The Metabolome in Different Sites of Gut Tract Regulates the Meat Quality of Longissimus Dorsi Muscle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
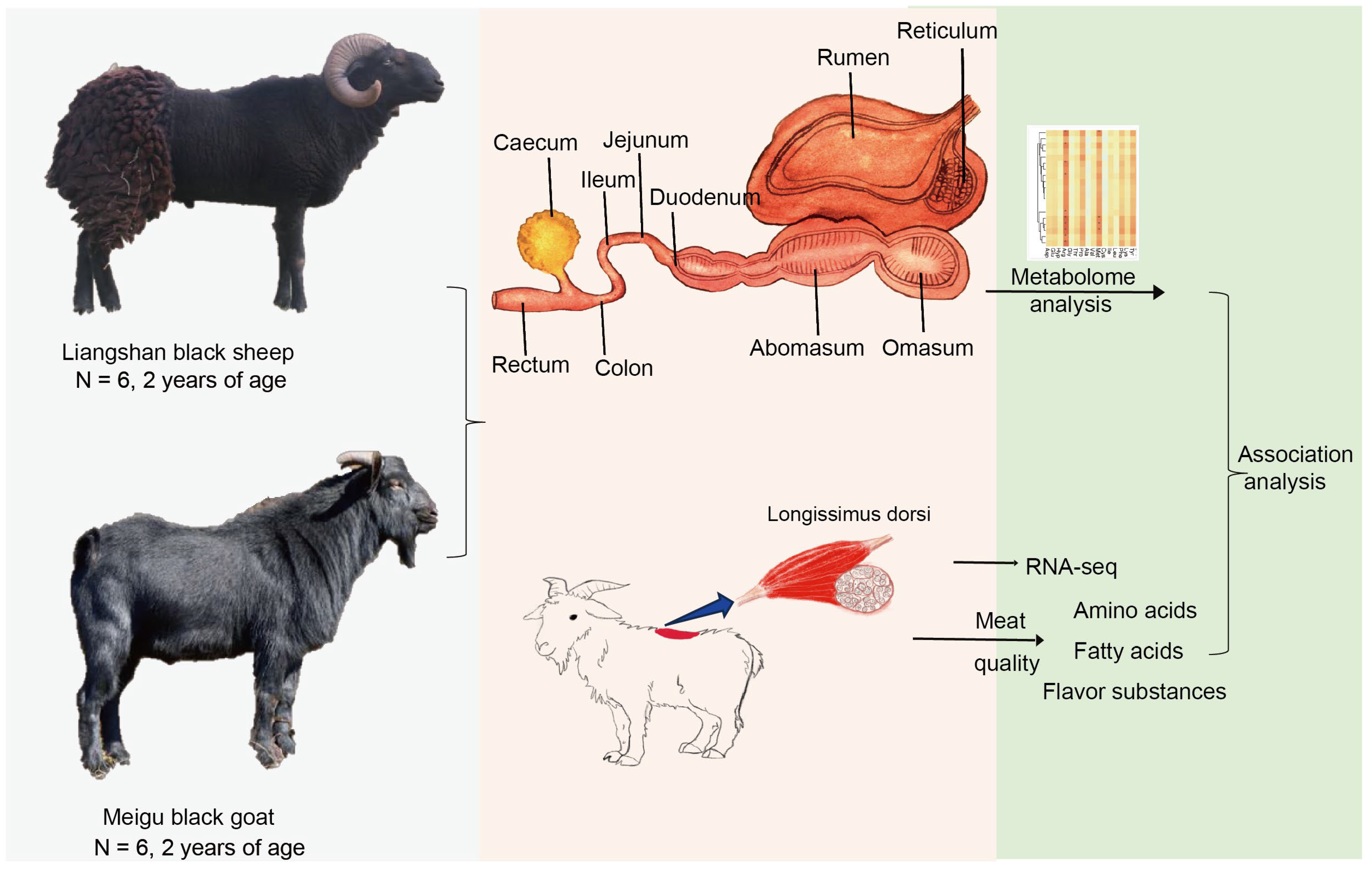
2.1. Sample Collection
2.2. Amino Acid Content
2.3. Fatty Acid Content
2.4. Flavor Content
2.4.1. Solid-Phase Microextraction
2.4.2. Gas Chromatography Conditions
2.5. Total RNA-Seq and Data Analysis
2.6. Gut Metabolite Analysis
2.6.1. Metabolite Extraction
2.6.2. Quality Control Sample
2.6.3. UHPLC-MS/MS Analysis
2.6.4. MS Conditions
2.6.5. Data Analysis
3. Results
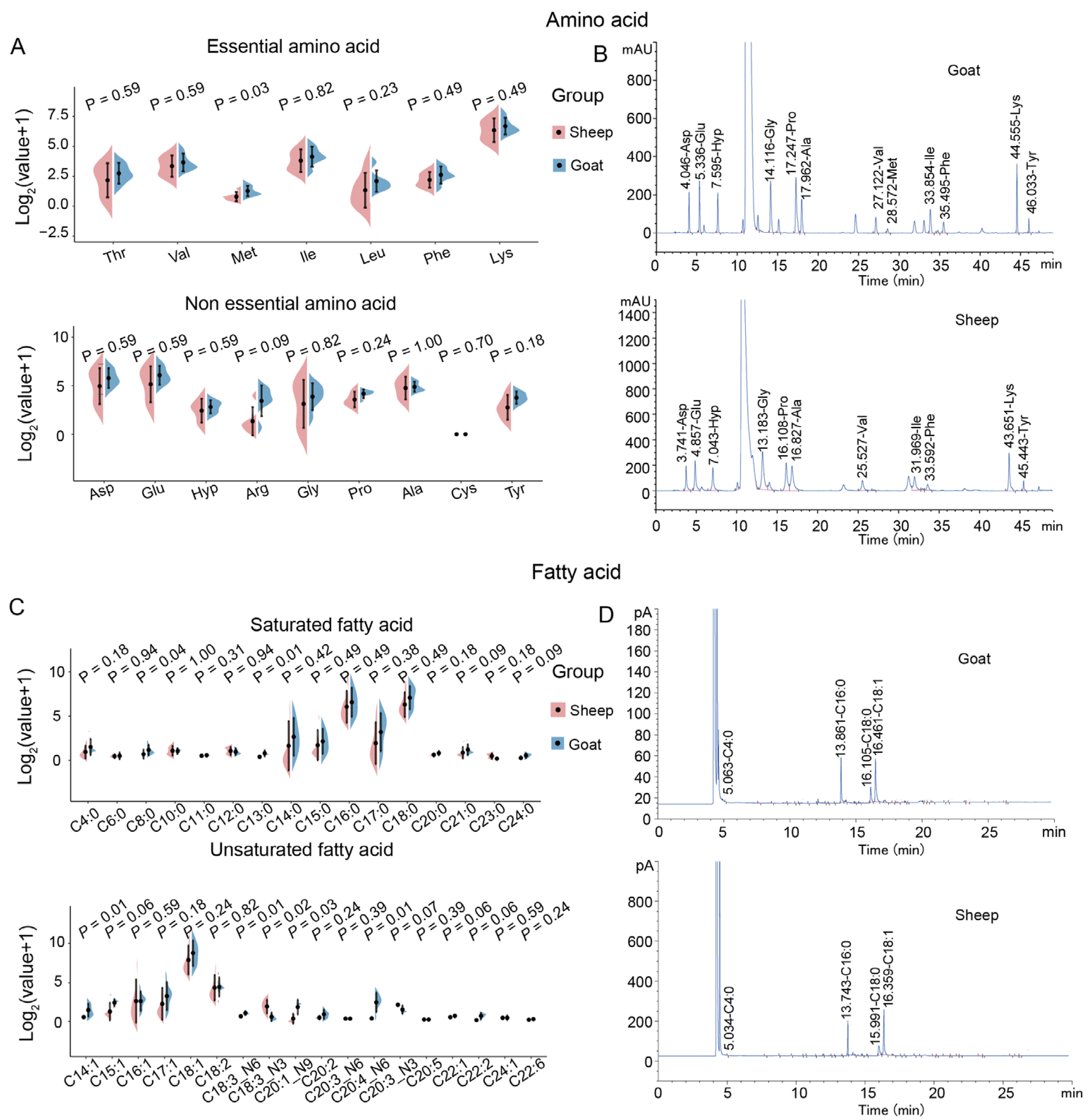
3.1. Amino Acid and Fatty Acid Composition of Sheep and Goats
3.2. Flavor Characteristics of Sheep and Goats
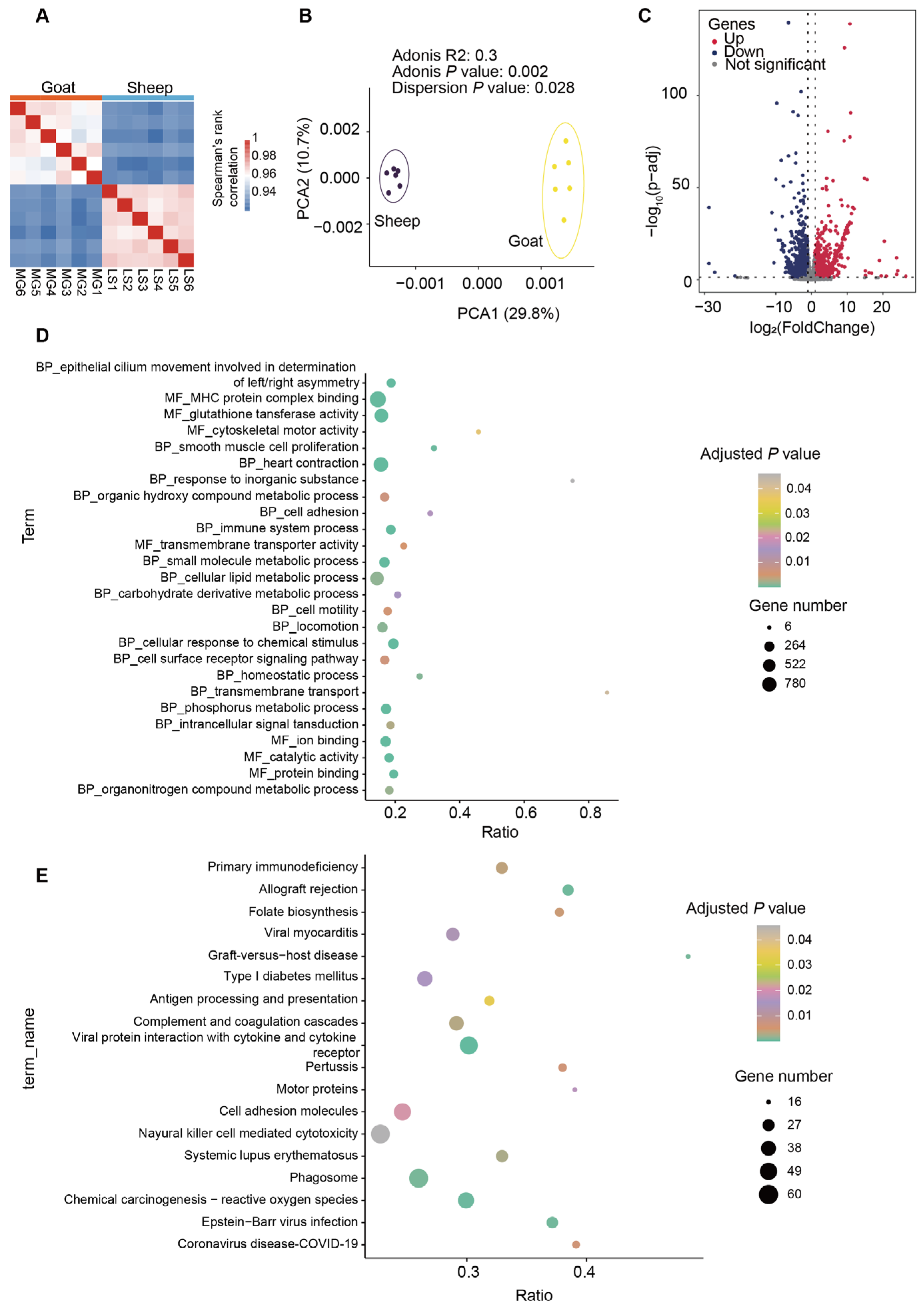
3.3. Differentially Expressed Genes of Longissimus Dorsi Muscle in Sheep and Goats
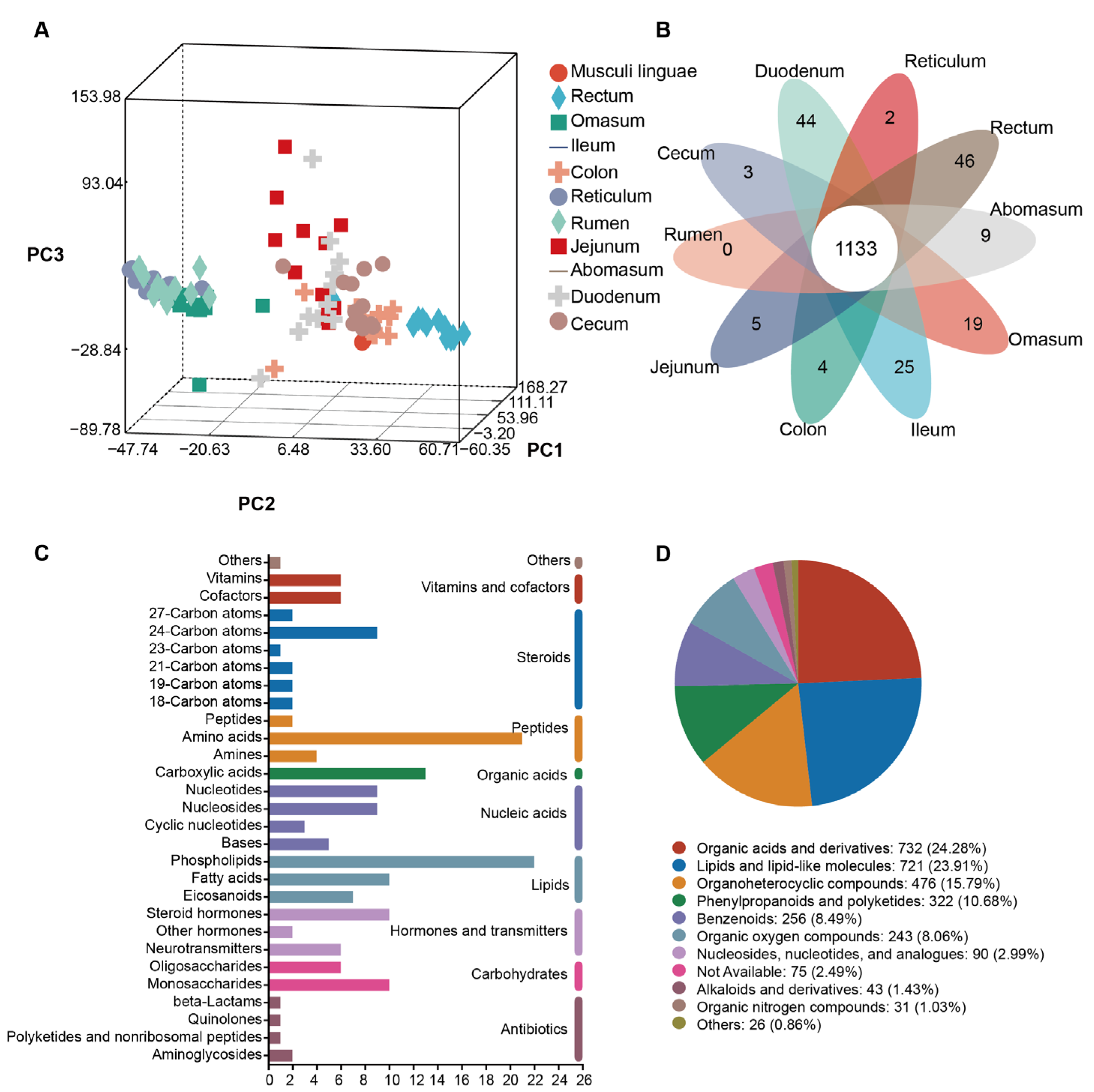
3.4. Gut Flora Metabolome Composition and Function of Sheep and Goats
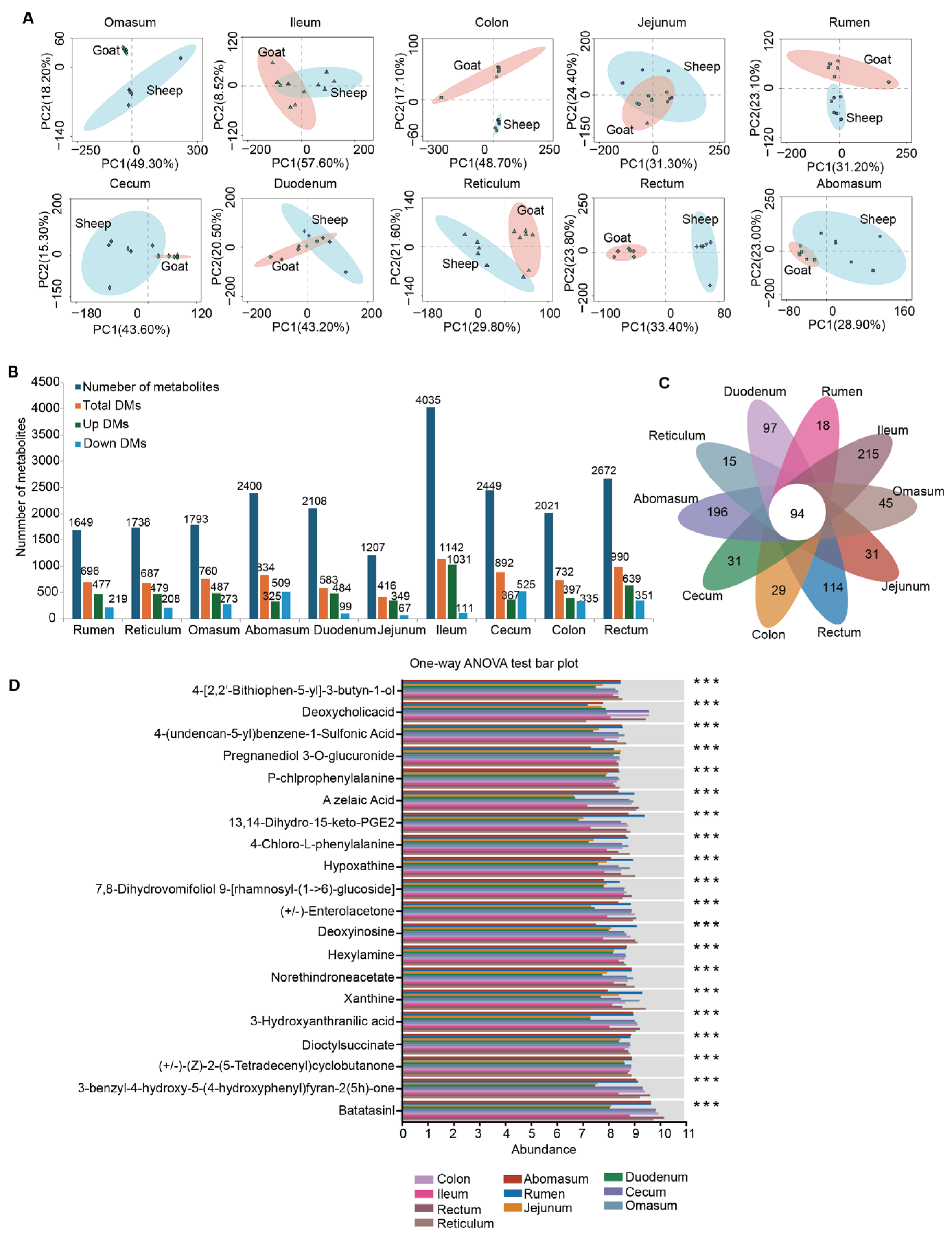
3.5. Differential Metabolites in Ten Gut Sites of Sheep and Goats
3.6. Weighted Gene Co-Expression Network Analysis of Metabolites
3.7. Contribution of the Changes of Longissimus Dorsi Muscle Gene Expression and Gut Metabolites to the Shifts of Meat Quality
4. Discussion
4.1. Goat Meat Exhibits Superior Nutritional and Flavor Profiles Compared to Sheep
4.2. Distinct Gut Metabolite Profiles Suggest Site-Specific Regulation of Muscle Traits
4.3. Muscle Gene Expression Is Coordinated with Gut-Derived Metabolite Signals
4.4. Coordinated Action of Gut Metabolites and Muscle Genes Drives Meat Quality
4.5. Implications for Breeding, Conservation, and Local Meat Production
