Selection Signatures in the Genome of Dzhalgin Merino Sheep Breed
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Animals and Sample Collection
2.3. Genotyping and Genotyping Quality Control
2.4. Principal Component Analysis
2.5. Search for Selection Signatures
2.6. Gene Annotation and Construction of Gene Networks
2.7. Visualization
3. Results
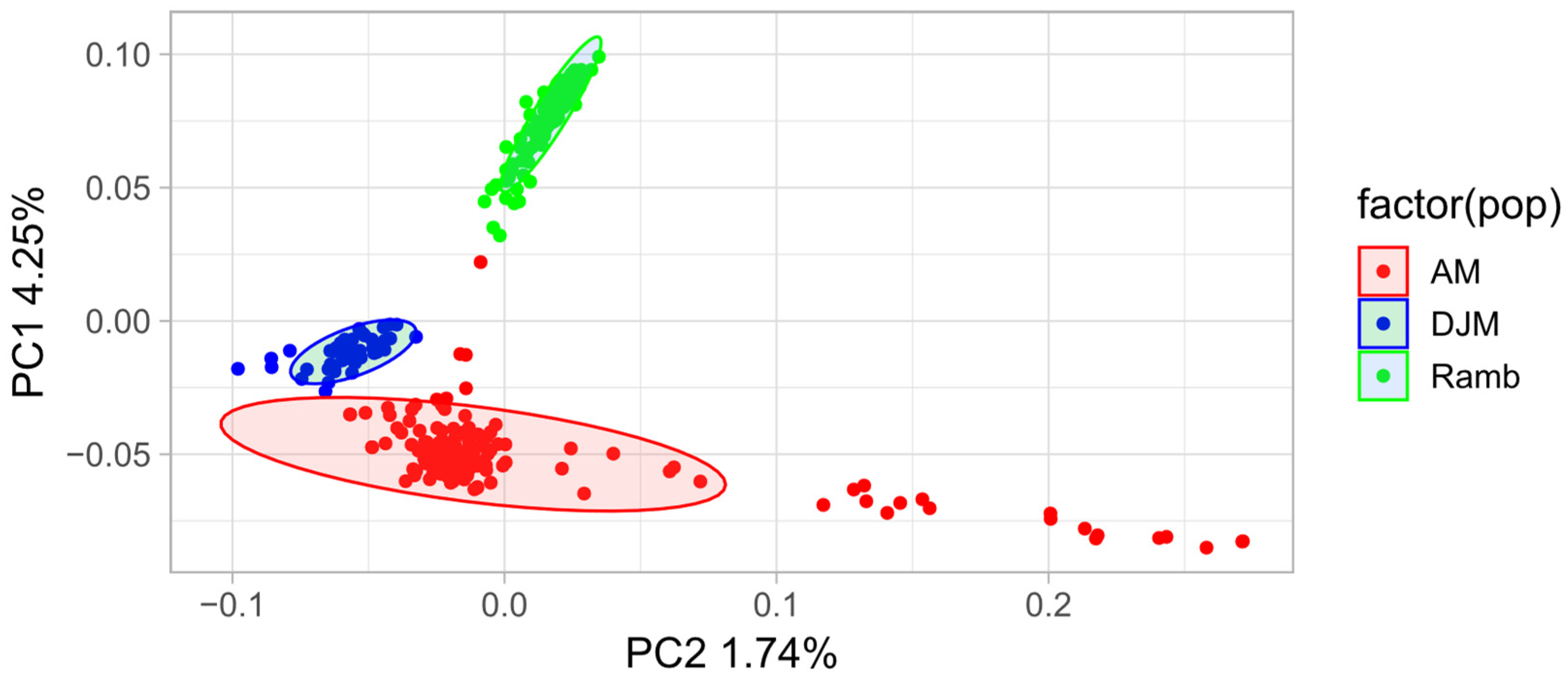
3.1. Genetic Differentiation of the Studied Groups Using PCA and FST Methods
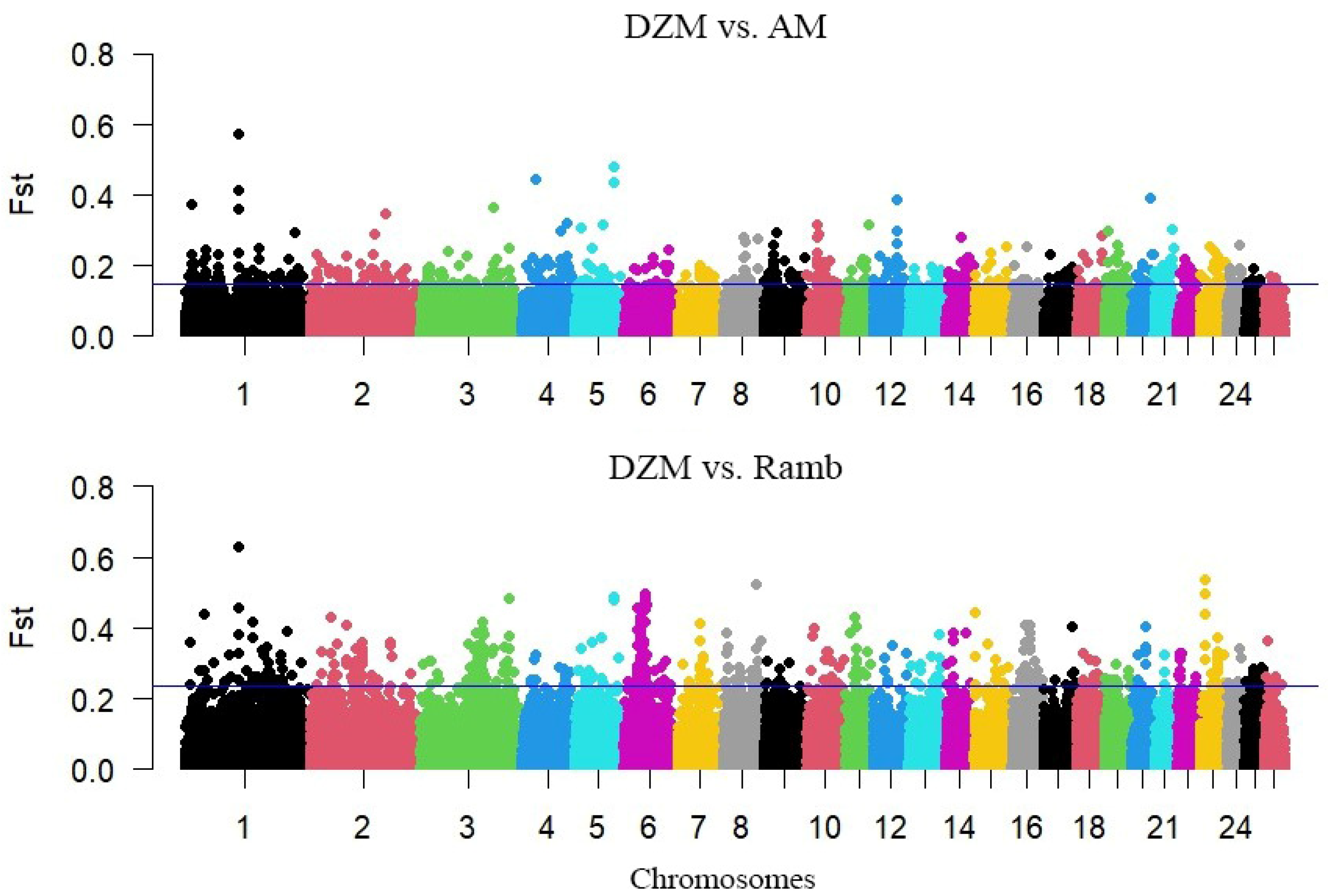
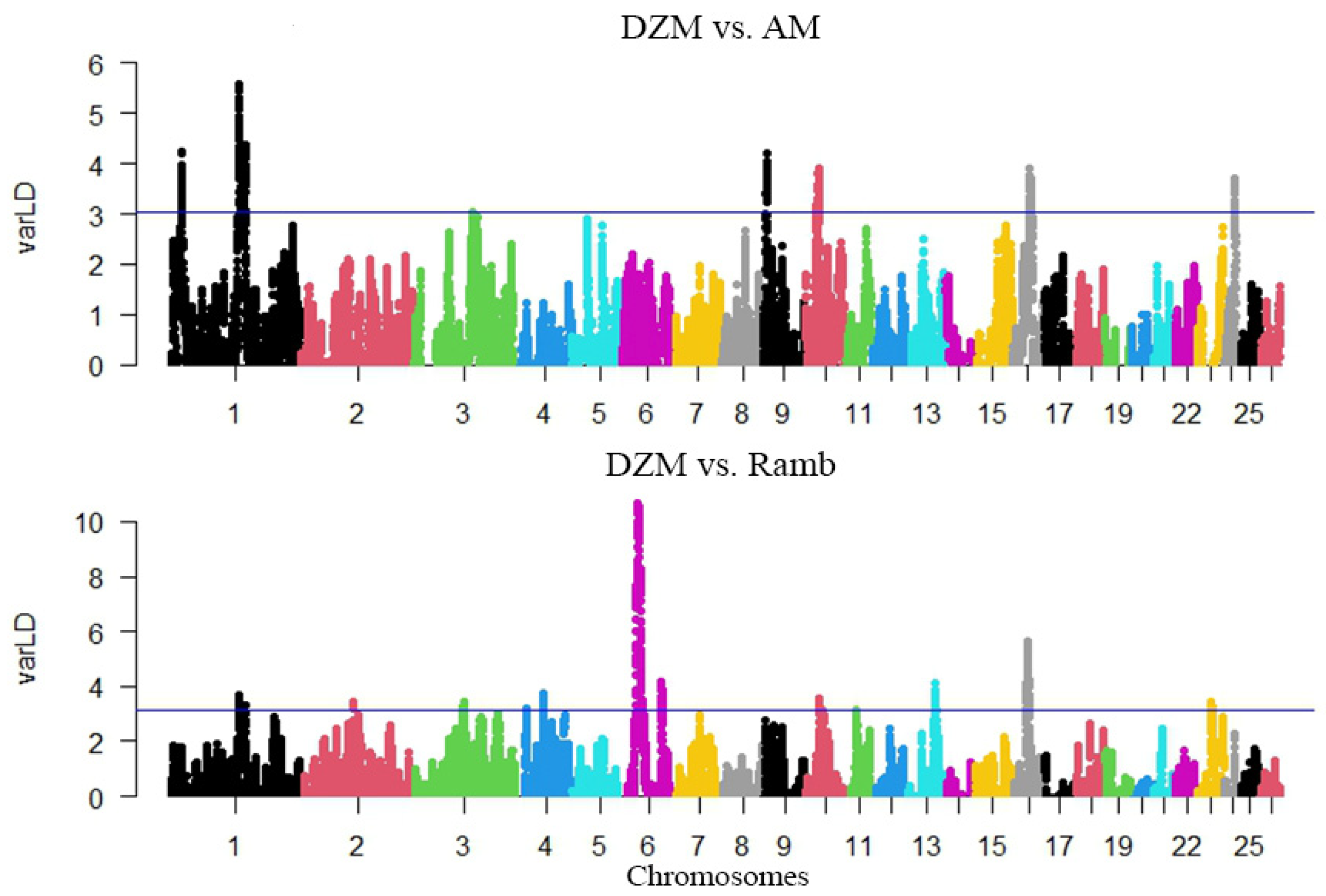
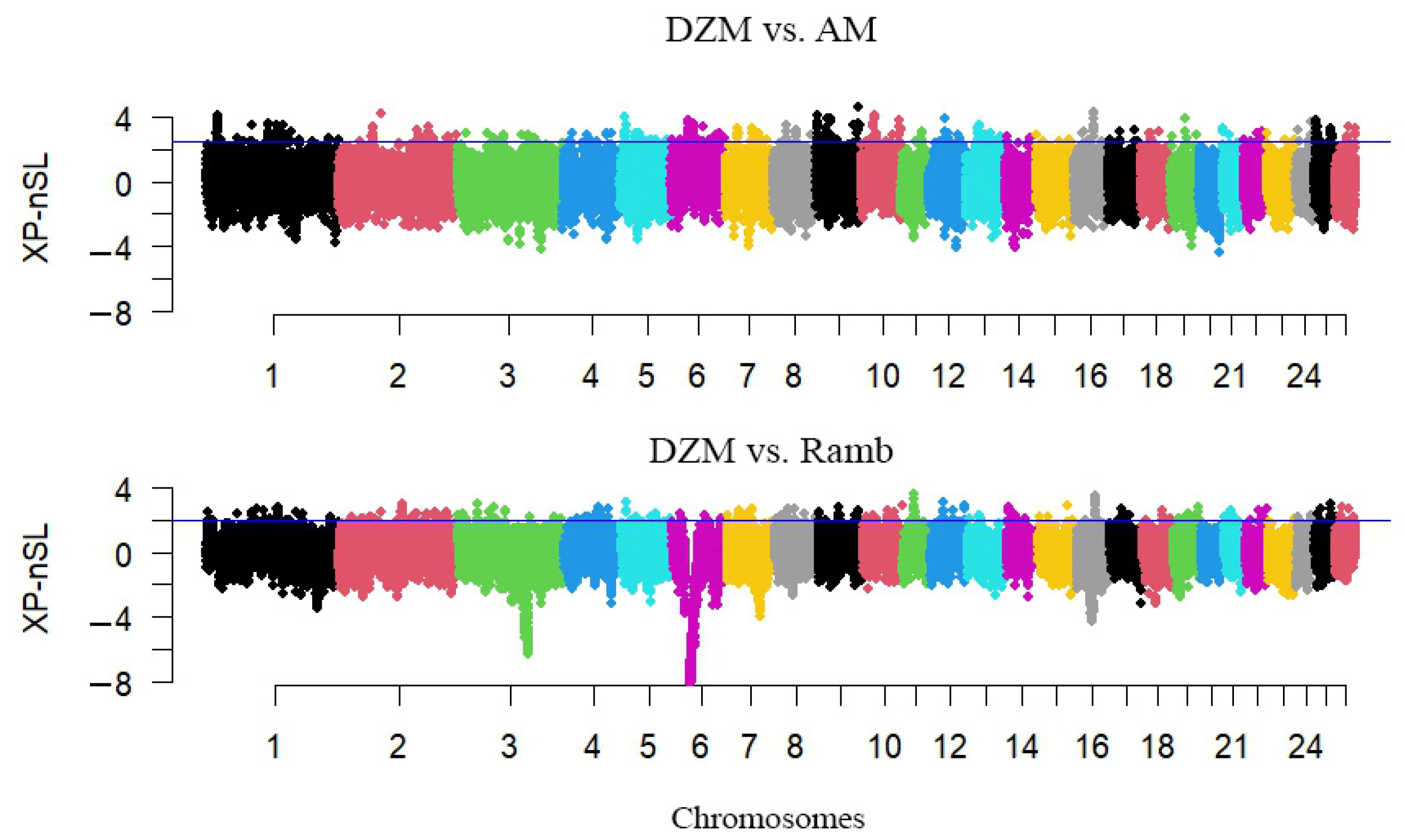
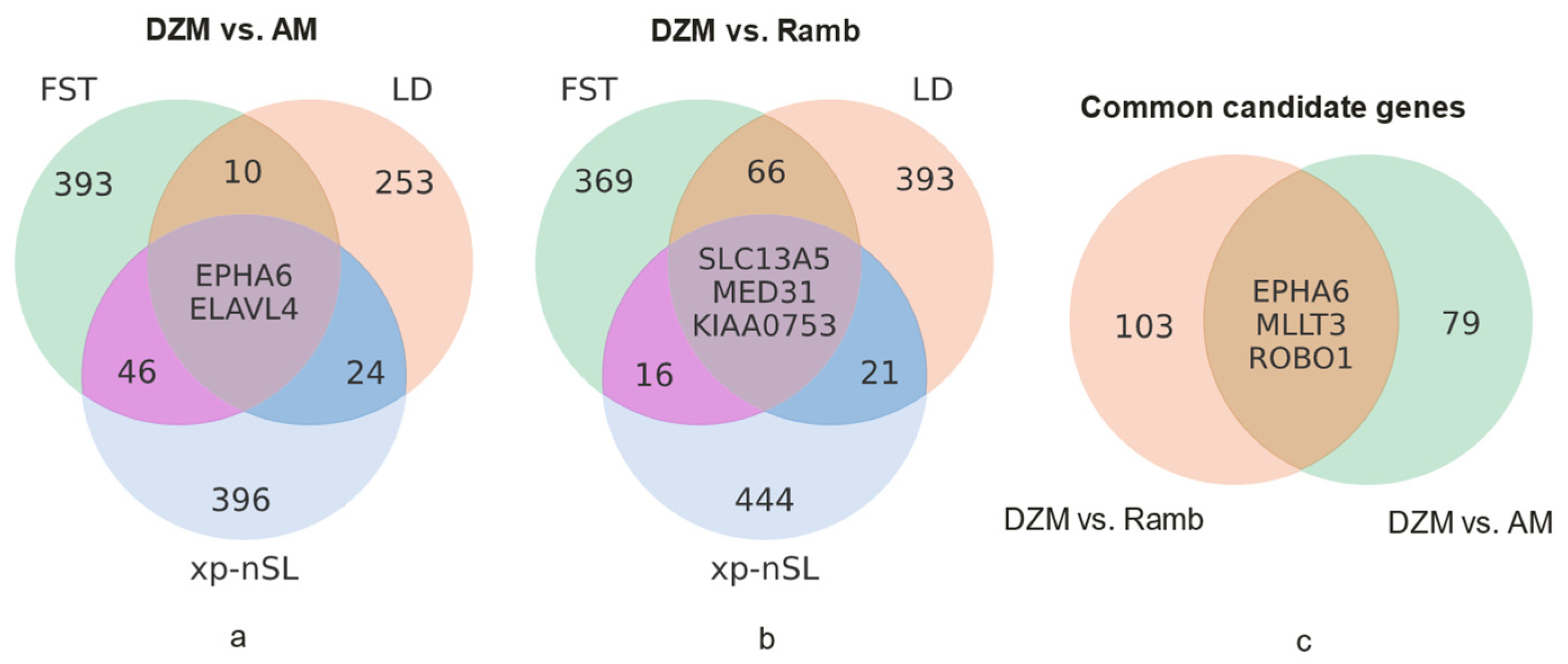
3.2. Search for Selection Signatures
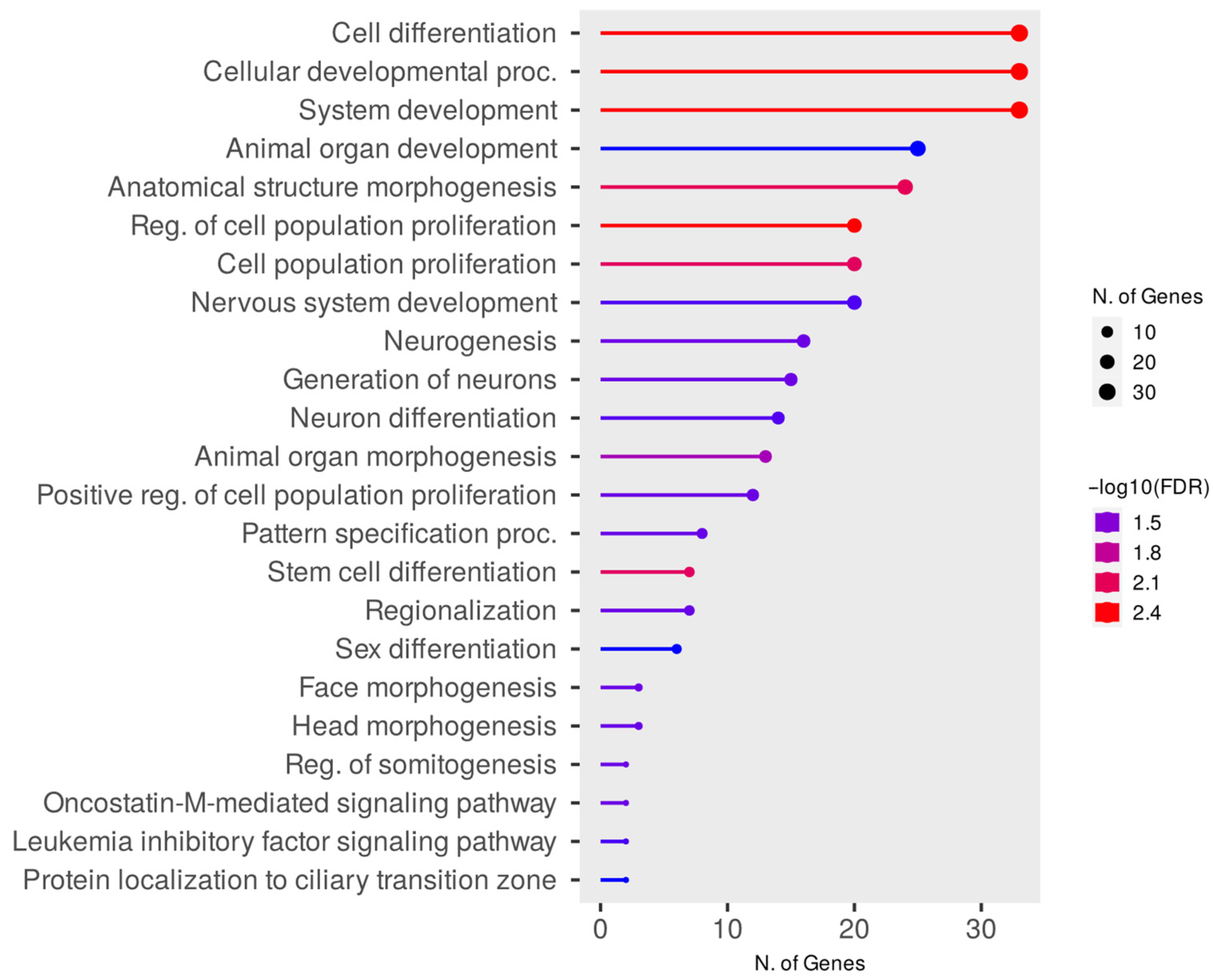
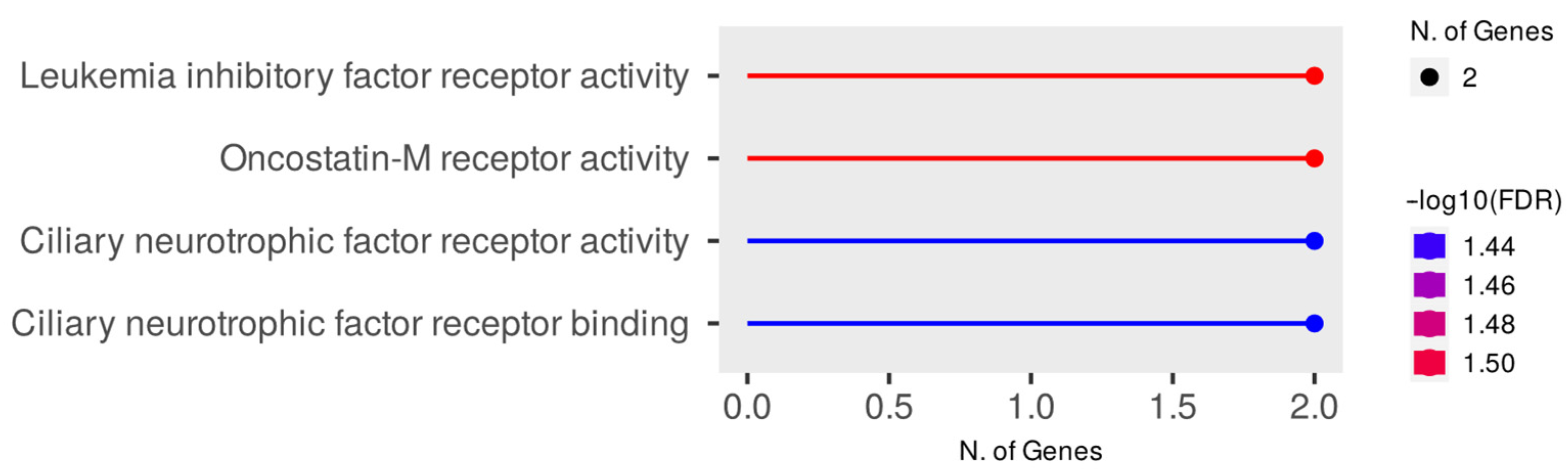
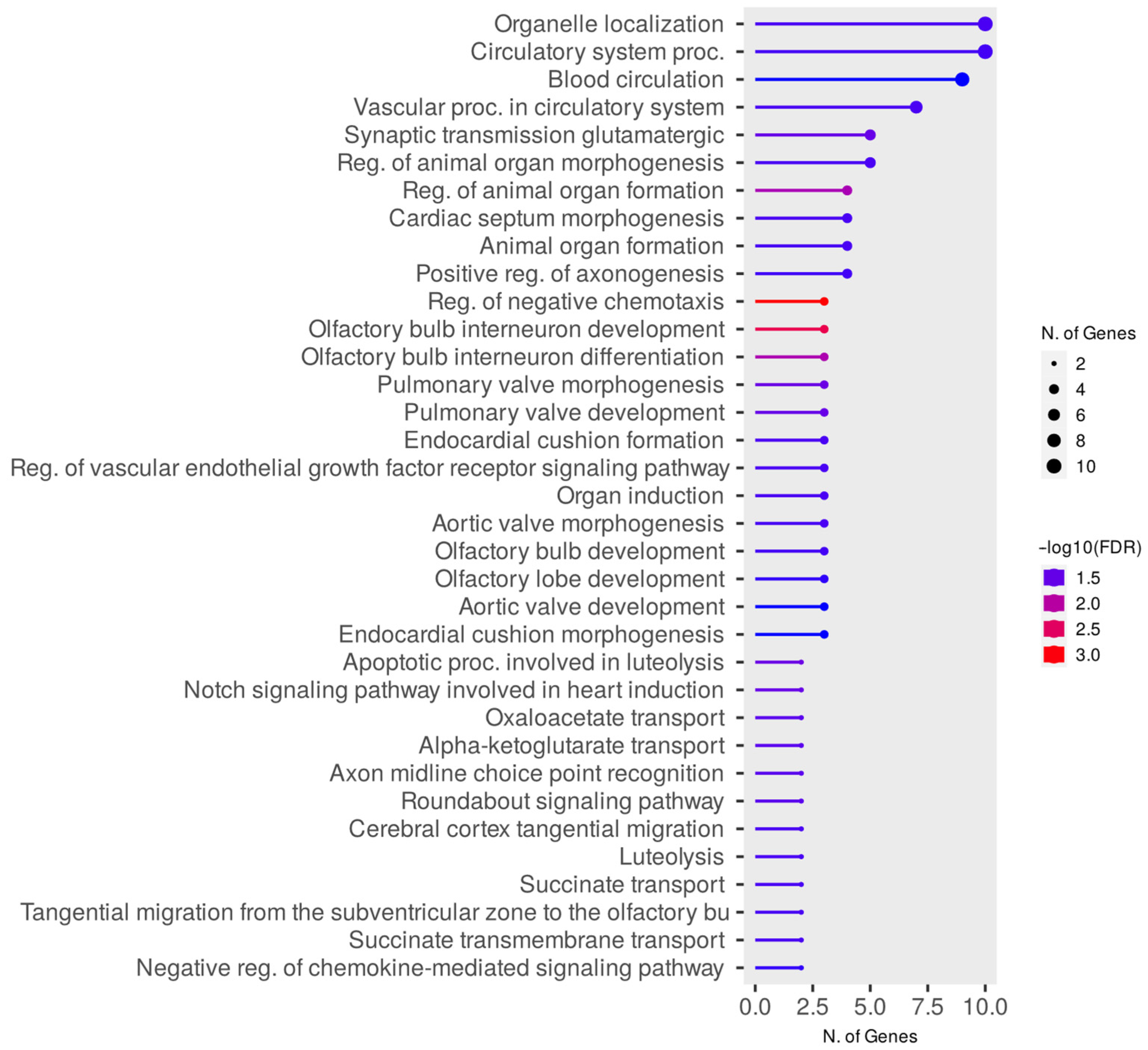
3.3. Evaluation of PPI Interactions and Functional Enrichment of GO Terms
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| iHS | Integrated haplotype score |
| xp-EHH | Cross-population extended haplotype homozygosity |
| LD | Linkage disequilibrium |
| ROH | Runs of homozygosity |
| DZM | Dzhalgin Merino |
| AM | Australian Merino |
| Ramb | Rambouillet |
| GWAS | Genome-Wide Association Study |
| xp-nSL | Cross-population number of Segregating sites by Length |
References
- Abondio, P.; Cilli, E.; Luiselli, D. Inferring Signatures of Positive Selection in Whole-Genome Sequencing Data: An Overview of Haplotype-Based Methods. Genes 2022, 13, 926. