Single Nucleotide Polymorphisms of Leptin and Calpain/Calpastatin in Key Traits of Pork Meat Quality
Simple Summary
Abstract
1. Introduction
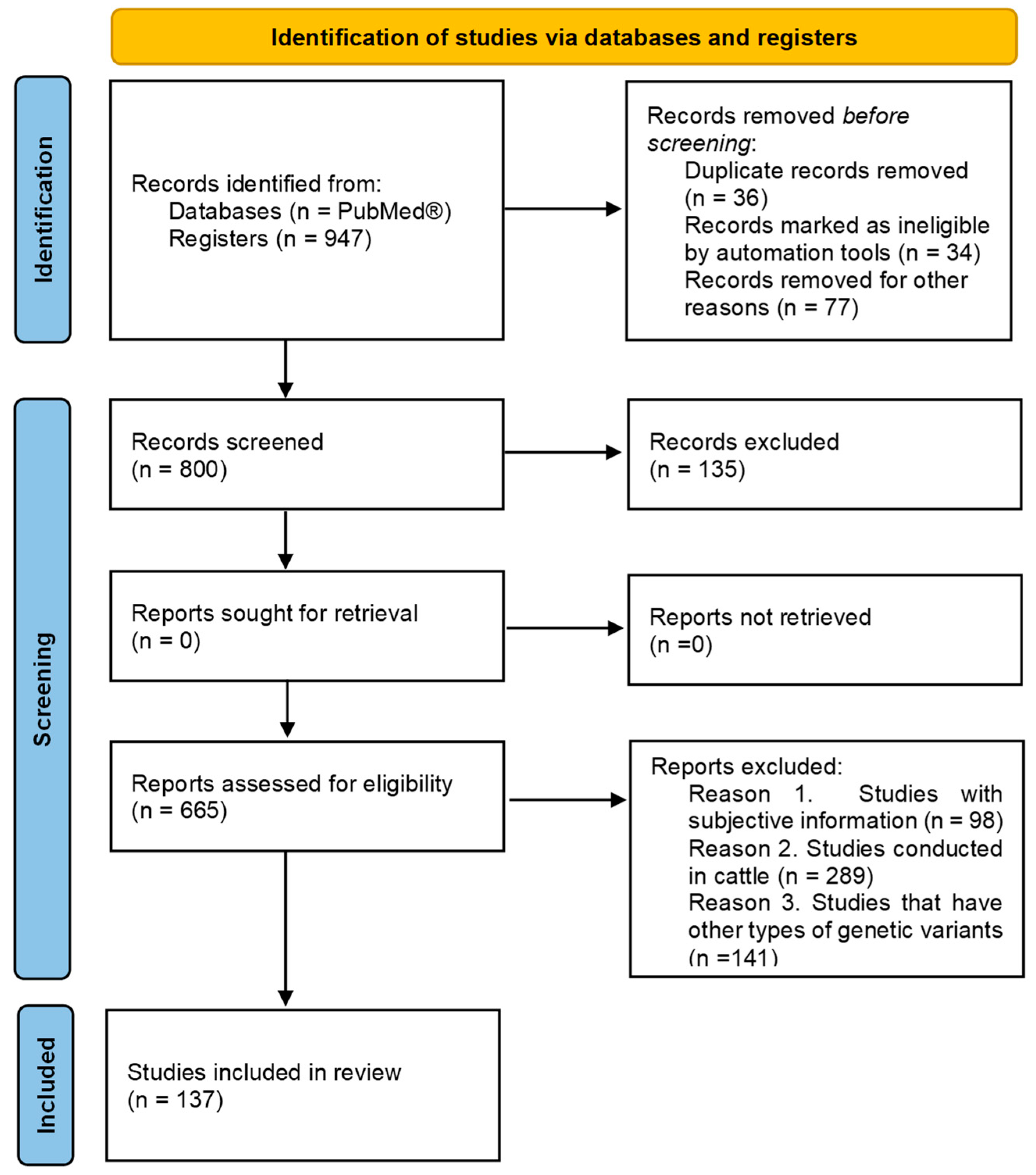
2. Materials and Methods
2.1. Exclusion Criteria
2.2. Information Sources, Search, and Selection
3. Results
3.1. Importance of SNPs to Meat Quality
3.2. Leptin as a Marker of Marbling and Meat Quality in Pork
3.2.1. Structure
3.2.2. Action Mechanism
- –
- Y985 activates Src homology domain protein-2 (SHP-2) and mitogen-activated protein kinase (MAPK) signaling to provide negative feedback in the leptin signaling pathway.
- –
- Y1077 triggers the signal transducer and activator of transcription-5 (STAT5) signaling to mediate leptin’s reproductive effects.
- –
- Y1138 activates STAT3 signaling, which plays a central role in leptin’s effects on energy balance and neuroendocrine functions [45]. It is well known that other pathways can be activated, depending on the type of tissue and its specific function.
