Paternal Lineage and Genetic Diversity of Jiuzhi Yaks Revealed by Y-Chromosome SRY Sequencing
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Sample Collection
2.2. Main Reagents and Instruments
Main Reagents
2.3. Genomic DNA Extraction
2.4. Primer Design, PCR Amplification, and Sequence Determination
2.5. Data Analysis
3. Results
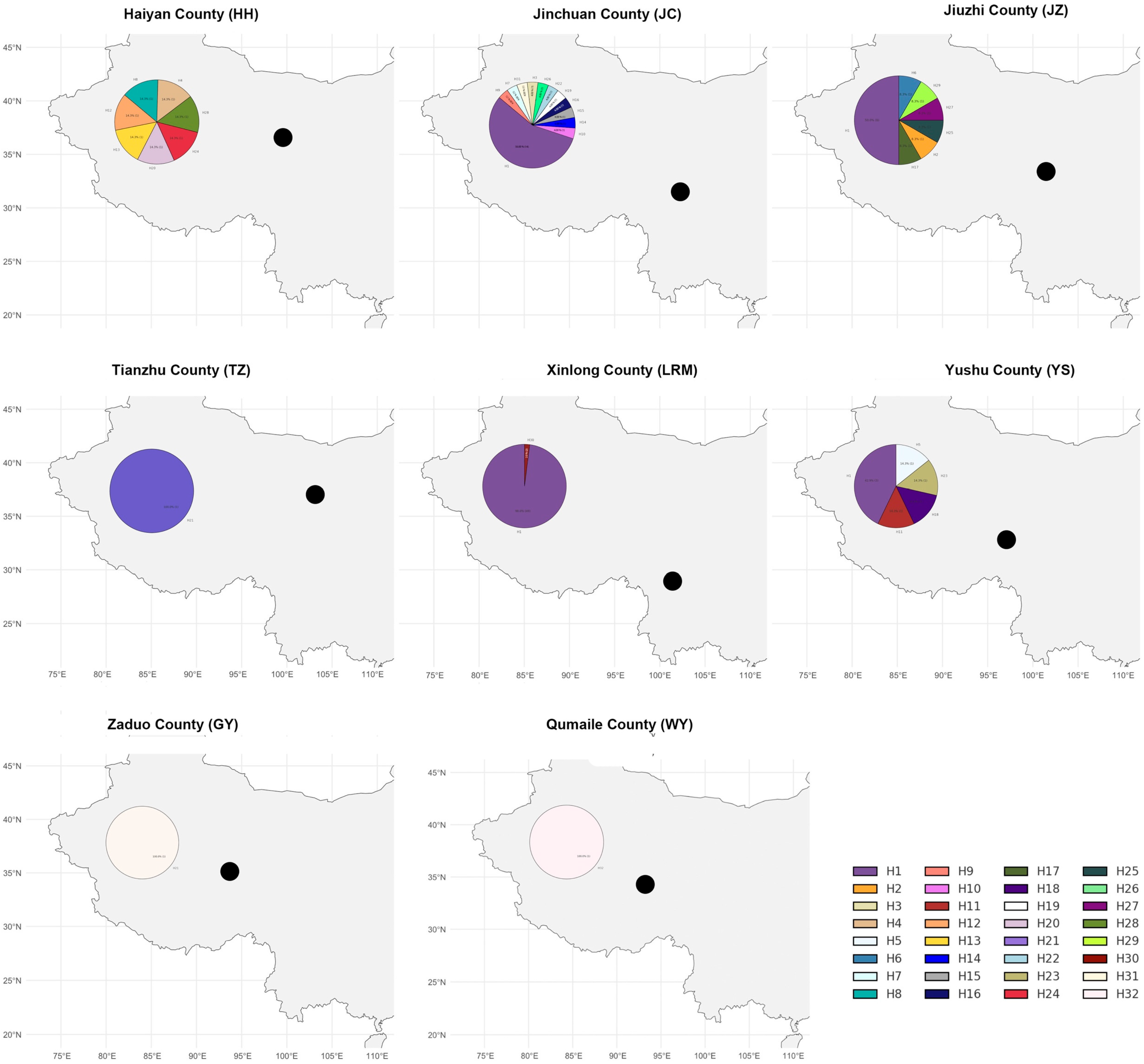
3.1. The Frequency Distribution Among the Eight Yak Breeds (Populations)
3.2. Base Composition of SRY Gene Fragments of Yak Y Chromosome
3.3. Haplotype Diversity Analysis of the SRY Region of the Yak Y Chromosome
3.4. Analysis of Paternal Genetic Differentiation Among Yak Breeds
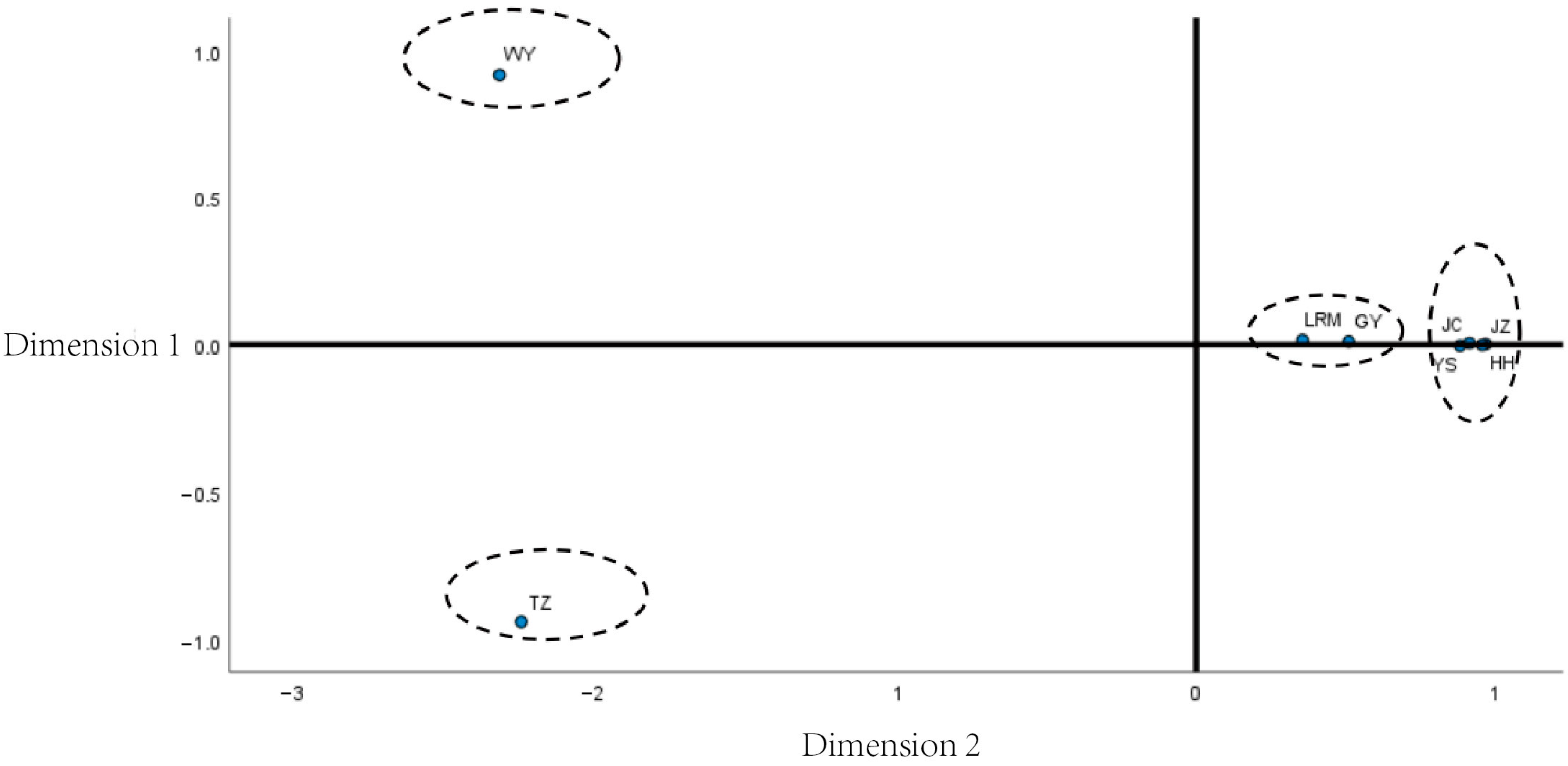
3.5. Multidimensional Scaling Analysis Among Yak Breeds
3.6. Analysis of Molecular Variation in Yak
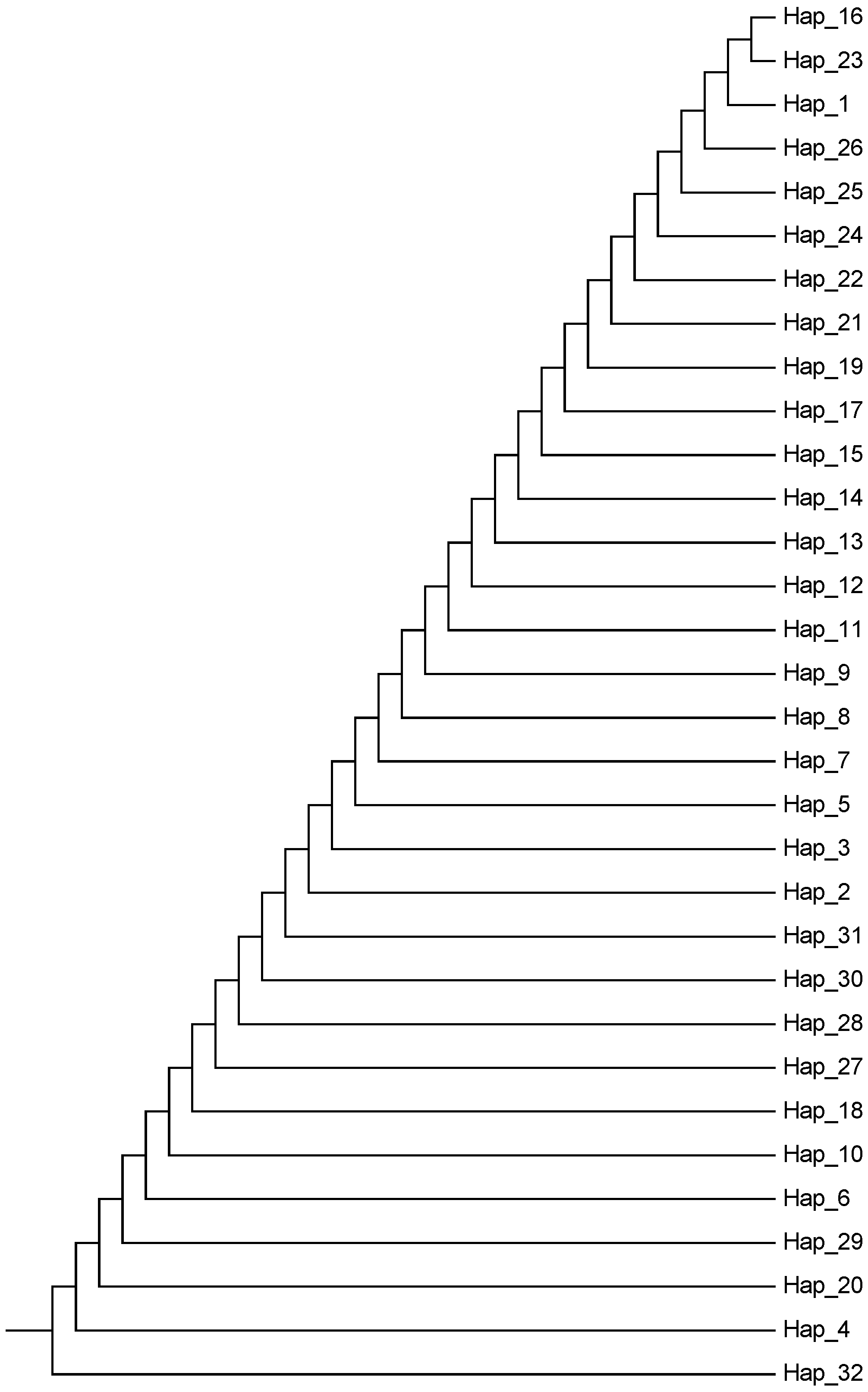
3.7. Phylogenetic Network Diagram Analysis of Y Chromosome Haplotypes in Six Yak Breeds (Populations)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lluís, Q.; Marc, F. The Human Y Chromosome. The Biological Role of a “Functional Wasteland”. J. Biomed. Biotechnol. 2001, 1, 18–24. [Google Scholar]
- Ma, Z.J.; Zhong, J.C.; Han, J.L.; Xu, J.T.; Liu, Z.N.; Bai, W.L. Research progress on molecular genetic diversity of the yak (Bos grunniens). Yi Chuan 2013, 35, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D. The puzzling guppy Y chromosome. Nat. Rev. Genet. 2021, 22, 480–481. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H.; Marszalek, J.D.; Minx, P.J.; Cordum, H.S.; Waterston, R.H.; Wilson, R.K.; Page, D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 2003, 423, 873–876. [Google Scholar] [CrossRef]