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Ding, X.; Qanbari, S.; Weigend, S.; Zhang, Q.; Simianer, H. Properties of Different Selection Signature Statistics and a New Strategy for Combining Them. Heredity 2015, 115, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bai, C.; Shi, L.; He, Y.; Hu, M.; Sun, H.; Peng, H.; Lai, W.; Jiao, S.; Zhao, Z.; et al. Detection of Selection Signatures in South African Mutton Merino Sheep Using Whole-Genome Sequencing Data. Anim. Genet. 2022, 53, 224–229. [Google Scholar] [CrossRef]
- Mészárosová, M.; Mészáros, G.; Moravčíková, N.; Pavlík, I.; Margetín, M.; Kasarda, R. Within-and between-Breed Selection Signatures in the Original and Improved Valachian Sheep. Animals 2022, 12, 1346. [Google Scholar] [CrossRef]
- Eydivandi, S.; Roudbar, M.A.; Ardestani, S.S.; Momen, M.; Sahana, G. A Selection Signatures Study among Middle Eastern and European Sheep Breeds. J. Anim. Breed. Genet. 2021, 138, 574–588. [Google Scholar] [CrossRef]
- Saravanan, K.A.; Panigrahi, M.; Kumar, H.; Bhushan, B.; Dutt, T.; Mishra, B.P. Genome-Wide Analysis of Genetic Diversity and Selection Signatures in Three Indian Sheep Breeds. Livest. Sci. 2021, 243, 104367. [Google Scholar] [CrossRef]
- Tao, L.; Wang, X.; Zhong, Y.; Liu, Q.; Xia, Q.; Chen, S.; He, X.; Di, R.; Chu, M. Combined Approaches Identify Known and Novel Genes Associated with Sheep Litter Size and Non-Seasonal Breeding. Anim. Genet. 2021, 52, 857–867. [Google Scholar] [CrossRef]
- Yurchenko, A.A.; Deniskova, T.E.; Yudin, N.S.; Dotsev, A.V.; Khamiruev, T.N.; Selionova, M.I.; Egorov, S.V.; Reyer, H.; Wimmers, K.; Brem, G.; et al. High-Density Genotyping Reveals Signatures of Selection Related to Acclimation and Economically Important Traits in 15 Local Sheep Breeds from Russia. BMC Genom. 2019, 20, 294. [Google Scholar] [CrossRef]