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fu, L.; Shi, J.; Meng, Q.; Tang, Z.; Liu, T.; Zhang, Q.; Cheng, S. Verification of Key Target Molecules for Intramuscular Fat Deposition and Screening of SNP Sites in Sheep from Small-Tail Han Sheep Breed and Its Cross with Suffolk. Int. J. Mol. Sci. 2024, 25, 2951. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, G.; Chen, M.; Pang, Q.; Jiang, S.; Zeng, J.; Du, D.; Yang, P.; Wu, W.; Zhao, H. Genetic diversity and population structure of sheep (Ovis aries) in Sichuan, China. PLoS ONE 2021, 16, e0257974. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Tao, H.; Li, P.; Li, L.I.; Zhong, T.; Wang, L.; Ma, J.; Chen, X.; Song, T.; Zhang, H. Whole-genome sequencing reveals selection signatures associated with important traits in six goat breeds. Sci. Rep. 2018, 8, 10405. [Google Scholar] [CrossRef]
- Weijie, Y. Breeding Technology and Demonstration Promotion of Meigu goat. Livest. Sci. 2022, 16, 43–45. (In Chinese) [Google Scholar]
- Quzhe, E.J.W.; Cuo, P.; Muqie, B.; Lei, R.; Zhangjia, B.; Shizhong, Y.; Amei, J. Study on the Measurement of Body Weight, Body Size, and Feed Intake of Liangshan Black Sheep at Different Growth Stages. Sichuan Anim. Vet. Sci. 2024, 51, 24–26. [Google Scholar]
- Chen, B.; Li, D.; Leng, D.; Kui, H.; Bai, X.; Wang, T. Gut microbiota and meat quality. Front. Microbiol. 2022, 13, 951726. [Google Scholar] [CrossRef]
- Joo, S.T.; Kim, G.D.; Hwang, Y.H.; Ryu, Y.C. Control of fresh meat quality through manipulation of muscle fiber characteristics. Meat Sci. 2013, 95, 828–836. [Google Scholar] [CrossRef]
- Nkrumah, J.D.; Okine, E.K.; Mathison, G.W.; Schmid, K.; Li, C.; Basarab, J.A.; Price, M.A.; Wang, Z.; Moore, S.S. Relationships of feedlot feed efficiency, performance, and feeding behavior with metabolic rate, methane production, and energy partitioning in beef cattle. J. Anim. Sci. 2006, 84, 145–153. [Google Scholar] [CrossRef]
