Genetic Diversity and Population Structure of Nine Local Sheep Populations Bred in the Carpathia Area of Central Europe Revealed by Microsatellite Analysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Isolation
2.2. Microsatellite Markers
2.3. PCR and Fragmentation Analysis
2.4. General Statistical Analysis
3. Results
3.1. Genetic Variability of Microsatellite Markers
3.2. Genetic Variability Among Populations
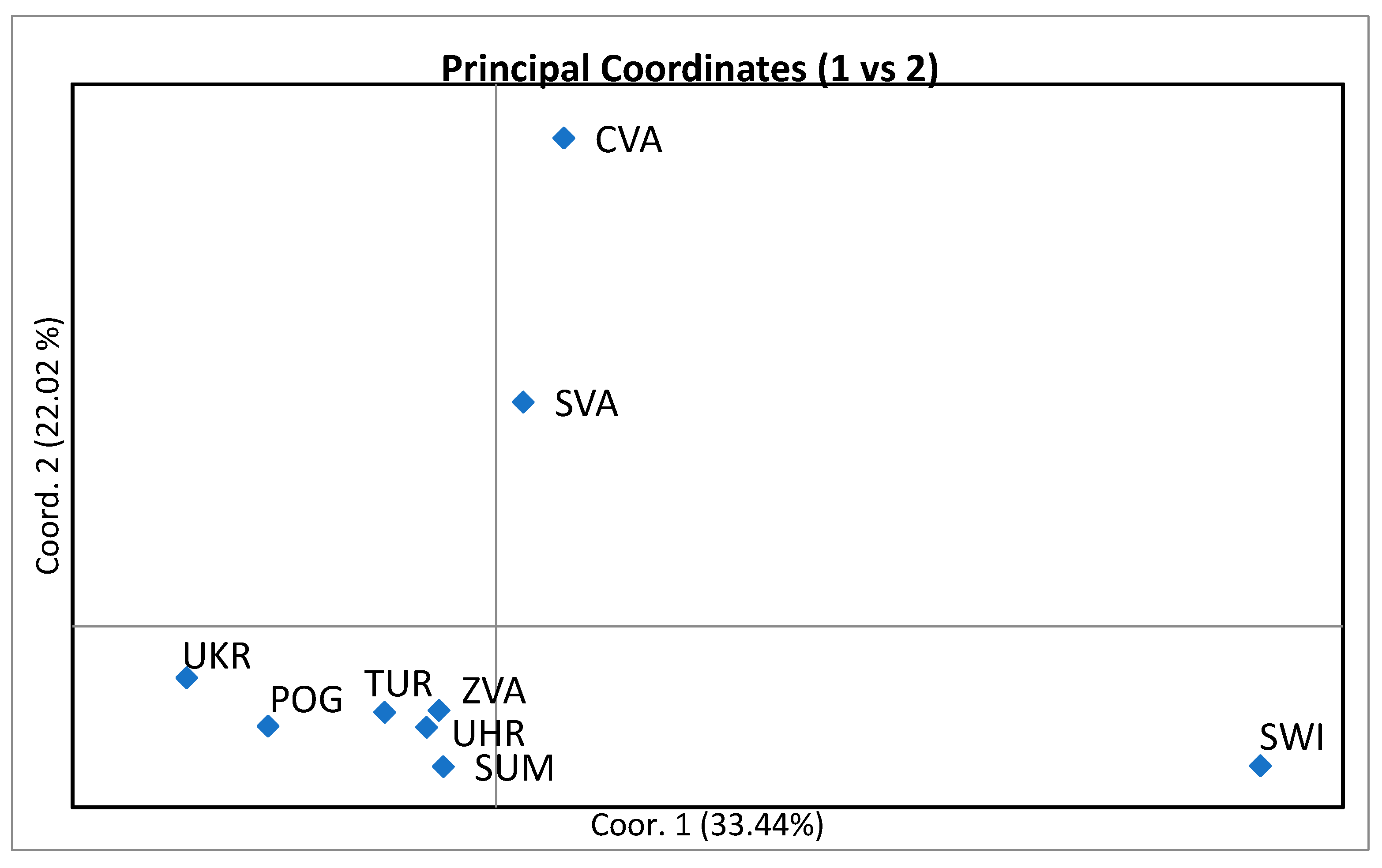
3.3. Genetic Distance Among Populations
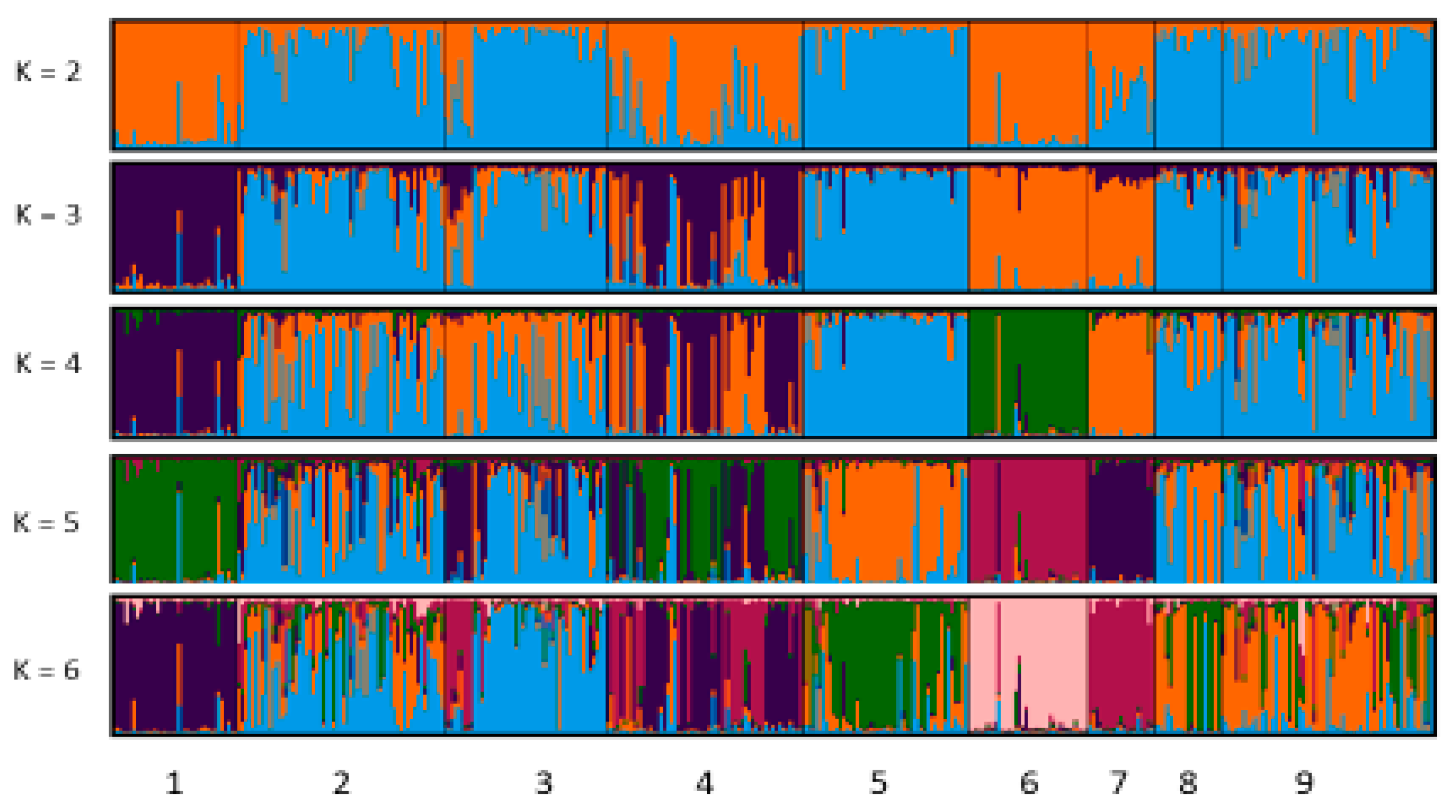
3.4. Genetic Structure and Admixture Analysis
4. Discussion
4.1. Population Genetic Diversity Based on Microsatellite Markers
4.2. Population Genetic Diversity Among Breeds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lawson Handley, L.; Byrne, K.; Santucci, F.; Townsend, S.; Taylor, M.; Bruford, M.W.; Hewitt, G. Genetic structure of European sheep breeds. Heredity 2007, 99, 620–631. [Google Scholar] [CrossRef]
- Ciani, E.; Mastrangelo, S.; Da Silva, A.; Marroni, F.; Ferenčaković, M.; Ajmone-Marsan, P.; Baird, H.; Barbato, M.; Colli, L.; Delvento, C.; et al. On the Origin of European Sheep as Revealed by the Diversity of the Balkan Breeds and by Optimizing Population-Genetic Analysis Tools. Genet. Sel. Evol. 2020, 52, 25. [Google Scholar] [CrossRef] [PubMed]
- Gurgul, A.; Jasielczuk, I.; Miksza-Cybulska, A.; Kawęcka, A.; Szmatoła, T.; Krupiński, J. Evaluation of Genetic Differentiation and Genome-Wide Selection Signatures in Polish Local Sheep Breeds. Livest. Sci. 2021, 251, 104635. [Google Scholar] [CrossRef]