- Dunin, I.M.; Serdyukov, I.G.; Pavlov, M.B. New Achievement in Breeding—The Fine-Wooled Dzhalginsky Merino Sheep Breed. Farm. Anim. 2013, 3, 46–48. [Google Scholar]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-Generation PLINK: Rising to the Challenge of Larger and Richer Datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Zheng, X.; Levine, D.; Shen, J.; Gogarten, S.M.; Laurie, C.; Weir, B.S. A High-Performance Computing Toolset for Relatedness and Principal Component Analysis of SNP Data. Bioinformatics 2012, 28, 3326–3328. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The Variant Call Format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef] [PubMed]
- Ong, R.T.-H.; Teo, Y.-Y. VarLD: A Program for Quantifying Variation in Linkage Disequilibrium Patterns between Populations. Bioinformatics 2010, 26, 1269–1270. [Google Scholar] [CrossRef]
- Szpiech, Z.A.; Hernandez, R.D. Selscan: An Efficient Multithreaded Program to Perform EHH-Based Scans for Positive Selection. Mol. Biol. Evol. 2014, 31, 2824–2827. [Google Scholar] [CrossRef]
- Delaneau, O.; Zagury, J.F.; Robinson, M.R.; Marchini, J.L.; Dermitzakis, E.T. Accurate, Scalable and Integrative Haplotype Estimation. Nat. Commun. 2019, 10, 24–29. [Google Scholar] [CrossRef]
- Drost, H.-G.; Paszkowski, J. Biomartr: Genomic Data Retrieval with R. Bioinformatics 2017, 33, 1216–1217. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, A.; Cotrina, M.; Wilkinson, D.G. Diverse Roles of Eph Receptors and Ephrins in the Regulation of Cell Migration and Tissue Assembly. Dev. Cell 2004, 7, 465–480. [Google Scholar] [CrossRef]