3.2.3. Leptin in Marbling
3.2.4. SNPs in Leptin (LEP) and the Leptin Receptor (LEPR) in Relation to Meat Quality
| Single Nucleotide Polymorfism | Population | Productive Trait | Method | Significance | Ref. |
|---|---|---|---|---|---|
| Leptin | Chromosome 18 | ||||
| g. 867 C>T | Duroc, Hampshire, Landrace and Large White pigs. | Backfat thicknes | Bi-PASA and PCR-RFLP assays | p < 0.001 | [65] |
| Exon 3 C>T | Landrace and Yorkshire | Average daily weight gain and feed efficiency | PCR-RFLP | p < 0.05 | [44] |
| Intron 1 C>T | Duroc | Backfat thicknes | PCR-RFLP | p < 0.05 | [44] |
| 2845 A>T | Landrace | Total feed consumption during growth and weight gain | PCR-RFLP | p < 0.0078 | [13] |
| 3996 T>C | Landrace | Total feed consumption during growth and weight gain | PCR-RFLP | p < 0.0078 | [13] |
| rs324640280 (c.339C>T) | Duroc | Subcutaneous fat | RFLP and SSCP | Without association | [69] |
| g.3469 T>C | Duroc, Hampshire, Landrace and Large White | Abdominal fat, backfat thickness, intramuscular fat, meat content, loin weight, loin muscle area, ham weight, and ham cut weight | RFLP and SSCP | p < 0.0078 | [13,69] |
| c. 2863 G>A | Duroc, Yorkshire, Laiwu, Lulai Black and Landrace/Yorkshire crossbreds | Leptin levels in serum and backfat | PCR-SSCP | p < 0.01 | [22] |
| LEP g.1387C>T LEP g.1382C>T | Experimental Iberian/Landrace crossbred | Weight (live and carcass), backfat thickness and saturated fatty acid content in fat, increased growth, increased voluntary feed intake | Pyrosequencing | p < 0.05 | [71] |
| rs45431504 (c.289T>C) | Polish Large White | Changes in fat deposits in skeletal muscle | Pyrosequencing | p < 0.001 | [70] |
| LEPR | Chromosome 6 | ||||
| c.2002C>T | Landrace, Yorkshire and Duroc | MS-PCR and PCR-RFLP | p < 0.05 | [44] | |
| Iberian × Meishan and Landrace | Backfat thickness | Pyrosequencing, and PCR-RFLP | p < 0.01 | [75,76,77] | |
| c.2002C>T | Duroc Duroc | Backfat thickness, fat area ratios, serum leptin concentration Growth rate and fat deposition | PCR–RFLP and SSCP | p < 0.01 | [34,78] |
| c.232A>T | Polish Landrace | Backfat thickness | Sequenom MassARRAY | p < 0.01 | [79] |
| c.232A>T | Duroc × Landrace Yorkshire × Maternal Landrace dams with Duroc | Serum leptin concentrations | Sequenom MassARRAY | p < 0.01 | [80] |
| c.2856C>T | Ukrainian white pigs | Backfat thickness and average daily weight gain | PCR–RFLP | p < 0.05 | [81] |
3.3. The Calpain–Calpastatin System as a Marker of Tenderness and Quality in Pork
3.3.1. Structure
- Domain I, the N-terminal domain, is hydrophobic and participates in triggering proteinase activity.
- -
- Type I begins at exon 1xa, 1y, and 1z and is expressed in the brain, liver, and testes of mice.
- -
- Type II begins at exon 1xb, 1y, and 1z and is primarily expressed in skeletal and cardiac muscle.
- -
- Type III begins at exon 1u and encodes the prototypical calpastatin, which is widely present in mouse tissues, but is also observed in the cardiac and skeletal muscle of pigs [106].
- -
- Type IV begins at exon 14t and is unique, as it is found only in the testes with no expression in other calpastatins.
3.3.2. Action Mechanism
3.3.3. The Calpain–Calpastatin System and Meat Tenderness
3.3.4. SNPs in Calpain (CAPN1) and Calpastatin (CAST) in Relation to Meat Quality
| Single Nucleotide Polymorfism | Population | Productive Trait | Method | Significance | Reference |
|---|---|---|---|---|---|
| CAPN1 | Chromosome 6 | ||||
| Not reported | Yorkshire pig, Min pig and wild boar | Not reported | Not reported | Not reported | [126] |
| g.157T>C | Italian Duroc × (Landrace × Large White) crossbred | Larger myofibril diameter, meat redness | PCR-RFLP | p < 0.0001 | [83] |
| rs81358667G>A | Iberian pigs | Shear force and cooking losses | KASP-PCR | p < 0.05 | [8] |
| CAST | Chromosome 2 | ||||
| CAST 66 Ser > Arg CAST 249 Arg>249Lys CAST 638Ser>638Arg | Berkshire × Yorkshire crossbreed Duroc–Landrace–Yorkshire swine lines Mexican creole pigs, as well as in the Yorkshire breed | Cooking loss, juiciness and tenderness Shear force (tenderness) Soft and juicy meat | PCR-RFLP and Sequenom MassARRAY | p < 0.05–p < 0.001 | [129,130] |
| g.76872 G>A | Italian Duroc × (Landrace × Large White) crossbred | Drip loss and pH | PCR-RFLP | p < 0.0001 | [83] |
| g.5669 T>C g.49346 C>T | Duroc × Iberian crosses and pure Iberian pigs | Thickness and shear force in muscle | Sanger sequencing | p < 0.05–p< 0.01 | [135] |
| rs196949783 G>A | Polish Landrace, Polish Large White, Pietrain and Duroc Iberian pigs | Intramuscular fat content, water holding capacity, pH, firmness and toughness and the weight of the Longissimus thoracis et lumbar | KASP-PCR | p < 0.05 | [8] |
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godfray, H.C.; Beddington, J.R.; Crute, I.R. Food security: The challenge of feeding 9 billion people. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- USDA-FAS. Livestock and Poultry: World Markets and Trade; United States Department of Agriculture: Washington, WA, USA, April 2024. [Google Scholar]
- FIRA. Panorama Agroalimentario Carne de Cerdo; FIRA: Morelia, México, 2024. [Google Scholar]
- Carne de Cerdo: Tradición y Versatilidad en México. Secretaría de Agricultura y Desarrollo Rural. 2025. Available online: https://www.gob.mx/agricultura/articulos/carne-de-cerdo-tradicion-y-versatilidad-en-mexico (accessed on 1 March 2025).
- Chen, K.; Baxter, T.; Muir, W.M.; Groenen, M.A.; Schook, L.B. Genetic resources, genome mapping and evolutionary genomics of the pig (Sus scrofa). Int. J. Biol. Sci. 2007, 3, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Swindle, M.M.; Makin, A.; Herron, A.J.; Clubb, F.J.; Frazier, K.S. Swine as models in biomedical research and toxicology testing. Vet. Pathol. 2012, 49, 344–356. [Google Scholar] [CrossRef] [PubMed]
- Blasco, A.; Gou, P.; Gispert, M.; Estany, J.; Soler, Q.; Distre, A.; Tibau, J. Comparison of five types of pig crosses. I. Growth and carcass trais. Livest. Prod. Sci. 1994, 40, 171–178. [Google Scholar] [CrossRef]
- Palma-Granados, P.; Muñoz, M.; Delgado-Gutierrez, M.A.; Óvilo, C.; Nuñez, Y.; Fernández-Barroso, M.A.; Sánchez-Esquiliche, F.; Ramírez, L.; García-Casco, J.M. Candidate SNPs for meat quality and carcass composition in free-range Iberian pigs. Meat Sci. 2024, 207, 109373. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.S.; Jin, S.K.; Jeong, Y.H.; Jung, Y.C.; Jung, J.H.; Shim, K.S.; Choi, Y.I. Relationships between Single Nucleotide Polymorphism Markers and Meat Quality Traits of Duroc Breeding Stocks in Korea. J. Anim. Sci. 2016, 29, 1229–1238. [Google Scholar] [CrossRef] [PubMed]
- Koopaee, H.K.; Koshkoiyeh, A.E. SNPs Genotyping technologies and their applications in farm animals breeding programs. Braz. Arch. Biol. Technol. 2014, 57, 87–95. [Google Scholar] [CrossRef]
- Page, M.J.; Moher, D.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; McKenzie, J.E. PRISMA 2020 explanation and elaboration: Updated guidance and exemplars for reporting systematic reviews. BMJ 2021, 372, n160. [Google Scholar] [CrossRef] [PubMed]
- Colombani, C.; Croiseau, P.; Hoze, C.; Fritz, S.; Guillaume, F.; Boichard, D.; Legarra, A.; Ducrocq, V.; Robert-Granié, C. Could Genomic Selection Methods be Efficient to Detect QTLs? Application in French Dairy Cattle; Hal Open Science: Lyon, France, 2011; Available online: https://hal.science/hal-01190269v1 (accessed on 1 March 2025).
