F.; Costa, E.D.; Chincarini, M.; Ferri, N.; Palestrini, C.; Minero, M.; Mazzola, S.; Piccinini, R.; Vignola, G.; et al. Analysis of Hindgut Microbiome of Sheep and Effect of Different Husbandry Conditions. Animals 2020, 11, 4. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Zhang, X.X.; Li, F.D.; Li, C.; Li, G.Z.; Zhang, D.Y.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Wang, W.M. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef]
- Rawal, S.; Kaur, H.; Bhathan, S.; Mittal, D.; Kaur, G.; Ali, S.A. Ruminant Gut Microbiota: Interplay, Implications, and Innovations for Sustainable Livestock Production. In Sustainable Agriculture Reviews: Animal Biotechnology for Livestock Production 4; Springer: Cham, Switzerland, 2024; pp. 205–228. [Google Scholar]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Dias, J.; Marcondes, M.I.; de Souza, S.M.; da Mata, E.S.B.C.; Noronha, M.F.; Resende, R.T.; Machado, F.S.; Mantovani, H.C.; Dill-McFarland, K.A.; Suen, G. Bacterial Community Dynamics across the Gastrointestinal Tracts of Dairy Calves during Preweaning Development. Appl. Environ. Microbiol. 2018, 84, e02675-17. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS ONE 2013, 8, e83424. [Google Scholar] [CrossRef]
- Jensen, R.B.; Walslag, I.H.; Marcussen, C.; Thorringer, N.W.; Junghans, P.; Nyquist, N.F. The effect of feeding order of forage and oats on metabolic and digestive responses related to gastric emptying in horses. J. Anim. Sci. 2025, 103, skae368. [Google Scholar] [CrossRef]
- Li, Y.; Ma, Q.; Shi, X.; Liu, G.; Wang, C. Integrated multi-omics reveals novel microbe-host lipid metabolism and immune interactions in the donkey hindgut. Front. Immunol. 2022, 13, 1003247. [Google Scholar] [CrossRef]
- Durazzi, F.; Sala, C.; Castellani, G.; Manfreda, G.; Remondini, D.; De Cesare, A. Comparison between 16S rRNA and shotgun sequencing data for the taxonomic characterization of the gut microbiota. Sci. Rep. 2021, 11, 3030. [Google Scholar] [CrossRef]
- Plancade, S.; Clark, A.; Philippe, C.; Helbling, J.C.; Moisan, M.P.; Esquerré, D.; Le Moyec, L.; Robert, C.; Barrey, E.; Mach, N. Unraveling the effects of the gut microbiota composition and function on horse endurance physiology. Sci. Rep. 2019, 9, 9620. [Google Scholar] [CrossRef]
- Zhang, G.; Wang, Y.; Luo, H.; Qiu, W.; Zhang, H.; Hu, L.; Wang, Y.; Dong, G.; Guo, G. The Association Between Inflammaging and Age-Related Changes in the Ruminal and Fecal Microbiota Among Lactating Holstein Cows. Front. Microbiol. 2019, 10, 1803. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Qin, C.; Xu, Y.; Song, X.; Fu, Y.; Li, R.; Liu, Q.; Shi, D. Multi-omics reveals the mechanism of rumen microbiome and its metabolome together with host metabolome participating in the regulation of milk production traits in dairy buffaloes. Front. Microbiol. 2024, 15, 1301292. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, H.F.; Zhou, Z.; Gomes, M.S.; Peixoto, P.M.G.; Bonsaglia, E.C.R.; Canisso, I.F.; Weimer, B.C.; Lima, F.S. Rumen and lower gut microbiomes relationship with feed efficiency and production traits throughout the lactation of Holstein dairy cows. Sci. Rep. 2022, 12, 4904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, C.; Huo, D.; Hu, Q.; Peng, Q. Comparative study of the gut microbiome potentially related to milk protein in Murrah buffaloes (Bubalus bubalis) and Chinese Holstein cattle. Sci. Rep. 2017, 7, 42189. [Google Scholar] [CrossRef]
- Sato, Y.; Sato, R.; Fukui, E.; Yoshizawa, F. Impact of rumen microbiome on cattle carcass traits. Sci. Rep. 2024, 14, 6064. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The rumen microbiome: A crucial consideration when optimising milk and meat production and nitrogen utilisation efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef]