- Machová, K.; Marina, H.; Arranz, J.J.; Pelayo, R.; Rychtářová, J.; Milerski, M.; Vostrý, L.; Suárez-Vega, A. Genetic diversity of two native sheep breeds by genome-wide analysis of single nucleotide polymorphisms. Animal 2023, 17, 100690. [Google Scholar] [CrossRef] [PubMed]
- Ptáček, M.; Ducháček, J.; Stádník, L.; Fantová, M. Analysis of Genotype, Dam’s Litter Size and Their Interaction on Selected Productive Traits of Origin Wallachian and Sumava Sheep in the Czech Republic. Acta Univ. Agric. Silvic. Mendel. Brun. 2017, 65, 473–479. [Google Scholar] [CrossRef]
- Peter, C.; Bruford, M.; Perez, T.; Dalamitra, S.; Hewitt, G. Genetic diversity and subdivision of 57 European and Middle-Eastern sheep breeds. Anim. Genet. 2007, 38, 37–44. [Google Scholar] [CrossRef]
- Kijas, J.W.; Townley, D.; Dalrymple, B.P.; Heaton, M.P.; Maddox, J.F.; McGrath, A.; Wilson, P.; Ingersoll, R.G.; McCulloch, R.; McWilliam, S.; et al. A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS ONE 2009, 4, e4668. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Porto Neto, L.R.; San Cristobal, M.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Kusza, S.; Ivankovic, A.; Ramljak, J.; Nagy, I.; Jávor, A.; Kukovics, S. Genetic structure of Tsigai, Ruda, Pramenka and other local sheep in Southern and Eastern Europe. Small Rumin. Res. 2011, 99, 130–134. [Google Scholar] [CrossRef]