- Kennes, Y.M.; Murphy, B.D.; Pothier, F.; Palin, M.F. Characterization of swine leptin (LEP) polymorphisms and their association with production traits. Anim. Genet. 2001, 32, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Vignal, A.; Milan, D.; San Cristobal, M.; Eggen, A. A review on SNP and other types of molecular markers and their use in animal genetics. Genet. Sel. Evol. 2002, 34, 275–305. [Google Scholar] [CrossRef] [PubMed]
- Mignon-Grasteau, S.A.; Boissy, J.; Bouix, J.M.; Faure, A.D.; Fisher, G.N.; Hinch, P.; Jensen, P.; Le Neindre, P.; Prunet, P. Genetics of adaptation and domestication in livestock. Livest. Prod. Sci. 2005, 93, 3–14. [Google Scholar] [CrossRef]
- Renaville, B.; Piasentier, E.; Fan, B.; Vitale, M.; Prandi, A.; Rothschild, M.F. Candidate gene markers involved in San Daniele ham quality. Meat Sci. 2010, 85, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Renaville, B.; Bacciu, N.; Lanzoni, M.; Corazzin, M.; Piasentier, E. Polymorphism of fat metabolism genes as candidate markers for meat quality and production traits in heavy pigs. Meat Sci. 2015, 110, 220–223. [Google Scholar] [CrossRef] [PubMed]
- Cepica, S.; Zambonelli, P.; Weisz, F.; Bigi, M.; Knoll, A.; Vykoukalová, Z.; Masopust, M.; Gallo, M.; Buttazzoni, L.; Davoli, R. Association mapping of quantitative trait loci for carcass and meat quality traits at the central part of chromosome 2 in Italian Large White pigs. Meat Sci. 2013, 95, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Pena, R.N.; Ros-Freixedes, R.; Tor, M.; Estany, J. Genetic marker discovery in complex traits: A field example on fat content and composition in pigs. Int. J. Mol. Sci. 2016, 17, 2100. [Google Scholar] [CrossRef] [PubMed]
- Tyra, M.; Ropka-Molik, K.; Terman, A.; Piórkowska, K.; Oczkowicz, M.; Bereta, A. Association between subcutaneous and intramuscular fat content in porcine ham and loin depending on age, breed and FABP3 and LEPR genes transcript abundance. Mol. Biol. Rep. 2013, 40, 2301–2308. [Google Scholar] [CrossRef] [PubMed]
- van Wijk, H.J.; Arts, D.J.; Matthews, J.O.; Webster, M.; Ducro, B.J.; Knol, E.F. Genetic parameters for carcass composition and pork quality estimated in a commercial production chain. J. Anim. Sci. 2005, 83, 324–333. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Hu, Y.; Yang, X.; Liu, Y.; Wei, S.; Jiang, Y. Identification and genetic effects of a novel polymorphism in the distal promoter region of porcine leptin gene. Mol. Biol. Rep. 2011, 38, 2051–2057. [Google Scholar] [CrossRef] [PubMed]
- Fortin, A.; Robertson, W.M.; Tong, A.K. The eating quality of Canadian pork and its relationship with intramuscular fat. Meat Sci. 2005, 69, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Renaudeau, D.; Mourot, J. A comparison of carcass and meat quality characteristics of Creole and Large White pigs slaughtered at 90 kg BW. Meat Sci. 2007, 76, 165–171. [Google Scholar] [CrossRef] [PubMed]
- Hocquette, J.F.; Gondret, F.; Baéza, E.; Médale, F.; Jurie, C.; Pethick, D.W. Intramuscular fat content in meat-producing animals: Development, genetic and nutritional control, and identification of putative markers. Anim. Int. J. Anim. Biosci. 2010, 4, 303–319. [Google Scholar] [CrossRef] [PubMed]
- Malgwi, I.H.; Halas, V.; Grünvald, P.; Schiavon, S.; Jócsák, I. Genes related to fat metabolism in pigs and intramuscular fat content of pork: A focus on nutrigenetics and nutrigenomics. Animals 2022, 12, 150. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Rui, L. Leptin signaling and leptin resistance. Front. Med. 2013, 7, 207–222. [Google Scholar] [CrossRef] [PubMed]
- Bell, C.G.; Walley, A.J.; Froguel, P. The genetics of human obesity. Nat. Rev. Genet. 2005, 6, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, M.W.; Peskind, E.; Raskind, M.; Boyko, E.J.; Porte, D. Cerebrospinal fluid leptin levels: Relationship to plasma levels and to adiposity in humans. Nat. Med. 1996, 2, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Ahima, R.S.; Prabakaran, D.; Mantzoros, C.; Qu, D.; Lowell, B.; Maratos-Flier, E.; Flier, J.S. Role of leptin in the neuroendocrine response to fasting. Nature 1996, 382, 250–252. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Hernandez, M.E.; Ramos, M.F.R.; Escobar, H.J.B.H.; López, M.J.J.D.; Escobar, C.P.; García, R.R.V.; Ortega, P.C.B.; Croda-Todd, M.T.; Gasca, P.E. Leptin biochemical mechanisms involved in the development of obesity. Rev. Médica Univ. Veracruzana 2015, 15, 103–113. [Google Scholar]
- Mazor, R.; Friedmann-Morvinski, D.; Alsaigh, T.; Kleifeld, O.; Kistler, E.B.; Rousso-Noori, L.; Huang, C.; Li, J.B.; Verma, I.M.; Schmid-Schönbein, G.W. Cleavage of the leptin receptor by matrix metalloproteinase-2 promotes leptin resistance and obesity in mice. Sci. Transl. Med. 2018, 10, eaah6324. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Mesa, R.; Ros-Freixedes, R.; Pena, R.N.; Reixach, J.; Estany, J. Impact of the leptin receptor gene on pig performance and quality traits. Sci. Rep. 2024, 14, 10652. [Google Scholar] [CrossRef] [PubMed]