- Sim, S.; Lee, H.; Yoon, S.; Seon, H.; Park, C.; Kim, M. The impact of different diets and genders on fecal microbiota in Hanwoo cattle. J. Anim. Sci. Technol. 2022, 64, 897–910. [Google Scholar] [CrossRef]
- Xie, L.; Xing, J.; Qi, X.; Lu, T.; Jin, Y.; Akhtar, M.F.; Li, L.; Liu, G. Effects of Concentrate Feeding Sequence on Growth Performance, Nutrient Digestibility, VFA Production, and Fecal Microbiota of Weaned Donkeys. Animals 2023, 13, 2893. [Google Scholar] [CrossRef]
- Li, L.; Guo, X.; Zhao, Y.; Guo, Y.; Shi, B.; Zhou, Y.; Zhang, Y.; Yan, S. Cecal Microbial Diversity and Metabolome Reveal a Reduction in Growth Due to Oxidative Stress Caused by a Low-Energy Diet in Donkeys. Antioxidants 2024, 13, 1377. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Miller, M.E.B.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 2018, 96, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- McGovern, E.; McGee, M.; Byrne, C.J.; Kenny, D.A.; Kelly, A.K.; Waters, S.M. Investigation into the effect of divergent feed efficiency phenotype on the bovine rumen microbiota across diet and breed. Sci. Rep. 2020, 10, 15317. [Google Scholar] [CrossRef] [PubMed]
- Myer, P.R.; Freetly, H.C.; Wells, J.E.; Smith, T.P.L.; Kuehn, L.A. Analysis of the gut bacterial communities in beef cattle and their association with feed intake, growth, and efficiency. J. Anim. Sci. 2017, 95, 3215–3224. [Google Scholar] [CrossRef]
- Huang, S.; Ji, S.; Suen, G.; Wang, F.; Li, S. The Rumen Bacterial Community in Dairy Cows Is Correlated to Production Traits During Freshening Period. Front. Microbiol. 2021, 12, 630605. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.B.; Bernal-Córdoba, C.; Fausak, E.D.; Silva-Del-Río, N. Effect of prebiotics on growth and health of dairy calves: A protocol for a systematic review and meta-analysis. PLoS ONE 2021, 16, e0253379. [Google Scholar] [CrossRef]
- Markowiak, P.; Śliżewska, K. Effects of Probiotics, Prebiotics, and Synbiotics on Human Health. Nutrients 2017, 9, 1021. [Google Scholar] [CrossRef]
- Cangiano, L.; Yohe, T.T.; Steele, M.; Renaud, D. Invited Review: Strategic use of microbial-based probiotics and prebiotics in dairy calf rearing. Appl. Anim. Sci. 2020, 36, 630–651. [Google Scholar]
- Bouwhuis, M.A.; McDonnell, M.J.; Sweeney, T.; Mukhopadhya, A.; O’Shea, C.J.; O’Doherty, J.V. Seaweed extracts and galacto-oligosaccharides improve intestinal health in pigs following Salmonella Typhimurium challenge. Animal 2017, 11, 1488–1496. [Google Scholar] [CrossRef]
- Sun, P.; Wang, J.Q.; Zhang, H.T. Effects of Bacillus subtilis natto on performance and immune function of preweaning calves. J. Dairy Sci. 2010, 93, 5851–5855. [Google Scholar] [CrossRef]
- Oikonomou, G.; Teixeira, A.G.; Foditsch, C.; Bicalho, M.L.; Machado, V.S.; Bicalho, R.C. Fecal microbial diversity in pre-weaned dairy calves as described by pyrosequencing of metagenomic 16S rDNA. Associations of Faecalibacterium species with health and growth. PLoS ONE 2013, 8, e63157. [Google Scholar]
- Song, Y.; Malmuthuge, N.; Steele, M.A.; Guan, L.L. Shift of hindgut microbiota and microbial short chain fatty acids profiles in dairy calves from birth to pre-weaning. FEMS Microbiol. Ecol. 2018, 94, fix179. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.N.; Chaudhary, P.; Kumar, S.; Grover, C.R.; Mondal, G. Effect of synbiotics on growth performance, gut health, and immunity status in pre-ruminant buffalo calves. Sci. Rep. 2023, 13, 10184. [Google Scholar]