- Odjakova, T.; Todorov, P.; Radoslavov, G.; Hristov, P. Microsatellite Genotyping of Two Bulgarian Sheep Breeds. Diversity 2022, 14, 210. [Google Scholar] [CrossRef]
- Odjakova, T.; Todorov, P.; Kalaydzhiev, G.; Salkova, D.; Dundarova, H.; Radoslavov, G.; Hristov, P. A Study on the Genetic Diversity and Subpopulation Structure of Three Bulgarian Mountainous Sheep Breeds, Based on Genotyping of Microsatellite Markers. Small Rumin. Res. 2023, 226, 107034. [Google Scholar] [CrossRef]
- Mihailova, Y.; Rusanov, K.; Rusanova, M.; Vassileva, P.; Atanassov, I.; Nikolov, V.; Todorovska, E.G. Genetic Diversity and Population Structure of Bulgarian Autochthonous Sheep Breeds Revealed by Microsatellite Analysis. Animals 2023, 13, 1878. [Google Scholar] [CrossRef]
- Cortes, O.; Cañon, J.; Gama, L.T. Applications of Microsatellites and Single Nucleotide Polymorphisms for the Genetic Characterization of Cattle and Small Rumin.: An Overview. Ruminants 2022, 2, 456–470. [Google Scholar] [CrossRef]
- Laoun, A.; Harkat, S.; Lafri, M.; Gaouar, S.B.S.; Belabdi, I.; Ciani, E.; Groot, M.D.; Blanquet, V.; Leroy, G.; Rognon, X.; et al. Inference of breed structure in farm animals: Empirical comparison between SNP and microsatellite performance. Genes 2020, 11, 57. [Google Scholar] [CrossRef]
- Wanjala, G.; Astuti, P.K.; Bagi, Z.; Kichamu, N.; Strausz, P.; Kusza, S. Assessing the Genomics Structure of Dorper and White Dorper Variants, and Dorper Populations in South AfricaandHungary. Biology 2023, 12, 386. [Google Scholar] [CrossRef] [PubMed]