- Uemoto, Y.; Kikuchi, T.; Nakano, H.; Sato, S.; Shibata, T.; Kadowaki, H.; Katoh, K.; Kobayashi, E.; Suzuki, K. Effects of porcine leptin receptor gene polymorphisms on backfat thickness, fat area ratios by image analysis, and serum leptin concentrations in a Duroc purebred population. Anim. Sci. J. 2012, 83, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Ros-Freixedes, R.; Gol, S.; Pena, R.N.; Tor, M.; Ibáñez-Escriche, N.; Dekkers, J.C.; Estany, J. Genome-Wide Association Study Singles Out SCD and LEPR as the Two Main Loci Influencing Intramuscular Fat Content and Fatty Acid Composition in Duroc Pigs. PLoS ONE 2016, 11, e0152496. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, R.; Rout, P.K.; Singh, S.K. Leptin: A biomolecule for enhancing livestock productivity. Int. J. Burn. Trauma 2009, 8, 169–176. [Google Scholar]
- Barb, C.R.; Hausman, G.J.; Houseknecht, K.L. Biology of leptin in the pig. Domest. Anim. Endocrinol. 2001, 21, 297–317. [Google Scholar] [CrossRef] [PubMed]
- Cameron, N.D.; Penman, J.C.; McCullough, E. Serum leptin concentration in pigs selected for high or low daily food intake. Genet. Res. 2000, 75, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.P.; McFadin, E.L.; Maddock, K.R.; Goodwin, R.N.; Baas, T.J.; Keisler, D.H. Serum concentrations of leptin in six genetic lines of swine and relationship with growth and carcass. J. Anim. Sci. 2003, 81, 167–171. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Inomata, K.; Katoh, K.; Kadowaki, H.; Shibata, T. Genetic correlations among carcass cross sectional fat area ratios, production traits, intramuscular fat, and serum leptin concentration in Duroc pigs. J. Anim. Sci. 2009, 87, 2209–2215. [Google Scholar] [CrossRef] [PubMed]
- Münzberg, H.; Morrison, C.D. Structure, production and signaling of leptin. Metabolism 2015, 64, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Gaucher, E.A.; Miyamoto, M.M.; Benner, S.A. Evolutionary, structural and biochemical evidence for a new interaction site of the leptin obesity protein. Genetics 2003, 163, 1549–1553. [Google Scholar] [CrossRef] [PubMed]
- Denver, R.J.; Bonett, R.M.; Boorse, G.C. Evolution of leptin structure and function. Neuroendocrinology 2011, 94, 21–38. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chang, T.; Su, H.T. Characterization of porcine leptin receptor polymorphisms and their association with reproduction and production traits. Anim. Biotechnol. 2004, 15, 89–102. [Google Scholar] [CrossRef] [PubMed]
- Gorska, E.; Popko, E.; Stelmaszczyk-Emmel, A.; Ciepiela, O.; Kucharska, A.; Wasik, M. Leptin receptors. Eur. J. Med. Res. 2010, 15 (Suppl. 2), 50–54. [Google Scholar] [CrossRef] [PubMed]
- Myers, M.G.; Cowley, M.A.; Munzberg, H. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Elefteriou, F.; Levasseur, R.; Liu, X.; Zhao, L.; Parker, K.L.; Armstrong, D.; Ducy, P.; Karsenty, G. Leptin regulates bone formation via the sympathetic nervous system. Cell 2002, 111, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yue, R.; Zhou, B.O.; Shimada, I.S.; Zhao, Z.Y.; Morrison, S.J. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell 2016, 18, 782–796. [Google Scholar] [CrossRef] [PubMed]
- Taga, T.; Kishimoto, T. Gp130 and the interleukin-6 family of cytokines. Annu. Rev. Immunol. 1997, 15, 797–819. [Google Scholar] [CrossRef] [PubMed]
- Banks, A.S.; Davis, S.M.; Bates, S.H.; Myers, M.G. Activation of downstream signals by the long form of the leptin receptor. J. Biol. Chem. 2000, 275, 14563–14572. [Google Scholar] [CrossRef] [PubMed]
- Joo, S.T.; Hwang, Y.H.; Frank, D. Characteristics of Hanwoo cattle and health implications of consuming highly marbled Hanwoo beef. Meat Sci. 2017, 132, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Keogh, K.; Kelly, A.K.; Kenny, D.A. Effect of plane of nutrition in early life on the transcriptome of visceral adipose tissue in Angus heifer calves. Sci. Rep. 2021, 11, 9716. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Yu, H.; Wang, J.; Liu, H.; Guo, J.; Wang, S.; Zan, L. LEP inhibits intramuscular adipogenesis through the AMPK signaling pathway in vitro. FASEB J. 2024, 38, e23836. [Google Scholar] [CrossRef] [PubMed]
- Hausman, G.J.; Dodson, M.V.; Ajuwon, K.; Azain, M.; Barnes, K.M.; Guan, L.L.; Jiang, Z.; Poulos, S.P.; Sainz, R.D.; Smith, S.; et al. Board-invited review: The biology and regulation of preadipocytes and adipocytes in meat animals. J. Anim. Sci. 2009, 87, 1218–1246. [Google Scholar] [CrossRef] [PubMed]
- Muoio, D.M.; Dohm, G.L.; Fiedorek, F.T.; Tapscott, E.B., Jr.; Coleman, R.A. Leptin directly alters lipid partitioning in skeletal muscle. Diabetes 1997, 46, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, G.R.; Rush, J.W.; Dyck, D.J. AMPK expression and phosphorylation are increased in rodent muscle after chronic leptin treatment. Am. J. Physiol. Endocrinol. Metab. 2003, 284, E648-54. [Google Scholar] [CrossRef] [PubMed]
- Wagoner, B.; Hausman, D.B.; Harris, R.B. Direct and indirect effects of leptin on preadipocyte proliferation and differentiation. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2006, 290, R1557–R1564. [Google Scholar] [CrossRef] [PubMed]