- Chang, T.C.; Yang, Y.; Retzel, E.F.; Liu, W.S. Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development. Proc. Natl. Acad. Sci. USA 2013, 110, 12373–12378. [Google Scholar] [CrossRef]
- O’Neill, M.J.; O’Neill, R.J. Whatever happened to SRY? Cell. Mol. Life Sci. 1999, 56, 883–893. [Google Scholar] [CrossRef]
- Yang, H.; Fries, R.; Stranzinger, G. The sex-determining region Y (SRY) gene is mapped to p12-pl3 ofin pig (Sus scrofa domestica) by in situ hybridization. Anim. Genet. 1993, 24, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Vahidi, S.M.F.; Ma, Y.H.; Han, J.L. Limited polymorphisms of two Y-chromosomal SNPs in Chinese and Iranian sheep. Anim. Genet. 2012, 43, 479. [Google Scholar] [CrossRef]
- Liang, Z.; Fangdong, Z.; Sane, C.; Ermi, Z.; Bisong, Y. Cloning of S RY Gene from Moschus berezovskii and M. chrysogaster and Its Application in Phylogenetic Analysis. Zool. Res. 2004, 25, 334–340. [Google Scholar]
- Li, G.; Luo, J.; Wang, F.; Xu, D.; Ahmed, Z.; Chen, S.; Li, R.; Ma, Z. Whole-genome resequencing reveals genetic diversity, differentiation, and selection signatures of yak breeds/populations in Qinghai, China. Front. Genet. 2023, 13, 1034094. [Google Scholar] [CrossRef]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evol. Bioinform. 2005, 1, 117693430500100003. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Alejandro, A.; Rodrigo, M.; Mario, C. Population structure and genetic diversity in Colombian Simmental cattle. Trop. Anim. Health Prod. 2020, 52, 1133–1139. [Google Scholar]