- Kumar, S.; Dagar, S.S.; Puniya, A.K.; Upadhyay, R.C. Changes in methane emission, rumen fermentation in response to diet and microbial interactions. Res. Vet. Sci. 2013, 94, 263–268. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.F.; Bahram, M.; Han, J.L.; Ding, X.Z.; Salekdeh, G.H. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2021, 15, 1108–1120. [Google Scholar] [CrossRef]
- Leng, D.; Huang, Z.; Bai, X.; Wang, T.; Zhang, Y.; Chang, W.; Zhao, W.; Li, D.; Chen, B. Gene expression profiles in specific skeletal muscles and meat quality characteristics of sheep and goats. Sci. Data 2024, 11, 1390. [Google Scholar] [CrossRef]
- Lamichhane, S.; Sen, P.; Dickens, A.M.; Orešič, M.; Bertram, H.C. Gut metabolome meets microbiome: A methodological perspective to understand the relationship between host and microbe. Methods 2018, 149, 3–12. [Google Scholar] [CrossRef] [PubMed]
- Lamichhane, S.; Yde, C.C.; Schmedes, M.S.; Jensen, H.M.; Meier, S.; Bertram, H.C. Strategy for Nuclear-Magnetic-Resonance-Based Metabolomics of Human Feces. Anal. Chem. 2015, 87, 5930–5937. [Google Scholar] [CrossRef]
- Oliver, S.G.; Winson, M.K.; Kell, D.B.; Baganz, F. Systematic functional analysis of the yeast genome. Trends Biotechnol. 1998, 16, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Macel, M.; Van Dam, N.M.; Keurentjes, J.J. Metabolomics: The chemistry between ecology and genetics. Mol. Ecol. Resour. 2010, 10, 583–593. [Google Scholar] [CrossRef]
- Muller, E.; Algavi, Y.M.; Borenstein, E. The gut microbiome-metabolome dataset collection: A curated resource for integrative meta-analysis. npj Biofilms Microbiomes 2022, 8, 79. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Ge, Y.; Xu, Z.; Zhang, D.; Li, D. The digestive and reproductive tract microbiotas and their association with body weight in laying hens. Poult. Sci. 2021, 100, 101422. [Google Scholar] [CrossRef]