- Minokoshi, Y.; Kim, Y.B.; Peroni, O.D.; Fryer, L.G.; Müller, C.; Carling, D.; Kahn, B.B. Leptin stimulates fatty-acid oxidation by activating AMP-activated protein kinase. Nature 2002, 415, 339–343. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.D.; Sung, Y.Y.; Jung, W.H.; Cheon, H.G. Leptin inhibits rosiglitazone-induced adipogenesis in murine primary adipocytes. Mol. Cell. Endocrinol. 2008, 294, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Janovská, A.; Hatzinikolas, G.; Staikopoulos, V.; McInerney, J.; Mano, M.; Wittert, G.A. AMPK and ACC phosphorylation: Effect of leptin, muscle fibre type and obesity. Mol. Cell. Endocrinol. 2008, 284, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.H.; Han, J.H.; Lee, Y.S.; Jung, Y.S.; Roh, Y.S.; Yun, J.S.; Han, S.B.; Hong, J.T. Chitinase-3-like-1 deficiency attenuates ethanol-induced liver injury by inhibition of sterol regulatory element binding protein 1-dependent triglyceride synthesis. Metab. Clin. Exp. 2019, 95, 46–56. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Hawley, S.A.; Scott, J.W.J. AMP-activated protein kinase—Development of the energy sensor concept. Physiology 2006, 574 Pt 1, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Bai, T.; Yang, Y.; Yao, Y.L.; Sun, P.; Lian, L.H.; Wu, Y.L.; Nan, J.X. Betulin alleviated ethanol-induced alcoholic liver injury via SIRT1/AMPK signaling pathway. Pharmacol. Res. 2016, 105, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Chang, T.; Su, H.Y. Genetic polymorphisms in porcine leptin gene and their association with reproduction and production traits. Austr. J. Agric. Res. 2004, 55, 699–704. [Google Scholar] [CrossRef]
- Jiang, Z.H.; Gibson, J.P. Genetic polymorphisms in the leptin gene and their association with fatness in four pig breeds. Mamm. Genome. 1999, 10, 191–193. [Google Scholar] [CrossRef] [PubMed]
- Kulig, H.; Grzesiak, W.; Szatkowska, I. Effect of leptin gene polymorphism on growth and carcass traits in pigs. Arch. Anim. Breed. 2001, 44, 291–296. [Google Scholar] [CrossRef]
- Szydlowski, M.; Stachowiak, M.; Mackowski, M.; Kamyczek, M.; Eckert, R.; Rozycki, M.; Switonski, M. No major effect of the leptin gene polymorphism on porcine production traits. J. Anim. Breed. Genet. 2004, 121, 149–155. [Google Scholar] [CrossRef]
- Urban, T.; Kuciel, J.; Mikolasova, R. Polymorphism of genes encoding for ryanodine receptor, growth hormone, leptin and MYC protooncogene protein and meat production in Duroc pigs. Czech J. Anim. Sci. 2002, 47, 411–417. [Google Scholar]
- Stachowiak, M.; Mackowski, M.; Madeja, Z.; Szydlowski, M.; Buszka, A.; Kaczmarek, P.; Rubis, B.; Mackowiak, P.; Nowak, K.W.; Switonski, M. Polymorphism of the porcine leptin gene promoter and analysis of its association with gene expression and fatness traits. Biochem. Genet. 2007, 45, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Stachowiak, M.; Flisikowski, K. Analysis of allele-specific expression of seven candidate genes involved in lipid metabolism in pig skeletal muscle and fat tissues reveals allelic imbalance of ACACA, LEP, SCD, and TNF. J. Appl. Genet. 2019, 60, 97–101. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montarelo, D.; Fernández, A.; Folch, J.M.; Pena, R.N.; Ovilo, C.; Rodríguez, C.; Silió, L.; Fernández, A.I. Joint effects of porcine leptin and leptin receptor polymorphisms on productivity and quality traits. Anim. Genet. 2012, 43, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Orrù, L.; Cifuni, G.F.; Piasentier, E.; Corazzin, M.; Bovolenta, S.; Moioli, B. Association analyses of single nucleotide polymorphisms in the and genes on the fatty acid profile of muscle fat in Simmental bulls. Meat Sci. 2011, 87, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, L.A. The leptin receptor. J. Biol. Chem. 1997, 272, 6093–6096. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Montarelo, D.; Fernández, A.; Barragán, C.; Noguera, J.L.; Folch, J.M.; Rodríguez, M.C.; Ovilo, C.; Silió, L.; Fernández, A.I. Transcriptional Characterization of Porcine Leptin and Leptin Receptor Genes. PLoS ONE 2013, 8, e66398. [Google Scholar] [CrossRef] [PubMed]
- Ovilo, C.; Fernandez, A.; Noguera, J.L.; Barragan, C.; Leton, R.; Rodriguez, C.; Mercade, A.; Alves, E.; Folch, J.M.; Varona, L.; et al. Fine mapping of porcine chromosome 6 QTL and LEPR effects on body composition in multiple generations of an Iberian by Landrace intercross. Genet. Res. 2015, 85, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, G.; Ovilo, C.; Silió, L.; Tomás, A.; Noguera, J.L.; Rodríguez, M.C. Single- and joint-population analyses of two experimental pig crosses to confirm quantitative trait loci on Sus scrofa chromosome 6 and leptin receptor effects on fatness and growth traits. J. Anim. Sci. 2009, 87, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, G.; Alcázar, E.; Fernández, A.; Barragán, C.; Carrasco, A.; De Pedro, E.; Rodríguez, M.C. Effects of porcine MC4R and LEPR polymorphisms, gender and Duroc sire line on economic traits in Duroc× Iberian crossbred pigs. Meat Sci. 2011, 88, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Hirose, K.; Ito, T.; Fukawa, K.; Arakawa, A.; Mikawa, S.; Hayashi, Y.; Tanaka, K. Evaluation of effects of multiple candidate genes (LEP, LEPR, MC4R, PIK3C3, and VRTN) on production traits in Duroc pigs. Anim. Sci. J. 2014, 85, 198–206. [Google Scholar] [CrossRef] [PubMed]