- Olschewsky, A.; Hinrichs, D. An overview of the use of genotyping techniques for assessing genetic diversity in local farm animal breeds. Animals 2021, 11, 2016. [Google Scholar] [CrossRef] [PubMed]
- Kawęcka, A.; Pasternak, M.; Miksza-Cybulska, A.; Puchała, M. Native Sheep Breeds in Poland-Importance and Outcomes of Genetic Resources Protection Programmes. Animals 2022, 12, 1510. [Google Scholar] [CrossRef] [PubMed]
- Dudu, A.; Popa, G.-O.; Ghiță, E.; Pelmuș, R.; Lazăr, C.; Costache, M.; Georgescu, S.E. Assessment of genetic diversity in main local sheep breeds from Romania using microsatellite markers. Arch. Anim. Breed. 2020, 63, 53–59. [Google Scholar] [CrossRef]
- Kusza, S.; Dimov, D.; Nagy, I.; Bõsze, Z.; Jávor, A.; Kukovics, S. Microsatellite analysis to estimate genetic relationships among five bulgarian sheep breeds. Genet. Mol. Biol. 2010, 33, 51–56. [Google Scholar] [CrossRef]
- Meyermans, R.; Gorssen, W.; Aerts, N.; Hooyberghs, K.; Chakkingal Bhaskaran, B.; Chapard, L.; Buys, N.; Janssens, S. Genomic characterisation and diversity assessment of eight endangered Belgian sheep breeds. Animal 2024, 18, 101315. [Google Scholar] [CrossRef]
- FAO. The Second Report on the State of the World’s Animal Genetic Resources for Food and Agriculture; Scherf, B.D., Pilling, D., Eds.; FAO Commission on Genetic Resources for Food and Agriculture Assess-Ments: Rome, Italy, 2015; Available online: http://reliefweb.int/report/world/second-report-state-worlds-animal-genetic-resources-food-and-agriculture (accessed on 12 March 2025).