- Van Treuren, W.; Dodd, D. Microbial Contribution to the Human Metabolome: Implications for Health and Disease. Annu. Rev. Pathol. 2020, 15, 345–369. [Google Scholar] [CrossRef]
- Postler, T.S.; Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef]
- Zhou, X.; Zhang, H.; Li, S.; Jiang, Y.; Kang, L.; Deng, J.; Yang, C.; Zhao, X.; Zhao, J.; Jiang, L.; et al. The effects of fermented feedstuff derived from Citri Sarcodactylis Fructus by-products on growth performance, intestinal digestive enzyme activity, nutrient utilization, meat quality, gut microbiota, and metabolites of broiler chicken. Front. Vet. Sci. 2023, 10, 1231996. [Google Scholar] [CrossRef]
- Ma, Y.; Yang, X.; Hua, G.; Deng, X.; Xia, T.; Li, X.; Feng, D.; Deng, X. Contribution of gut microbiomes and their metabolomes to the performance of Dorper and Tan sheep. Front. Microbiol. 2022, 13, 1047744. [Google Scholar] [CrossRef]
- Goethals, S.; Rombouts, C.; Hemeryck, L.Y.; Van Meulebroek, L.; Van Hecke, T.; Vossen, E.; Van Camp, J.; De Smet, S.; Vanhaecke, L. Untargeted Metabolomics to Reveal Red versus White Meat-Associated Gut Metabolites in a Prudent and Western Dietary Context. Mol. Nutr. Food Res. 2020, 64, e2000070. [Google Scholar] [CrossRef]
- Rikimaru, K.; Takahashi, H. Evaluation of the meat from Hinai-jidori chickens and broilers: Analysis of general biochemical components, free amino acids, inosine 5′-monophosphate, and fatty acids. J. Appl. Poult. Res. 2010, 19, 327–333. [Google Scholar] [CrossRef]
- Zhao, H.; Chong, J.; Tang, R.; Li, L.; Xia, J.; Li, D. Metabolomics investigation of dietary effects on flesh quality in grass carp (Ctenopharyngodon idellus). Gigascience 2018, 7, giy111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Tang, C.; Zhao, Q.; Yang, Y.; Li, F.; Qin, Y.; Liu, X.; Yue, X.; Zhang, J. Integrated lipidomics and targeted metabolomics analyses reveal changes in flavor precursors in psoas major muscle of castrated lambs. Food Chem. 2020, 333, 127451. [Google Scholar] [CrossRef]