- Yang, C.; He, J.; Mao, J.; Ren, Y.; Liu, G.; Wei, C.; Zhang, G.; Tian, K.; Huang, X. Genome-Wide DNA Methylation Analysis and Functional Validation of Litter Size Traits in Jining Grey Goats. Genes 2024, 15, 353. [Google Scholar] [CrossRef]
- Smołucha, G.; Gurgul, A.; Jasielczuk, I.; Kawęcka, A.; Miksza-Cybulska, A. A Genome-Wide Association Study for Prolificacy in Three Polish Sheep Breeds. J. Appl. Genet. 2021, 62, 323–326. [Google Scholar] [CrossRef]
- Forutan, M.; Engle, B.; Goddard, M.E.; Hayes, B.J. 262. A Conditional Multi-Trait Sequence GWAS of Heifer Fertility in Tropically Adapted Beef Cattle; Wageningen Academic Publishers: Wageningen, The Netherlands, 2022; pp. 1106–1109. [Google Scholar]
- Dos Santos, F.C.; Peixoto, M.G.C.D.; De Souza Fonseca, P.A.; De Fátima Ávila Pires, M.; Ventura, R.V.; Da Cruz Rosse, I.; Bruneli, F.A.T.; Machado, M.A.; Carvalho, M.R.S. Identification of Candidate Genes for Reactivity in Guzerat (Bos Indicus) Cattle: A Genome-Wide Association Study. PLoS ONE 2017, 12, e0169163. [Google Scholar] [CrossRef]
- Huang, C.; Zhao, Q.; Chen, Q.; Su, Y.; Ma, Y.; Ye, S.; Zhao, Q. Runs of Homozygosity Detection and Selection Signature Analysis for Local Goat Breeds in Yunnan, China. Genes 2024, 15, 313. [Google Scholar] [CrossRef]
- Mudadu, M.A.; Porto-Neto, L.R.; Mokry, F.B.; Tizioto, P.C.; Oliveira, P.S.N.; Tullio, R.R.; Nassu, R.T.; Niciura, S.C.M.; Tholon, P.; Alencar, M.M.; et al. Genomic Structure and Marker-Derived Gene Networks for Growth and Meat Quality Traits of Brazilian Nelore Beef Cattle. BMC Genom. 2016, 17, 235. [Google Scholar] [CrossRef]