- Mackowski, M.; Szymoniak, K.; Szydlowski, M.; Kamyczek, M.; Eckert, R.; Rozycki, M.; Switonski, M. Missense mutations in exon 4 of the porcine LEPR gene encoding extracellular domain and their association with fatness traits. Anim. Genet. 2025, 36, 135–137. [Google Scholar] [CrossRef] [PubMed]
- Kuehn, L.A.; Nonneman, D.J.; Klindt, J.M.; Wise, T.H. Genetic relationships of body composition, serum leptin, and age at puberty in gilts. J. Anim. Sci. 2009, 87, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Balatsky, V.; Oliinychenko, Y.; Sarantseva, N.; Getya, A.; Saienko, A.; Vovk, V.; Doran, O. Association of single nucleotide polymorphisms in leptin (LEP) and leptin receptor (LEPR) genes with backfat thickness and daily weight gain in Ukrainian Large White pigs. Livest. Sci. 2018, 217, 157–161. [Google Scholar] [CrossRef]
- Moeller, S.J.; Miller, R.K.; Edwards, K.K.; Zerby, H.N.; Logan, K.E.; Aldredge, T.L. Consumer perceptions of pork eating quality as affected by pork quality attributes and end-point cooked temperature. Meat Sci. 2010, 84, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Gandolfi, G.; Pomponio, L.; Ertbjerg, P.; Karlsson, A.H.; Nanni Costa, L.; Lametsch, R.; Russo, V.; Davoli, R. Investigation on CAST, CAPN1 and CAPN3 porcine gene polymorphisms and expression in relation to post-mortem calpain activity in muscle and meat quality. Meat Sci. 2011, 88, 694–700. [Google Scholar] [CrossRef] [PubMed]
- Parr, T.; Sensky, P.L.; Scothern, G.P.; Bardsley, R.G.; Buttery, P.J.; Wood, J.D.; Warkup, C. Relationship between skeletal muscle-specific calpain and tenderness of conditioned porcine longissimus muscle. J. Anim. Sci. 1999, 77, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Hao, L.Y.; Kameyama, A.; Kuroki, S.; Takano, J.; Takano, E.; Maki, M.; Kameyama, M. Calpastatin domain L is involved in the regulation of L-type Ca2+ channels in guinea pig cardiac myocytes. Biochem. Biophys. Res. Commun. 2000, 279, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Huff-Lonergan, E.; Lonergan, S.M. Mechanisms of water-holding capacity of meat: The role of postmortem biochemical and structural changes. Meat Sci. 2005, 71, 194–204. [Google Scholar] [CrossRef] [PubMed]
- Koohmaraie, M. The role of Ca2+ -dependent proteases (calpains) in post mortem proteolysis and meat tenderness. Biochimie 1992, 74, 239–245. [Google Scholar] [CrossRef] [PubMed]
- Kemp, C.M.; Sensky, P.L.; Bardsley, R.G.; Buttery, P.J.; Parr, T. Tenderness—An enzymatic view. Meat Sci. 2010, 84, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Sorimachi, H.; Saido, T.C.; Suzuki, K. New era of calpain research. Discovery of tissue-specific calpains. FEBS Lett. 1994, 343, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Huston, R.B.; Krebs, E.G. Activation of skeletal muscle phosphorylase kinase by Ca2+. II. Identification of the kinase activating factor as a proteolytic enzyme. Biochemistry 1968, 7, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Kishimoto, A.; Takai, Y.; Nishizuka, Y. Studies on a cyclic nucleotide-independent protein kinase and its proenzyme in mammalian tissues. II. Proenzyme and its activation by calcium-dependent protease from rat brain. J. Biol. Chem. 1977, 252, 7610–7616. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Kawashima, S. Regulation of the calpain-calpastatin system by membranes (review). Mol. Membr. Biol. 1996, 13, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Goll, D.E.; Thompson, V.F.; Li, H.; Wei, W.; Cong, J. The calpain system. Physiol. Rev. 2003, 83, 731–801. [Google Scholar] [CrossRef] [PubMed]
- Murphy, R.M. Calpains, skeletal muscle function and exercise. Clin. Exp. Pharmacol. Physiol. 2010, 37, 385–391. [Google Scholar] [CrossRef] [PubMed]
- Imajoh, S.; Aoki, K.; Ohno, S.; Emori, Y.; Kawasaki, H.; Sugihara, H.; Suzuki, K. Molecular cloning of the eDNA for the large subunit of the high-Ca-requiring form of human Ca-activated neutral protease. Biochemistry 1988, 27, 8122–8128. [Google Scholar] [CrossRef] [PubMed]
- Sakihama, T.; Kakidani, H.; Zenita, K.; Yumoto, N.; Kikuchi, T.; Sasaki, T.; Kannagi, R.; Nakanishi, S.; Ohmori, M.; Takio, K. A putatixe Ca-binding protein: Structure of the light subunit of porcine calpain elucidated by molecular cloning and protein sequence analysis. Proc. Natl. Acad. Sci. USA 1985, 82, 6075–6079. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K. The structure of calpains and the calpain gene. In Httracelhdar Calcium-Dependent Proteolysis; Mellgren, R.L., Murachi, T., Eds.; CRC Press: Boca Raton, FL, USA, 1990; pp. 25–35. [Google Scholar]
- Melloni, E.; Salamino, F.; Sparatore, B. The calpain-calpastatin system in mammalian cells: Properties and possible functions. Biochimie 1992, 74, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Cong, J.Y.; Goll, D.E.; Peterson, A.M.; Kapprell, H.P. The role of autolysis in activity of the Ca2+-dependent proteinases (μ-Calpain and m-Calpain). J. Biol. Chem. 1989, 264, 10096–10103. [Google Scholar] [CrossRef] [PubMed]
- Tompa, P.; Emori, Y.; Sorimachi, H.; Suzuki, K.; Friedrich, P. Domain III of calpain is a Ca2+ -regulated phospholipid-binding domain. Biochem. Biophys. Res. Commun. 2001, 280, 1333–1339. [Google Scholar] [CrossRef] [PubMed]