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314. [Google Scholar] [PubMed]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Rogers, J.S. Measures of genetic similarity and genetic distance. Stud. Genet. 1972, 7, 145–153. [Google Scholar]
- Cavalli-Sforza, L.L.; Edwards, A.W. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 1967, 19 Pt 1, 233–257. [Google Scholar] [PubMed] [PubMed Central]
- Reynolds, J.; Weir, B.S.; Cockerham, C. Estimation of the co-ancestry coefficient basis for a shortterm genetic distance. Genetics 1983, 105, 767–779. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef]
- Kopelman, N.M.; Mayzel, J.; Jakobsson, M.; Rosenberg, N.A.; Mayrose, I. Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Mol. Ecol. Resour. 2015, 15, 1179–1191. [Google Scholar] [CrossRef]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Puechmaille, S. The program structure does not reliably recover the correct population structure when sampling is uneven: Subsampling and new estimators alleviate the problem. Mol. Ecol. Resour. 2016, 16, 608–627. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Liu, J.X. StructureSelector: A web-based software to select and visualize the optimal number of clusters using multiple methods. Mol. Ecol. Resour. 2018, 18, 176–177. [Google Scholar] [CrossRef] [PubMed]
- Kardos, M.; Taylor, H.R.; Ellegren, H.; Luikart, G.; Allendorf, F.W. Genomics advances the study of inbreeding depression in the wild. Evol. Appl. 2016, 9, 1205–1218. [Google Scholar] [CrossRef]
- Jandurová, O.M.; Kott, T.; Kottová, B.; Czerneková, V.; Milerski, M. Genetic relationships among Šumava, Valachian and Improved Valachian sheep. Small Rumin. Res. 2005, 57, 157–165. [Google Scholar] [CrossRef]
- Hristova, D.; Todorovska, E.; Vassilev, D.; Metodiev, S.; Popov, I.; Yablanski, T.; Zhelyazkov, E. Microsatellites based genetic diversity and population structure of seven Bulgarian indigenous sheep breeds. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 569–581. [Google Scholar]
- Mihailova, Y. Genetic diversity and structure of 2 indigenous sheep breeds (Kotel and Teteven) in Bulgaria using microsatellite markers. Biotechnol. Biotechnol. Equip. 2021, 35, 576–585. [Google Scholar] [CrossRef]
- Phookan, A.; Das, B.; Das, A.; Islam, R.; Sharma, M.; Bharali, K.; Basumatary, K. Morphology, morphometry and certain eggquality traits of indigenous ducks of North Easternregion of India. Int. J. Chem. Stud. 2018, 6, 3131–3133. [Google Scholar]
- Yilmaz, O.; Sezenler, T.; Sevim, S.; Cemal, I.; Karaca, O.; Yaman, Y.; Karadag, O. Genetic relationships among four Turkish sheep breeds using microsatellites. Turk. J. Vet. Anim. Sci. 2015, 39, 576–582. [Google Scholar] [CrossRef]