- Nayak, S.S.; Panigrahi, M.; Rajawat, D.; Ghildiyal, K.; Sharma, A.; Parida, S.; Bhushan, B.; Mishra, B.P.; Dutt, T. Comprehensive Selection Signature Analyses in Dairy Cattle Exploiting Purebred and Crossbred Genomic Data. Mamm. Genome 2023, 34, 615–631. [Google Scholar] [CrossRef]
- Vetokh, A.N. Genome-Wide Association Studies of Chicken (Gallus gallus L.) Breast Meat Color Characteristics. Sel’skokhozyaistvennaya Biol. 2023, 58, 1068–1078. [Google Scholar] [CrossRef]
- Loureiro, B.; Ereno, R.L.; Pupulim, A.G.R.; Tramontana, M.C.V.B.; Tabosa, H.P.; Barros, C.M.; Favoreto, M.G. Genome-Wide Association Study of Nelore and Angus Heifers with Low and High Ovarian Follicle Counts. Anim. Reprod. 2024, 21, e20230110. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Jin, M.; Li, T.; Lu, Z.; Wang, H.; Yuan, Z.; Wei, C. Whole-Genome Resequencing Reveals Selection Signal Related to Sheep Wool Fineness. Animals 2023, 13, 2944. [Google Scholar] [CrossRef]
- Germano, G.; Porazzi, P.; Felix, C.A. Leukemia-associated Transcription Factor Mllt3 Is Important for Primitive Erythroid Development in Zebrafish Embryogenesis. Dev. Dyn. 2022, 251, 1728–1740. [Google Scholar] [CrossRef] [PubMed]
- Büttner, N.; Johnsen, S.A.; Kügler, S.; Vogel, T. Af9/Mllt3 Interferes with Tbr1 Expression through Epigenetic Modification of Histone H3K79 during Development of the Cerebral Cortex. Proc. Natl. Acad. Sci. USA 2010, 107, 7042–7047. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sun, L.; Dai, J.; Lv, Y.; Liao, R.; Shen, X.; Gao, J. Characterization and Comparative Analysis of Whole-Transcriptome Sequencing in High- and Low-Fecundity Chongming White Goat Ovaries during the Estrus Phase. Animals 2024, 14, 988. [Google Scholar] [CrossRef] [PubMed]