- King, F.J.; Banga, C.B.; Visser, C. Genetic diversity and population structure of three native cattle populations in Mozambique. Trop. Anim. Health Prod. 2021, 53, 117. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.J.; Xia, X.T.; Chen, S.M.; Zhao, X.C.; Zeng, L.L.; Xie, Y.L.; Chao, S.Y.; Xu, J.T.; Sun, Y.G.; Li, R.Z.; et al. Identification and diversity of Y-chromosome haplotypes in Qinghai yak populations. Anim. Genet. 2018, 49, 618–622. [Google Scholar] [CrossRef]
- Fukui, E.; Koganezawa, M.; Yoshizawa, M. Determination of nucleotide sequence of SRY gene in sika deer (Cervus nippon). Anim. Sci. J. 2010, 77, 250–252. [Google Scholar] [CrossRef]
- Li, W.; Zhou, C.; Cheng, M.; Tu, H.; Wang, G.; Mao, Y.; Huang, Y.; Chen, M.; Price, M.; Meng, Y.; et al. Large-scale genetic surveys for main extant population of wild giant panda (Ailuropoda melanoleuca) reveals an urgent need of human management. Evol. Appl. 2023, 16, 738–749. [Google Scholar] [CrossRef]
- Kappeler, P.M.; Benhaiem, S.; Fichtel, C.; Fromhage, L.; Höner, O.P.; Jennions, M.D.; Kaiser, S.; Krüger, O.; Schneider, J.M.; Tuni, C.; et al. Sex roles and sex ratios in animals. Biol. Rev. 2023, 98, 462–480. [Google Scholar] [CrossRef] [PubMed]
- Strucken, E.M.; Gebrehiwot, N.Z.; Swaminathan, M.; Joshi, S.; Al Kalaldeh, M.; Gibson, J.P. Genetic diversity and effective population sizes of thirteen Indian cattle breeds. Genet. Sel. Evol. 2021, 53, 47. [Google Scholar] [CrossRef]
- Wright, S. Variability within and among natural populations. Evol. Genet. Popul. 1984, 4. [Google Scholar]
- Sato, K.; Sato, M. Multiple ways to prevent transmission of paternal mitochondrial DNA for maternal inheritance in animals. J. Biochem. 2017, 162, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Sun, Z.; Ding, F.; Wang, H.; Li, L.; Li, X.; Zheng, X.; Li, N.; Dai, Y.; Wu, C. Efficient TALEN-mediated gene knockin at the bovine Y chromosome and generation of a sex-reversal bovine. Cell. Mol. Life Sci. 2021, 78, 5415–5425. [Google Scholar] [CrossRef] [PubMed]