- Ligda, C.; Altarayrah, J.; Georgoudis, A. Genetic analysis of Greek sheep breeds using microsatellite markers for setting conservation priorities. Small Rumin. Res. 2009, 83, 42–48. [Google Scholar] [CrossRef]
- Kusza, S.; Nagy, I.; Sasvári, Z.; Stágel, A.; Németh, T.; Molnár, A.; Kume, K.; Bosze, Z.; Jávor, A.; Kukovics, S. Genetic diversity and population structure of Tsigai and Zackel type of sheep breeds in the Central-, Eastern-and Southern-European regions. Small Rumin. Res. 2008, 78, 13–23. [Google Scholar] [CrossRef]
- Ćurković, M.; Ramljak, J.; Ivanković, S.; Mioc, B.; Ivankovic, A.; Pavic, V.; Brka, M.; Veit-Kensch, C.; Medugorac, I. The genetic diversity and structure of 18 sheep breeds exposed to isolation and selection. J. Anim. Breed. Genet. 2016, 133, 71–80. [Google Scholar] [CrossRef]
- Marković, M.; Radonjić, D.; Zorc, M.; Đokić, M.; Marković, B. Genetic Diversity of Montenegrin Local Sheep Breeds Based on Microsatellite Markers. Animals 2022, 12, 3029. [Google Scholar] [CrossRef]
- Machová, K.; Hofmanová, B.; Rychtářová, J.; Vostrý, L.; Moravčíková, N.; Kasarda, R. Genetic Variability Analysis of 26 Sheep Breeds in the Czech Republic. Acta Fytotech. Zootech. 2020, 23, 38–45. [Google Scholar] [CrossRef]
- Moravcikova, N.; Kasarda, R.; Kukuckova, V.; Vostry, L.; Kadlecík, O. Genetic diversity of Old Kladruber and Nonius horse populations through microsatellite variation analysis. Acta. Agric. Slov. 2016, 107, 45–49. [Google Scholar]
- Mastranestasis, I.; Ekateriniadou, L.; Ligda, C.; Theodorou, K. Genetic diversity and structure of the Lesvos sheep breed. Small Rumin. Res. 2015, 130, 54–59. [Google Scholar] [CrossRef]
- Ocampo, R.; Cardona, H.; Martínez, R. Genetic diversity of Colombian sheep by microsatellite markers. Chil. J. Agric. Res. 2016, 76, 40–47. [Google Scholar] [CrossRef]
- Salamon, D.; Gutierrez-Gil, B.; Arranz, J.; Barreta, J.; Batinic, V.; Dzidic, A. Genetic Diversity and Differentiation of 12 Eastern Adriatic and Western Dinaric Native Sheep Breeds Using Microsatellites. Animal 2014, 8, 200–207. [Google Scholar] [CrossRef] [PubMed]


| Locus | K | HW | Na | Ne | PIC | Ho | He | Ht | Fis | Fit | Fst | I | Gst | Gis |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SRCSP23 | 18 | *** | 12.667 | 7.782 | 0.907 | 0.831 | 0.856 | 0.913 | 0.029 | 0.09 | 0.063 | 2.210 | 0.050 | 0.044 |
| SPS113 | 11 | ns | 6.647 | 3.298 | 0.687 | 0.712 | 0.683 | 0.720 | −0.043 | 0.011 | 0.052 | 1.371 | 0.040 | −0.029 |
| TGL53 | 13 | *** | 10.333 | 6.248 | 0.876 | 0.824 | 0.825 | 0.880 | 0.002 | 0.064 | 0.062 | 1.993 | 0.050 | 0.016 |
| INRA23 | 15 | *** | 9.667 | 5.628 | 0.863 | 0.804 | 0.789 | 0.860 | −0.019 | 0.066 | 0.083 | 1.870 | 0.071 | −0.004 |
| SRCSP9 | 13 | *** | 7.444 | 2.963 | 0.641 | 0.756 | 0.650 | 0.684 | −0.162 | −0.105 | 0.048 | 1.371 | 0.038 | −0.147 |