- Fu, Y.; Cao, S.; Yang, L.; Li, Z. Flavor formation based on lipid in meat and meat products: A review. J. Food Biochem. 2022, 46, e14439. [Google Scholar] [CrossRef] [PubMed]
- Davenport, K.M.; Bickhart, D.M.; Worley, K.; Murali, S.C.; Salavati, M.; Clark, E.L.; Cockett, N.E.; Heaton, M.P.; Smith, T.P.; Murdoch, B.M.; et al. An improved ovine reference genome assembly to facilitate in-depth functional annotation of the sheep genome. Gigascience 2022, 11, giab096. [Google Scholar] [CrossRef]
- Bickhart, D.M.; Rosen, B.D.; Koren, S.; Sayre, B.L.; Hastie, A.R.; Chan, S.; Lee, J.; Lam, E.T.; Liachko, I.; Sullivan, S.T.; et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017, 49, 643–650. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Ghosh, S.; Chan, C.K. Analysis of RNA-Seq Data Using TopHat and Cufflinks. Methods Mol. Biol. 2016, 1374, 339–361. [Google Scholar]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascenção, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Zhao, B.; Sun, B.; Wang, S.; Zhang, Y.; Zang, M.; Le, W.; Wang, H.; Wu, Q. Effect of different cooking water on flavor characteristics of mutton soup. Food Sci. Nutr. 2021, 9, 6047–6059. [Google Scholar] [CrossRef]
- Paulos, K.; Rodrigues, S.; Oliveira, A.F.; Leite, A.; Pereira, E.; Teixeira, A. Sensory Characterization and Consumer Preference Mapping of Fresh Sausages Manufactured with Goat and Sheep Meat. J. Food Sci. 2015, 80, S1568–S1573. [Google Scholar] [CrossRef]
- Whitfield, F.B. Volatiles from interactions of Maillard reactions and lipids. Crit. Rev. Food Sci. Nutr. 1992, 31, 1–58. [Google Scholar] [CrossRef]