- Soleymani, B.; Mansouri, K.; Rastegari-Pouyani, M.; Parvaneh, S.; Khademi, F.; Sharifi Tabar, M.; Mostafaie, A. Production of monoclonal antibody against recombinant bovine sex-determining region Y (SRY) and their preferential binding to Y chromosome-bearing sperm. Reprod. Domest. Anim. 2021, 56, 270–277. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef]
- Luo, J.; Wei, X.; Liu, W.; Chen, S.; Ahmed, Z.; Sun, W.; Lei, C.; Ma, Z. Paternal genetic diversity, differentiation and phylogeny of three white yak breeds/populations in China. Sci. Rep. 2022, 12, 19331. [Google Scholar] [CrossRef]

| Community | Sample Size |
|---|---|
| LARIMA YAK (LRM) | 50 |
| Jiuzhi Yak (JZ) | 12 |
| Huanhu Yak (HH) | 20 |
| Yushu Yak (YS) | 7 |
| Jinchuan Yak (JC) | 25 |
| Datong Yak (DT) | 31 |
| Qinghai Plateau Yak (GY) | 1 |
| Tianzhu White Yak (TZ) | 1 |
| Wild Yak (WY) | 1 |
| Assortment | Quantities | Login Number |
|---|---|---|
| Wild Yak (WY) | 1 | XM_005908336 |
| Qinghai Plateau Yak (GY) | 1 | EU547257 |
| Tianzhu White Yak (TZ) | 1 | FJ373272 |
| Yak Groups | Base Composition | Segment Length/bp | |||||
|---|---|---|---|---|---|---|---|
| A% | C% | G% | T% | A + T% | G + C% | ||
| Qinghai Plateau Yak (GY) | 30.00% | 20.87% | 24.78% | 24.35% | 54.35% | 45.65% | 690 |
| Huan hu Yak (HH) | 30.13% | 20.91% | 24.65% | 24.30% | 54.43% | 45.56% | 690 |
| Jinchuan Yak (JC) | 30.09% | 20.89% | 24.67% | 24.35% | 54.44% | 45.56% | 690 |
| Jiuzhi Yak (JZ) | 30.12% | 20.88% | 24.65% | 24.34% | 54.46% | 45.53% | 690 |