| MAF65 | 14 | ns | 8.222 | 4.456 | 0.782 | 0.824 | 0.770 | 0.805 | −0.070 | −0.023 | 0.043 | 1.700 | 0.032 | −0.055 |
| MCM527 | 12 | ns | 7.000 | 4.268 | 0.791 | 0.771 | 0.754 | 0.810 | −0.023 | 0.047 | 0.069 | 1.579 | 0.057 | −0.009 |
| CSRD247 | 22 | *** | 11.444 | 6.053 | 0.864 | 0.809 | 0.810 | 0.870 | 0.001 | 0.069 | 0.068 | 1.953 | 0.056 | 0.016 |
| ILST11 | 10 | *** | 8.000 | 4.953 | 0.820 | 0.837 | 0.788 | 0.836 | −0.062 | −0.002 | 0.057 | 1.744 | 0.045 | −0.048 |
| OARFCB20 | 14 | ns | 9.000 | 5.095 | 0.824 | 0.776 | 0.771 | 0.845 | −0.006 | 0.082 | 0.087 | 1.754 | 0.075 | 0.010 |
| SRCSP5 | 8 | *** | 5.222 | 2.836 | 0.648 | 0.480 | 0.635 | 0.680 | 0.244 | 0.294 | 0.066 | 1.241 | 0.050 | 0.258 |
| INRA63 | 20 | ns | 10.000 | 5.061 | 0.825 | 0.802 | 0.784 | 0.846 | −0.023 | 0.051 | 0.073 | 1.821 | 0.061 | −0.008 |
| SRCSP8 | 18 | *** | 9.222 | 3.185 | 0.687 | 0.712 | 0.666 | 0.703 | −0.069 | −0.013 | 0.052 | 1.506 | 0.040 | −0.046 |
| Mean | 14.54 | 8.883 | 4.756 | 0.786 | 0.765 | 0.753 | 0.804 | −0.015 | 0.048 | 0.063 | 1.701 | 0.052 | −0.001 |
| Breed | N | Na | Ne | Ho | He | I | Fis | PIC | PA |
|---|---|---|---|---|---|---|---|---|---|
| CVA | 94 | 7.231 | 4.248 | 0.747 | 0.729 | 1.559 | −0.026 | 0.697 | - |
| IVA | 141 | 10.84 | 5.916 | 0.817 | 0.797 | 1.915 | −0.02 | 0.775 | 4 |
| SUM | 118 | 9.077 | 5.057 | 0.769 | 0.771 | 1.775 | 0.006 | 0.745 | 1 |
| SVA | 122 | 9.385 | 4.846 | 0.778 | 0.767 | 1.722 | −0.01 | 0.736 | 3 |
| POG | 130 | 10.00 | 5.322 | 0.771 | 0.779 | 1.815 | 0.015 | 0.752 | 5 |
| SWI | 92 | 7.077 | 3.158 | 0.710 | 0.649 | 1.380 | −0.09 | 0.615 | 2 |
| UHR | 85 | 6.538 | 3.941 | 0.773 | 0.719 | 1.517 | −0.06 | 0.683 | 1 |
| UKR | 104 | 8.000 | 4.588 | 0.745 | 0.767 | 1.712 | 0.028 | 0.737 | - |
| TUR | 148 | 11.38 | 5.727 | 0.770 | 0.795 | 1.914 | 0.031 | 0.776 | 8 |
| Mean | 8.838 | 4.756 | 0.765 | 0.753 | 1.701 | −0.016 |
| Breed | CVA | IVA | SUM | SVA | POG | SWI | UHR | UKR | TUR |
|---|---|---|---|---|---|---|---|---|---|
| CVA | 0.000 | 7.667 | 5.680 | 12.273 | 6.276 | 3.448 | 4.412 | 5.666 | 6.854 |
| IVA | 0.032 | 0.000 | 14.856 | 13.420 | 18.058 | 4.752 | 7.017 | 10584 | 21.854 |
| SUM | 0.042 | 0.017 | 0.000 | 8.674 | 9.918 | 4.071 | 6.461 | 7.374 | 11.094 |
| SVA | 0.020 | 0.018 | 0.028 | 0.000 | 9.494 | 4.222 | 6.437 | 7.360 | 12.143 |
| POG | 0.038 | 0.014 | 0.025 | 0.026 | 0.000 | 3.607 | 5.495 | 11.419 | 20.946 |
| SWI | 0.068 | 0.050 | 0.058 | 0.056 | 0.065 | 0.000 | 3.127 | 3.041 | 4.321 |
| UHR | 0.054 | 0.034 | 0.037 | 0.037 | 0.044 | 0.074 | 0.000 | 5.113 | 5.940 |
| UKR | 0.042 | 0.023 | 0.033 | 0.033 | 0.021 | 0.076 | 0.047 | 0.000 | 11.438 |
| TUR | 0.035 | 0.011 | 0.022 | 0.020 | 0.012 | 0.055 | 0.040 | 0.021 | 0.000 |