- Leng, D.; Yang, M.; Miao, X.; Huang, Z.; Li, M.; Liu, J.; Wang, T.; Li, D.; Feng, C. Dynamic changes in the skin transcriptome for the melanin pigmentation in embryonic chickens. Poult. Sci. 2024, 104, 104210. [Google Scholar] [CrossRef]
- Watkins, P.J.; Jaborek, J.R.; Teng, F.; Day, L.; Castada, H.Z.; Baringer, S.; Wick, M. Branched chain fatty acids in the flavour of sheep and goat milk and meat: A review. Small Rumin. Res. 2021, 200, 106398. [Google Scholar] [CrossRef]
- Domínguez, R.; Pateiro, M.; Gagaoua, M.; Barba, F.J.; Zhang, W.; Lorenzo, J.M. A Comprehensive Review on Lipid Oxidation in Meat and Meat Products. Antioxidants 2019, 8, 429. [Google Scholar] [CrossRef]
- Aghwan, Z.A.; Alimon, A.R.; Goh, Y.M.; Nakyinsige, K.; Sazili, A.Q. Fatty Acid Profiles of Supraspinatus, Longissimus lumborum and Semitendinosus Muscles and Serum in Kacang Goats Supplemented with Inorganic Selenium and Iodine. Asian-Australas. J. Anim. Sci. 2014, 27, 543–550. [Google Scholar] [CrossRef]
- Zhang, N.; Teng, Z.; Qi, Q.; Hu, G.; Lian, H.; Gao, T. Carcass traits, meat quality characteristics, and lipid metabolism-related gene expression pattern of Yaoshan white goats raised in traditional extensive production system: Effects of slaughter age and meat cuts. Small Rumin. Res. 2020, 182, 29–36. [Google Scholar] [CrossRef]
- Wang, K.; Wu, P.; Wang, S.; Ji, X.; Chen, D.; Xiao, W.; Gu, Y.; Zeng, Y.; Xu, X.; Tang, G. Differential DNA methylation analysis reveals key genes in Chinese Qingyu and Landrace pigs. Genome 2022, 65, 19–26. [Google Scholar] [CrossRef]
- Khannpnavar, B.; Mehta, V.; Qi, C.; Korkhov, V. Structure and function of adenylyl cyclases, key enzymes in cellular signaling. Curr. Opin. Struct. Biol. 2020, 63, 34–41. [Google Scholar] [CrossRef]