- Fonseca, P.A.S.; Schenkel, F.S.; Cánovas, A. Genome-Wide Association Study Using Haplotype Libraries and Repeated-Measures Model to Identify Candidate Genomic Regions for Stillbirth in Holstein Cattle. J. Dairy. Sci. 2022, 105, 1314–1326. [Google Scholar] [CrossRef]
- Popovitchenko, T.; Park, Y.; Page, N.F.; Luo, X.; Krsnik, Z.; Liu, Y.; Salamon, I.; Stephenson, J.D.; Kraushar, M.L.; Volk, N.L.; et al. Translational Derepression of Elavl4 Isoforms at Their Alternative 5′ UTRs Determines Neuronal Development. Nat. Commun. 2020, 11, 1674. [Google Scholar] [CrossRef]
- Mulligan, M.R.; Bicknell, L.S. The Molecular Genetics of NELAVL in Brain Development and Disease. Eur. J. Hum. Genet. 2023, 31, 1209–1217. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic Studies of Body Mass Index Yield New Insights for Obesity Biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef]
- Akhtar, M.J.; Khan, S.A.; Kumar, B.; Chawla, P.; Bhatia, R.; Singh, K. Role of Sodium Dependent SLC13 Transporter Inhibitors in Various Metabolic Disorders. Mol. Cell. Biochem. 2023, 478, 1669–1687. [Google Scholar] [CrossRef]
- Wang, L.Y.; Jiang, J.; Ma, H.M. Molecular Characterization, Tissue Expression Profile, and SNP Analysis of Porcine SLC13A5. Genet. Mol. Res. 2015, 14, 16090–16101. [Google Scholar] [CrossRef]
- Beadle, E.P.; Straub, J.A.; Bunnell, B.A.; Newman, J.J. MED31 Involved in Regulating Self-Renewal and Adipogenesis of Human Mesenchymal Stem Cells. Mol. Biol. Rep. 2018, 45, 1545–1550. [Google Scholar] [CrossRef] [PubMed]
- Risley, M.D.; Clowes, C.; Yu, M.; Mitchell, K.; Hentges, K.E. The Mediator Complex Protein Med31 Is Required for Embryonic Growth and Cell Proliferation during Mammalian Development. Dev. Biol. 2010, 342, 146–156. [Google Scholar] [CrossRef]