- Huang, B.; Khan, M.Z.; Chen, Y.; Liang, H.; Kou, X.; Wang, X.; Ren, W.; Wang, C.; Zhang, Z. Yeast polysaccharide supplementation: Impact on lactation, growth, immunity, and gut microbiota in Dezhou donkeys. Front. Microbiol. 2023, 14, 1289371. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Ma, Z.; Du, X.; Wang, C.; Liu, G.; Zhou, M. Methionine Alters the Fecal Microbiota and Enhances the Antioxidant Capacity of Lactating Donkeys. Animals 2025, 15, 648. [Google Scholar] [CrossRef]
- Li, C.; Li, X.Y.; Li, X.B.; Ma, C.; Chen, H.; Yang, F. Growth performance, nutrient digestibility, fecal microbial diversity and volatile fatty acid, and blood biochemical indices of suckling donkeys fed diets supplemented with multienzymes. BMC Vet. Res. 2024, 20, 61. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
- Liu, K.; Zhang, Y.; Yu, Z.; Xu, Q.; Zheng, N.; Zhao, S.; Huang, G.; Wang, J. Ruminal microbiota-host interaction and its effect on nutrient metabolism. Anim. Nutr. 2021, 7, 49–55. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Durunna, O.N.; Moore, S.S.; Guan, L.L. Impact of feed efficiency and diet on adaptive variations in the bacterial community in the rumen fluid of cattle. Appl. Environ. Microbiol. 2012, 78, 1203–1214. [Google Scholar] [CrossRef]
- Amin, N.; Schwarzkopf, S.; Kinoshita, A.; Tröscher-Mußotter, J.; Dänicke, S.; Camarinha-Silva, A.; Huber, K.; Frahm, J.; Seifert, J. Evolution of rumen and oral microbiota in calves is influenced by age and time of weaning. Anim. Microbiome 2021, 3, 31. [Google Scholar] [CrossRef]
- Hagey, J.V.; Laabs, M.; Maga, E.A.; DePeters, E.J. Rumen sampling methods bias bacterial communities observed. PLoS ONE 2022, 17, e0258176. [Google Scholar]
- Xiang, Q.; Su, Q.; Li, Q.; Liu, J.; Du, Y.; Shi, H.; Li, Z.; Ma, Y.; Niu, Y.; Chen, L.; et al. Microbial community analyses provide a differential diagnosis for the antemortem and postmortem injury of decayed cadaver: An animal model. J. Forensic Leg. Med. 2023, 93, 102473. [Google Scholar] [CrossRef]
- Yang, F.; Zhang, X.; Hu, S.; Nie, H.; Gui, P.; Zhong, Z.; Guo, Y.; Zhao, X. Changes in Microbial Communities Using Pigs as a Model for Postmortem Interval Estimation. Microorganisms 2023, 11, 2811. [Google Scholar] [CrossRef]
- Turner, P.V.; Kloeze, H.; Dam, A.; Ward, D.; Leung, N.; Brown, E.E.; Whiteman, A.; Chiappetta, M.E.; Hunter, D.B. Mass depopulation of laying hens in whole barns with liquid carbon dioxide: Evaluation of welfare impact. Poult. Sci. 2012, 91, 1558–1568. [Google Scholar] [CrossRef]
- Kittelmann, S.; Kirk, M.R.; Jonker, A.; McCulloch, A.; Janssen, P.H. Buccal swabbing as a noninvasive method to determine bacterial, archaeal, and eukaryotic microbial community structures in the rumen. Appl. Environ. Microbiol. 2015, 81, 7470–7483. [Google Scholar] [CrossRef]
- Tapio, I.; Shingfield, K.J.; McKain, N.; Bonin, A.; Fischer, D.; Bayat, A.R.; Vilkki, J.; Taberlet, P.; Snelling, T.J.; Wallace, R.J. Oral Samples as Non-Invasive Proxies for Assessing the Composition of the Rumen Microbial Community. PLoS ONE 2016, 11, e0151220. [Google Scholar] [CrossRef]
- Young, J.; Skarlupka, J.H.; Cox, M.S.; Resende, R.T.; Fischer, A.; Kalscheur, K.F.; McClure, J.C.; Cole, J.B.; Suen, G.; Bickhart, D.M. Validating the Use of Bovine Buccal Sampling as a Proxy for the Rumen Microbiota by Using a Time Course and Random Forest Classification Approach. Appl. Environ. Microbiol. 2020, 86, e00861-20. [Google Scholar] [CrossRef]
- Miura, H.; Takeda, M.; Yamaguchi, M.; Ohtani, Y.; Endo, G.; Masuda, Y.; Ito, K.; Nagura, Y.; Iwashita, K.; Mitani, T.; et al. Application of MinION Amplicon Sequencing to Buccal Swab Samples for Improving Resolution and Throughput of Rumen Microbiota Analysis. Front. Microbiol. 2022, 13, 783058. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).