| Larima YAK (LRM) | 30.15% | 20.87% | 24.63% | 24.34% | 54.49% | 45.50% | 690 |
| Tianzhu White Yak (TZ) | 30.16% | 20.48% | 26.35% | 23.02% | 53.18% | 46.83% | 690 |
| Wild Yak (WY) | 29.59% | 25.95% | 23.32% | 21.14% | 50.73% | 49.27% | 690 |
| Yushu Yak (YS) | 30.00% | 20.87% | 24.78% | 24.35% | 54.35% | 45.65% | 690 |
| on average | 30.03% | 21.47% | 24.73% | 23.77% | 53.80% | 46.19% | 690 |
| Yak Population | Sample Size | No. of Haplotypes | Haplotype Gene Diversity | Nucleotide Diversity | Mean Number of Pairwise Differences |
|---|---|---|---|---|---|
| Qinghai Plateau Yak (GY) | 1 | 1 | 1.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 |
| Huan hu Yak (HH) | 20 | 10 | 0.711 ± 0.114 | 0.003 ± 0.002 | 1.800 ± 1.084 |
| Jinchuan Yak (JC) | 25 | 12 | 0.697 ± 0.105 | 0.002 + 0.001 | 1.440 ± 0.907 |
| Jiuzhi Yak (JZ) | 12 | 7 | 0.773 ± 0.128 | 0.003 ± 0.002 | 2.000 ± 1.211 |
| Larima YAK (LRM) | 50 | 2 | 0.040 ± 0.038 | 0.000 ± 0.000 | 0.120 ± 0.191 |
| Tianzhu White Yak (TZ) | 1 | 1 | 1.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 |
| Wild Yak (WY) | 1 | 1 | 1.000 ± 0.000 | 0.000 ± 0.000 | 0.000 ± 0.000 |
| Yushu Yak (YS) | 7 | 6 | 0.952 ± 0.096 | 0.003 ± 0.002 | 2.000 ± 1.276 |
| Total/average | 117 | 40 | 0.772 ± 0.060 | 0.001 ± 0.001 | 0.920 ± 0.584 |
| Yak Population | GY | HH | JC | JZ | LRM | TZ | WY | YS |
|---|---|---|---|---|---|---|---|---|
| Qinghai Plateau Yak (GY) | 0.000 | |||||||
| Huan hu Yak (HH) | 0.053 | 0.000 | ||||||
| Jinchuan Yak (JC) | 0.163 | −0.001 | 0.000 | |||||
| Jiuzhi Yak (JZ) | 0.000 | 0.002 | 0.008 | 0.000 | ||||
| Larima YAK (LRM) | 0.887 | 0.042 | 0.024 | 0.115 | 0.000 | |||