- Hammarsjö, A.; Wang, Z.; Vaz, R.; Taylan, F.; Sedghi, M.; Girisha, K.M.; Chitayat, D.; Neethukrishna, K.; Shannon, P.; Godoy, R.; et al. Novel KIAA0753 Mutations Extend the Phenotype of Skeletal Ciliopathies. Sci. Rep. 2017, 7, 15585. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, Y.; Wu, X.; Chen, Q.; Huang, J.; Zhu, H.; Yang, S.; Wang, J.; Li, L.T.; Liu, X.; et al. KIAA0753 Enhances Osteoblast Differentiation Suppressed by Diabetes. J. Cell Mol. Med. 2024, 28, e70035. [Google Scholar] [CrossRef]
- Adeniyi, O.O.; Simon, R.; Bytyqi, H.; Kugler, W.; Mehmeti, H.; Berisha, K.; Simčič, M.; Magdy, M.; Lühken, G. Capturing Genetic Diversity and Selection Signatures of the Endangered Kosovar Balusha Sheep Breed. Genes 2022, 13, 866. [Google Scholar] [CrossRef]
- Chen, Y.; Wu, X.; Wang, J.; Hou, Y.; Liu, Y.; Wang, B.; Hu, X.; Zheng, X.; Zhang, X.; Ding, Y.; et al. Detection of Selection Signatures in Anqing Six-End-White Pigs Based on Resequencing Data. Genes 2022, 13, 2310. [Google Scholar] [CrossRef]
- Waineina, R.W.; Okeno, T.O.; Ilatsia, E.D.; Ngeno, K. Selection Signature Analyses Revealed Genes Associated With Adaptation, Production, and Reproduction in Selected Goat Breeds in Kenya. Front. Genet. 2022, 13, 858923. [Google Scholar] [CrossRef] [PubMed]
- Dzomba, E.F.; Van Der Nest, M.A.; Mthembu, J.N.T.; Soma, P.; Snyman, M.A.; Chimonyo, M.; Muchadeyi, F.C. Selection Signature Analysis and Genome-Wide Divergence of South African Merino Breeds from Their Founders. Front. Genet. 2023, 13, 932272. [Google Scholar] [CrossRef] [PubMed]