- Liu, M.; Woodward-Greene, J.; Kang, X.; Pan, M.G.; Rosen, B.; Van Tassell, C.P.; Chen, H.; Liu, G.E. Genome-wide CNV analysis revealed variants associated with growth traits in African indigenous goats. Genomics 2020, 112, 1477–1480. [Google Scholar] [CrossRef]
- Yarwood, S.J.; Kilgour, E.; Anderson, N.G. Cyclic AMP potentiates growth hormone-dependent differentiation of 3T3-F442A preadipocytes: Possible involvement of the transcription factor CREB. Mol. Cell Endocrinol. 1998, 138, 41–50. [Google Scholar] [CrossRef]
- Harwood, J.P.; Grewe, C.; Aguilera, G. Actions of growth hormone-releasing factor and somatostatin on adenylate cyclase and growth hormone release in rat anterior pituitary. Mol. Cell Endocrinol. 1984, 37, 277–284. [Google Scholar] [CrossRef]
- Shi, Q.; Padmanabhan, R.; Villegas, C.J.; Gu, S.; Jiang, J.X. Membrane topological structure of neutral system N/Aamino acid transporter 4 (SNAT4) protein. J. Biol. Chem. 2011, 286, 38086–38094. [Google Scholar] [CrossRef]
- Kondou, H.; Kawai, M.; Tachikawa, K.; Kimoto, A.; Yamagata, M.; Koinuma, T.; Yamazaki, M.; Nakayama, M.; Mushiake, S.; Ozono, K.; et al. Sodium-coupled neutral amino acid transporter 4 functions as a regulator of protein synthesis during liver development. Hepatol. Res. 2013, 43, 1211–1223. [Google Scholar] [CrossRef]
- Bröer, S. The SLC38 family of sodium-amino acid co-transporters. Pflugers Arch. 2014, 466, 155–172. [Google Scholar] [CrossRef] [PubMed]
- Pizzagalli, M.D.; Bensimon, A.; Superti-Furga, G. A guide to plasma membrane solute carrier proteins. FEBS J. 2021, 288, 2784–2835. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Dominguez Gutierrez, G.; Xin, Y.; Cavino, K.; Sung, B.; Sipos, B.; Kloeppel, G.; Gromada, J.; Okamoto, H. Increased SLC38A4 Amino Acid Transporter Expression in Human Pancreatic α-Cells After Glucagon Receptor Inhibition. Endocrinology 2019, 160, 979–988. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Leng, D.; Cai, Z.; Chen, B.; Li, J.; Kui, H.; Li, D.; Li, Z. Insights into left-right asymmetric development of chicken ovary at the single-cell level. J. Genet. Genom. 2024, 51, 1265–1277. [Google Scholar] [CrossRef]