- Inomata, M.; Hayashi, M.; Ohno-Iwashita, Y.; Tsubuki, S.; Saido, T.C.; Kawashima, S. Involvement of calpain in integrin-mediated signal transduction. Arch. Biochem. Biophys. 1996, 328, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Ernst, C.W.; Robic, A.; Yerle, M.; Wang, L.; Rothschild, M.F. Mapping of calpastatin and three microsatellites to porcine chromosome 2q2.1-q2.4. Anim. Genet. 1998, 29, 212–215. [Google Scholar] [CrossRef] [PubMed]
- Cong, M.; Thompson, V.F.; Goll, D.E.; Antin, P.B. The bovine calpastatin gene promoter and a new N-terminal region of the protein are targets for cAMP dependent protein kinase activity. J. Biol. Chem. 1998, 273, 660–666. [Google Scholar] [CrossRef] [PubMed]
- Imajoh, S.; Kawasaki, H.; Emori, Y.; Suzuki, K. Calcium-activated neutral protease inhibitor from rabbit erythrocytes lacks the N-terminal region of the liver inhibitor but retains three inhibitory units. Biochem. Biophys. Res. Commun. 1987, 146, 630–632. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.F.; Wei, S.G.; Miao, S.Y.; Liu, Q.Y.; Koide, S.S. Calpastatin genein human testis. Biochem. Mol. Biol. Int. 1994, 33, 245–252. [Google Scholar] [PubMed]
- Parr, T.; Sensky, P.L.; Bardsley, R.G.; Buttery, P.J. Calpastatin expression in porcine cardiac and skeletal muscle and partial gene structure. Arch. Biochem. Biophys. 2001, 395, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.J.; Hatanaka, M.; Maki, M. Multiple forms of rat calpastatin cDNA in the coding region of functionally unknown amino terminal domain. Biochim. Biophys. Acta 1992, 1129, 251–253. [Google Scholar] [CrossRef] [PubMed]
- Takano, J.; Watanabe, M.; Hitomi, K.; Maki, M. Four types of calpastatin isoforms with distinct amino-terminal sequences are identified by alternative first exons and differentially expressed in mouse tissues. J. Biochem. 2000, 128, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Sorimachi, H.; Ishiura, S.; Suzuki, K. Molecular cloning of a novel mammalian calcium dependent protease distinct from both m-and μ-types. J. Biol. Chem. 1989, 264, 20106–20111. [Google Scholar] [CrossRef] [PubMed]
- Mellgren, R.L. Calcium-dependent proteases: An enzyme system active at cellular membranes? FASEB J. 1987, 1, 110–115. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.A.; Campbell, R.L.; Davies, P.L. Calcium-bound structure of calpain and its mechanism of inhibition by calpastatin. Nature 2008, 456, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Imajoh, S.; Suzuki, K. Reversible interaction between Ca2+ activated neutral protease (CANP) and its endogenous inhibitor. FEBS Lett. 1985, 187, 47–50. [Google Scholar] [CrossRef] [PubMed]
- Geesink, G.H.; Koohmaraie, M. Postmortem proteolysis and calpain/calpastatin activity in callipyge and normal lamb biceps femoris during extended postmortem storage. J. Anim. Sci. 1999, 77, 1490–1501. [Google Scholar] [CrossRef] [PubMed]
- Melody, J.L.; Lonergan, S.M.; Rowe, L.J.; Huiatt, T.W.; Mayes, M.S.; Huff-Lonergan, E. Early post-mortem biochemical factors influence tenderness and water-holding capacity of three porcine muscles. J. Anim. Sci. 2004, 82, 1195–1205. [Google Scholar] [CrossRef] [PubMed]
- Geesink, G.H.; Kuchay, S.; Chishti, A.H.; Koohmaraie, M. Micro-calpain is essential for postmortem proteolysis of muscle proteins. J. Anim. Sci. 2006, 84, 2834–2840. [Google Scholar] [CrossRef] [PubMed]
- Pomponio, L.; Lametsch, R.; Karlsson, A.H.; Costa, L.N.; Grossi, A.; Ertbjerg, P. Evidence for post-mortem m-calpain autolysis in porcine muscle. Meat Sci. 2008, 80, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Kent, M.P.; Spencer, M.J.; Koohmaraie, M. Postmortem proteolysis is reduced in transgenic mice overexpressing calpastatin. J. Anim. Sci. 2004, 82, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Koohmaraie, M.; Geesink, G.H. Contribution of postmortem muscle biochemistry to the delivery of consistent meat quality with particular focus on the calpain system. Meat Sci. 2006, 74, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Geesink, G.H.; Taylor, R.G.; Koohmaraie, M. Calpain 3/p94 is not involved in postmortem proteolysis. J. Anim. Sci. 2005, 83, 1646–1652. [Google Scholar] [CrossRef] [PubMed]
- Kapprell, H.P.; Goll, D.E. Effect of Ca2+ on binding of the calpains to calpastatin. J. Biol. Chem. 1989, 264, 17888–17896. [Google Scholar] [CrossRef] [PubMed]
- Maki, M.; Takano, E.; Osawa, T.; Ooi, T.; Murachi, T.; Hatanaka, M. Analysis of structure–function relationship of pig calpastatin by expression of mutated cDNAs in Escherichia coli. J. Biol. Chem. 1988, 263, 10254–10261. [Google Scholar] [CrossRef] [PubMed]
- Page, B.T.; Casas, E.; Heaton, M.P.; Cullen, N.G.; Hyndman, D.L.; Morris, C.A.; Crawford, A.M.; Wheeler, T.L.; Koohmaraie, M.; Keele, J.W.; et al. Evaluation of single-nucleotide polymorphisms in CAPN1 for association with meat tenderness in cattle. J. Anim. Sci. 2002, 80, 3077–3085. [Google Scholar] [CrossRef] [PubMed]