- Weir, B.S.; Cockerham, C.C. Estimating F-statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Pérez O’Brien, A.M.; Utsunomiya, Y.T.; Mészáros, G.; Bickhart, D.M.; Liu, G.E.; Van Tassell, C.P.; Sonstegard, T.S.; Da Silva, M.V.; Garcia, J.F.; Sölkner, J. Assessing Signatures of Selection through Variation in Linkage Disequilibrium between Taurine and Indicine Cattle. Genet. Sel. Evol. 2014, 46, 19. [Google Scholar] [CrossRef]
| DZM | AM | |
|---|---|---|
| DZM | - | - |
| AM | 0.023298 | - |
| Ramb | 0.041518 | 0.050841 |
| Dataset | PPI Enrichment p-Value | |
|---|---|---|
| DZM vs. AM | DZM vs. Ramb | |
| Ovis aries | 6.33 × 10−3 | 1.69 × 10−10 |
| Homo sapiens | 5.48 × 10−8 | 4.38 × 10−6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krivoruchko, A.; Yatsyk, O.; Skokova, A.; Safaryan, E.; Usai, L.; Kanibolotskaya, A. Selection Signatures in the Genome of Dzhalgin Merino Sheep Breed. Animals 2025, 15, 2871. https://doi.org/10.3390/ani15192871
Krivoruchko A, Yatsyk O, Skokova A, Safaryan E, Usai L, Kanibolotskaya A. Selection Signatures in the Genome of Dzhalgin Merino Sheep Breed. Animals. 2025; 15(19):2871. https://doi.org/10.3390/ani15192871
Chicago/Turabian StyleKrivoruchko, Alexander, Olesya Yatsyk, Antonina Skokova, Elena Safaryan, Ludmila Usai, and Anastasia Kanibolotskaya. 2025. "Selection Signatures in the Genome of Dzhalgin Merino Sheep Breed" Animals 15, no. 19: 2871. https://doi.org/10.3390/ani15192871
APA StyleKrivoruchko, A., Yatsyk, O., Skokova, A., Safaryan, E., Usai, L., & Kanibolotskaya, A. (2025). Selection Signatures in the Genome of Dzhalgin Merino Sheep Breed. Animals, 15(19), 2871. https://doi.org/10.3390/ani15192871