- Li, J.; Lin, Y.; Li, D.; He, M.; Kui, H.; Bai, J.; Chen, Z.; Gou, Y.; Zhang, J.; Wang, T.; et al. Building Haplotype-Resolved 3D Genome Maps of Chicken Skeletal Muscle. Adv. Sci. 2024, 11, 2305706. [Google Scholar] [CrossRef]
- Leng, D.; Zeng, B.; Wang, T.; Chen, B.L.; Li, D.Y.; Li, Z.J. Single nucleus/cell RNA-seq of the chicken hypothalamic-pituitary-ovarian axis offers new insights into the molecular regulatory mechanisms of ovarian development. Zool. Res. 2024, 45, 1088–1107. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, B.; Zheng, T.; Bai, X.; Chang, W.; Zhang, Y.; Yang, S.; Li, H.; Li, D.; Wang, T. The Metabolome in Different Sites of Gut Tract Regulates the Meat Quality of Longissimus Dorsi Muscle. Animals 2025, 15, 2399. https://doi.org/10.3390/ani15162399
Chen B, Zheng T, Bai X, Chang W, Zhang Y, Yang S, Li H, Li D, Wang T. The Metabolome in Different Sites of Gut Tract Regulates the Meat Quality of Longissimus Dorsi Muscle. Animals. 2025; 15(16):2399. https://doi.org/10.3390/ani15162399
Chicago/Turabian StyleChen, Binlong, Tingting Zheng, Xue Bai, Weihua Chang, Yi Zhang, Shizhong Yang, Hao Li, Diyan Li, and Tao Wang. 2025. "The Metabolome in Different Sites of Gut Tract Regulates the Meat Quality of Longissimus Dorsi Muscle" Animals 15, no. 16: 2399. https://doi.org/10.3390/ani15162399
APA StyleChen, B., Zheng, T., Bai, X., Chang, W., Zhang, Y., Yang, S., Li, H., Li, D., & Wang, T. (2025). The Metabolome in Different Sites of Gut Tract Regulates the Meat Quality of Longissimus Dorsi Muscle. Animals, 15(16), 2399. https://doi.org/10.3390/ani15162399