| Breed | CVA | POG | SUM | SVA | SWI | TUR | UHR | UKR | IVA |
|---|---|---|---|---|---|---|---|---|---|
| CVA | 0 | 0.277 | 0.299 | 0.125 | 0.377 | 0.257 | 0.362 | 0.305 | 0.230 |
| POG | 0.229 | 0 | 0.192 | 0.198 | 0.403 | 0.095 | 0.318 | 0.164 | 0.108 |
| SUM | 0.248 | 0.194 | 0 | 0.212 | 0.346 | 0.178 | 0.264 | 0.258 | 0.129 |
| SVA | 0.167 | 0.196 | 0.201 | 0 | 0.317 | 0.156 | 0.256 | 0.255 | 0.143 |
| SWI | 0.307 | 0.310 | 0.281 | 0.282 | 0 | 0.323 | 0.445 | 0.493 | 0.288 |
| TUR | 0.227 | 0.133 | 0.182 | 0.175 | 0.287 | 0 | 0.297 | 0.171 | 0.094 |
| UHR | 0.273 | 0.251 | 0.229 | 0.234 | 0.315 | 0.244 | 0 | 0.333 | 0.249 |
| UKR | 0.248 | 0.180 | 0.221 | 0.223 | 0.332 | 0.183 | 0.257 | 0 | 0.186 |
| ZVA | 0.211 | 0.140 | 0.158 | 0.163 | 0.272 | 0.131 | 0.222 | 0.188 | 0 |
| Breed | CVA | POG | SUM | SVA | SWI | TUR | UHR | UKR | IVA |
|---|---|---|---|---|---|---|---|---|---|
| CVA | 0 | 0.073 | 0.079 | 0.038 | 0.125 | 0.066 | 0.102 | 0.081 | 0.061 |
| POG | 0.412 | 0 | 0.048 | 0.050 | 0.123 | 0.024 | 0.085 | 0.042 | 0.027 |
| SUM | 0.417 | 0.331 | 0 | 0.054 | 0.112 | 0.043 | 0.075 | 0.064 | 0.033 |
| SVA | 0.289 | 0.341 | 0.342 | 0 | 0.106 | 0.039 | 0.073 | 0.064 | 0.036 |
| SWI | 0.446 | 0.488 | 0.451 | 0.430 | 0 | 0.104 | 0.144 | 0.142 | 0.096 |
| TUR | 0.390 | 0.265 | 0.306 | 0.317 | 0.446 | 0 | 0.078 | 0.042 | 0.022 |
| UHR | 0.447 | 0.461 | 0.396 | 0.396 | 0.479 | 0.428 | 0 | 0.090 | 0.068 |
| UKR | 0.413 | 0.335 | 0.376 | 0.381 | 0.498 | 0.322 | 0.458 | 0 | 0.046 |
| IVA | 0.363 | 0.291 | 0.279 | 0.290 | 0.442 | 0.263 | 0.407 | 0.349 | 0 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sztankoová, Z.; Milerski, M.; Vostrý, L.; Rychtářová, J. Genetic Diversity and Population Structure of Nine Local Sheep Populations Bred in the Carpathia Area of Central Europe Revealed by Microsatellite Analysis. Animals 2025, 15, 2400. https://doi.org/10.3390/ani15162400
Sztankoová Z, Milerski M, Vostrý L, Rychtářová J. Genetic Diversity and Population Structure of Nine Local Sheep Populations Bred in the Carpathia Area of Central Europe Revealed by Microsatellite Analysis. Animals. 2025; 15(16):2400. https://doi.org/10.3390/ani15162400
Chicago/Turabian StyleSztankoová, Zuzana, Michal Milerski, Luboš Vostrý, and Jana Rychtářová. 2025. "Genetic Diversity and Population Structure of Nine Local Sheep Populations Bred in the Carpathia Area of Central Europe Revealed by Microsatellite Analysis" Animals 15, no. 16: 2400. https://doi.org/10.3390/ani15162400
APA StyleSztankoová, Z., Milerski, M., Vostrý, L., & Rychtářová, J. (2025). Genetic Diversity and Population Structure of Nine Local Sheep Populations Bred in the Carpathia Area of Central Europe Revealed by Microsatellite Analysis. Animals, 15(16), 2400. https://doi.org/10.3390/ani15162400