| Tianzhu White Yak (TZ) | 1.000 | 0.971 | 0.977 | 0.968 | 0.998 | 0.000 | ||
| Wild Yak (WY) | 1.000 | 0.995 | 0.996 | 0.994 | 1.000 | 1.000 | 0.000 | |
| Yushu Yak (YS) | 0.000 | 0.005 | 0.019 | 0.000 | 0.266 | 0.968 | 0.994 | 0.000 |
| Average F_ST | 0.915 | 0.901 | 0.903 | 0.900 | 0.914 | 0.915 | 0.915 | 0.901 |
| Source of Variation | d.f. | Sum of Squares | Variance Components | Percentage of Variation | Fixed Fixation Index |
|---|---|---|---|---|---|
| Among populations | 7 | 417.868 | 4.86381 Va | 90.71 | |
| Within populations | 109 | 54.32 | 0.49835 Vb | 9.29 | |
| Total | 116 | 472.188 | 5.36215 | F_ST = 0.9070 × 10−6 *** |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Luo, X.; An, T.; Guan, J.; Zhang, X.; Bai, Q.; Sha, Q.; Zhao, H. Paternal Lineage and Genetic Diversity of Jiuzhi Yaks Revealed by Y-Chromosome SRY Sequencing. Animals 2025, 15, 2783. https://doi.org/10.3390/ani15192783
Yang B, Luo X, An T, Guan J, Zhang X, Bai Q, Sha Q, Zhao H. Paternal Lineage and Genetic Diversity of Jiuzhi Yaks Revealed by Y-Chromosome SRY Sequencing. Animals. 2025; 15(19):2783. https://doi.org/10.3390/ani15192783
Chicago/Turabian StyleYang, Boxuan, Xiaolin Luo, Tianwu An, Jiuqiang Guan, Xiangfei Zhang, Qin Bai, Quan Sha, and Hongwen Zhao. 2025. "Paternal Lineage and Genetic Diversity of Jiuzhi Yaks Revealed by Y-Chromosome SRY Sequencing" Animals 15, no. 19: 2783. https://doi.org/10.3390/ani15192783
APA StyleYang, B., Luo, X., An, T., Guan, J., Zhang, X., Bai, Q., Sha, Q., & Zhao, H. (2025). Paternal Lineage and Genetic Diversity of Jiuzhi Yaks Revealed by Y-Chromosome SRY Sequencing. Animals, 15(19), 2783. https://doi.org/10.3390/ani15192783