- Casas, E.; White, S.N.; Wheeler, T.L.; Shackelford, S.D.; Koohmaraie, M.; Riley, D.G.; Chase, C.C.; Johnson, D.D., Jr.; Smith, T.P. Effects of calpastatin and micro-calpain markers in beef cattle on tenderness traits. J. Anim. Sci. 2006, 84, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Curi, R.A.; Chardulo, L.A.; Mason, M.C.; Arrigoni, M.D.; Silveira, A.C.; de Oliveira, H.N. Effect of single nucleotide polymorphisms of CAPN1 and CAST genes on meat traits in Nellore beef cattle (Bos indicus) and in their crosses with Bos taurus. Anim. Genet. 2009, 40, 456–462. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Kim, S.C.; Chai, H.H.; Cho, S.H.; Kim, H.C.; Lim, D.; Choi, B.H.; Dang, C.G.; Sharma, A.; Gondro, C.; et al. Mutations in calpastatin and mu-calpain are associated with meat tenderness, flavor and juiciness in Hanwoo (Korean cattle): Molecular modeling of the effects of substitutions in the calpastatin/mu-calpain complex. Meat Sci. 2014, 96, 1501–1508. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.Q.; Liu, H.; Guo, L.J.; Xu, Y.; Liu, D. The mutation site analysis on CAPN1 gene of Wild boar, Min pig and Yorkshire. Yi Chuan 2007, 29, 581–586. [Google Scholar] [CrossRef] [PubMed]
- Nonneman, D.J.; Shackelford, S.D.; King, D.A.; Wheeler, T.L.; Wiedmann, R.T.; Snelling, W.M.; Rohrer, G.A. Genome-wide association of meat quality traits and tenderness in swine. J. Anim. Sci. 2013, 91, 4043–4050. [Google Scholar] [CrossRef] [PubMed]
- Koćwin-Podsiadła, M.; Kurył, J.; Krzȩcio, E.; Zybert, A.; Przybylski, W. The interaction between calpastatin and RYR1 genes for some pork quality traits. Meat Sci. 2003, 65, 731–735. [Google Scholar] [CrossRef] [PubMed]
- Ciobanu, D.C.; Bastiaansen, J.W.; Lonergan, S.M.; Thomsen, H.; Dekkers, J.C.; Plastow, G.S.; Rothschild, M.F. New alleles in calpastatin gene are associated with meat quality traits in pigs. J. Anim. Sci. 2004, 82, 2829–2839. [Google Scholar] [CrossRef] [PubMed]
- Nonneman, D.; Lindholm-Perry, A.K.; Shackelford, S.D.; King, D.A.; Wheeler, T.L.; Rohrer, G.A.; Bierman, C.D.; Schneider, J.F.; Miller, R.K.; Zerby, H.; et al. Predictive markers in calpastatin for tenderness in commercial pig populations. J. Anim. Sci. 2011, 89, 2663–2672. [Google Scholar] [CrossRef] [PubMed]
- Linholm-Perry, A.K.; Rohrer, G.A.; Holl, J.W.; Shackelford, S.D.; Wheeler, T.L.; Koohmaraie, M.; Nonneman, D. Relationships among calpastatin single nucleotide polymorphisms, calpastatin expression and tenderness in pork longissimus. Anim. Genet. 2009, 40, 713–721. [Google Scholar] [CrossRef] [PubMed]
- Kaminski, S.; Help, H.; Brym, P.; Rusc, A.; Wójcik, E. SNiPORK—A microarray of SNPs in candidate genes potentially associated with pork yield and quality-development and validation in commercial breeds. Anim. Biotechnol. 2008, 19, 43–69. [Google Scholar] [CrossRef] [PubMed]
- Rohrer, G.A.; Nonneman, D.J.; Miller, R.K.; Zerby, H.; Moeller, S.J. Association of single nucleotide polymorphism (SNP) markers in candidate genes and QTL regions with pork quality traits in commercial pigs. Meat Sci. 2012, 92, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Y.; Wang, Z.; Bruce, H.L.; Janz, J.; Goddard, E.; Moore, S.; Plastow, G.S. Associations between single nucleotide polymorphisms in 33 candidate genes and meat quality traits in commercial pigs. Anim. Genet. 2014, 45, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Alves, E.; Benítez, R.; García-Casco, J.M.; Muñoz, M.; Caraballo, C.; García, F.; Rodríguez, C. Polimorfismos en las regiones reguladoras del gen CAST: Efectos in vivo y postmortem en cerdos de tipo ibérico. In Proceedings of the XVII Jornadas Sobre Producción Animal, Zaragoza, Spain, 30–31 May 2017; pp. 486–488. [Google Scholar]
- Won, S.; Jung, J.; Park, E.; Kim, H. Identification of genes related to intramuscular fat content of pigs using genome-wide association study. Asian-Australas. J. Anim. Sci. 2018, 31, 157–162. [Google Scholar] [CrossRef] [PubMed]
- Zalewska, M.; Puppel, K.; Sakowski, T. Associations between gene polymorphisms and selected meat traits in cattle—A review. Anim. Biosci. 2021, 34, 1425–1438. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Limón-Morales, O.; Bonilla-Jaime, H.; Arteaga-Silva, M.; Roldán-Santiago, P.; Cruz-Cruz, L.A.d.l.; Orozco-Gregorio, H.; Cerbón, M.; Cortes-Altamirano, J.L. Single Nucleotide Polymorphisms of Leptin and Calpain/Calpastatin in Key Traits of Pork Meat Quality. Animals 2025, 15, 2270. https://doi.org/10.3390/ani15152270
Limón-Morales O, Bonilla-Jaime H, Arteaga-Silva M, Roldán-Santiago P, Cruz-Cruz LAdl, Orozco-Gregorio H, Cerbón M, Cortes-Altamirano JL. Single Nucleotide Polymorphisms of Leptin and Calpain/Calpastatin in Key Traits of Pork Meat Quality. Animals. 2025; 15(15):2270. https://doi.org/10.3390/ani15152270
Chicago/Turabian StyleLimón-Morales, Ofelia, Herlinda Bonilla-Jaime, Marcela Arteaga-Silva, Patricia Roldán-Santiago, Luis Alberto de la Cruz-Cruz, Héctor Orozco-Gregorio, Marco Cerbón, and José Luis Cortes-Altamirano. 2025. "Single Nucleotide Polymorphisms of Leptin and Calpain/Calpastatin in Key Traits of Pork Meat Quality" Animals 15, no. 15: 2270. https://doi.org/10.3390/ani15152270
APA StyleLimón-Morales, O., Bonilla-Jaime, H., Arteaga-Silva, M., Roldán-Santiago, P., Cruz-Cruz, L. A. d. l., Orozco-Gregorio, H., Cerbón, M., & Cortes-Altamirano, J. L. (2025). Single Nucleotide Polymorphisms of Leptin and Calpain/Calpastatin in Key Traits of Pork Meat Quality. Animals, 15(15), 2270. https://doi.org/10.3390/ani15152270






