Global Whole-Genome Resequencing of Beef Cattle Reveals Characteristic Traits Related Genes in Pinan Cattle
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Preparation
2.3. Read Mapping and SNP Calling
2.4. Population Genetic Analysis
2.5. Detection of Selective Signatures
2.6. Enrichment Analysis
2.7. Protein Structure and Function Prediction
3. Results
3.1. Data Analysis and SNP Calling
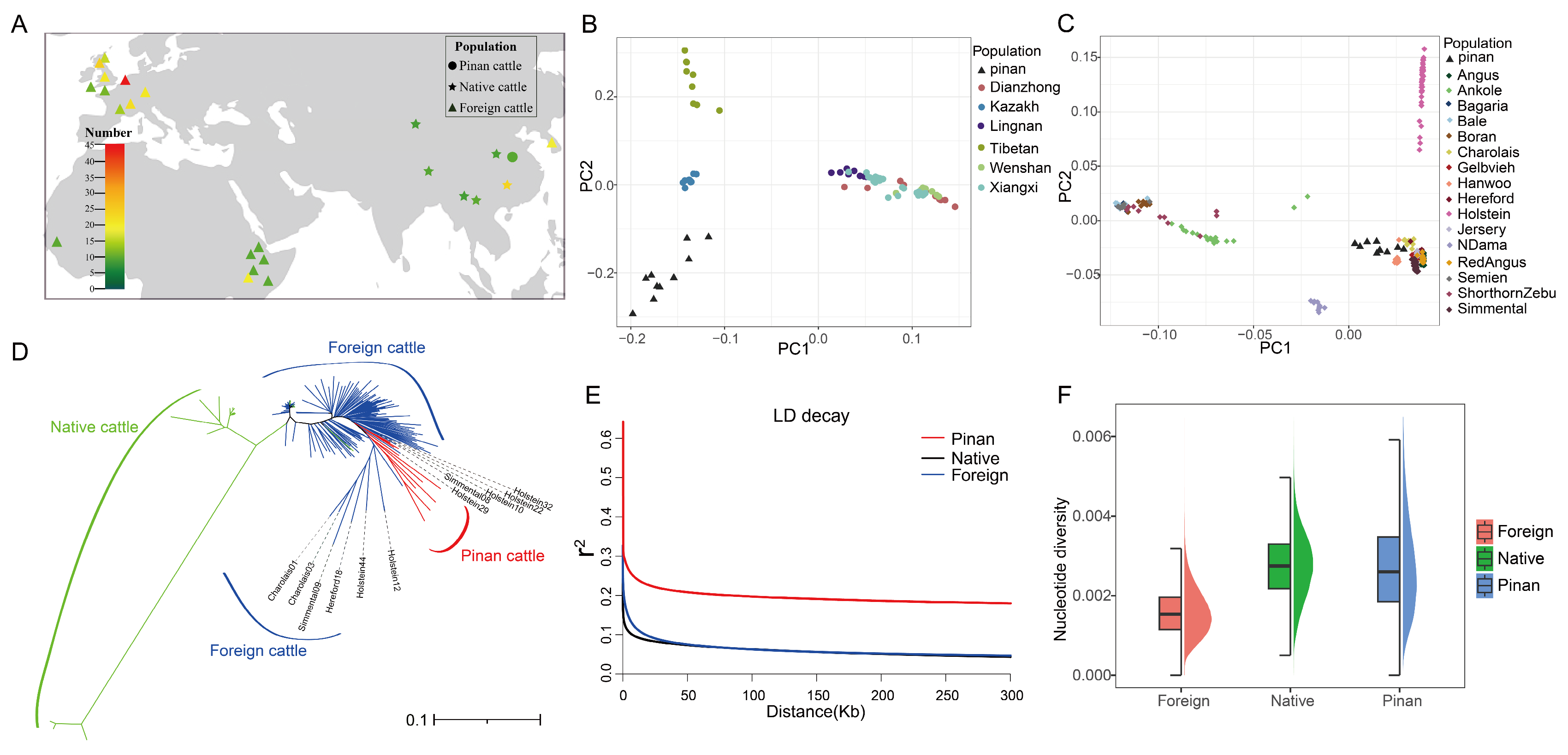
3.2. Population Genetic Structure and Diversity Analysis
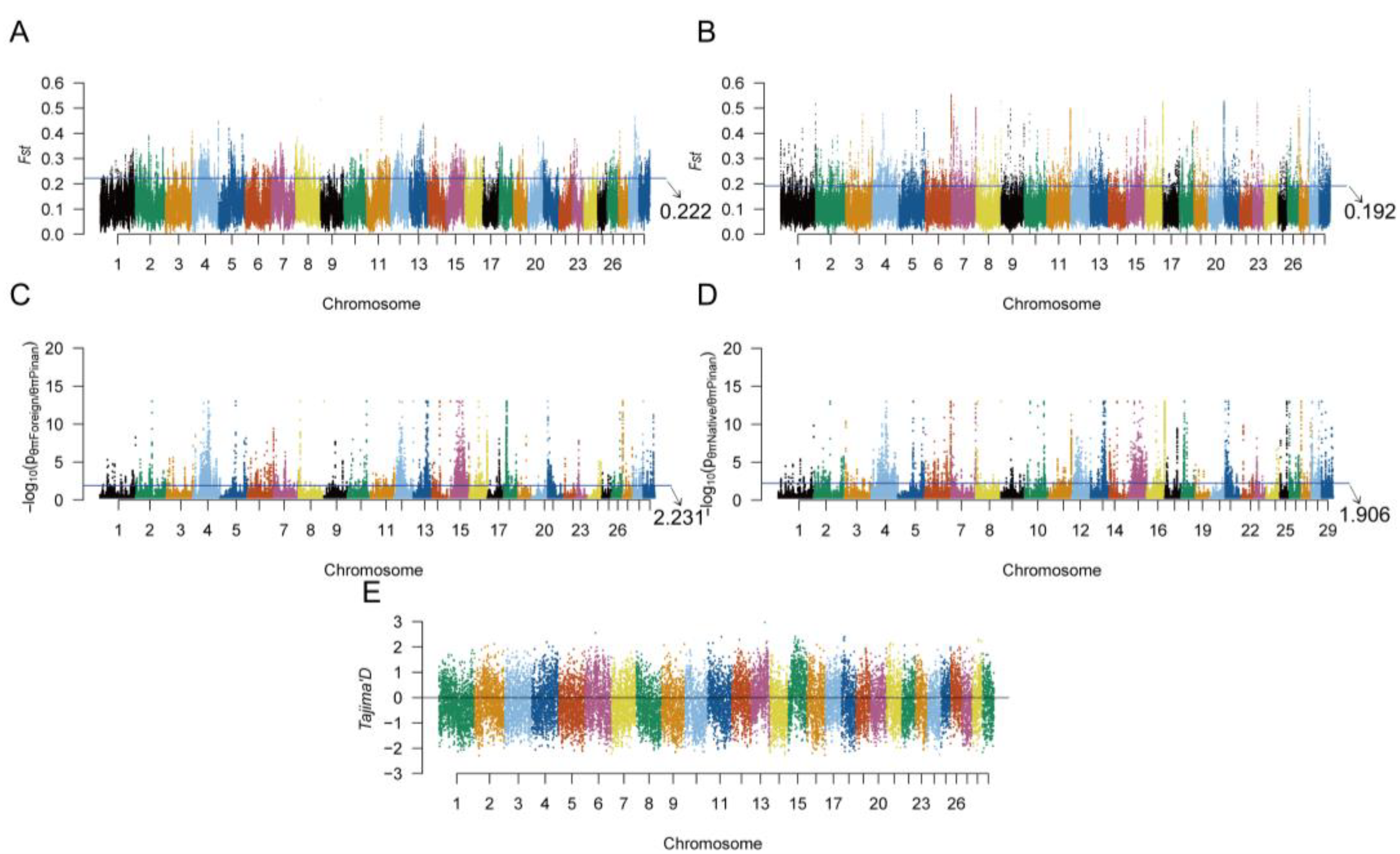
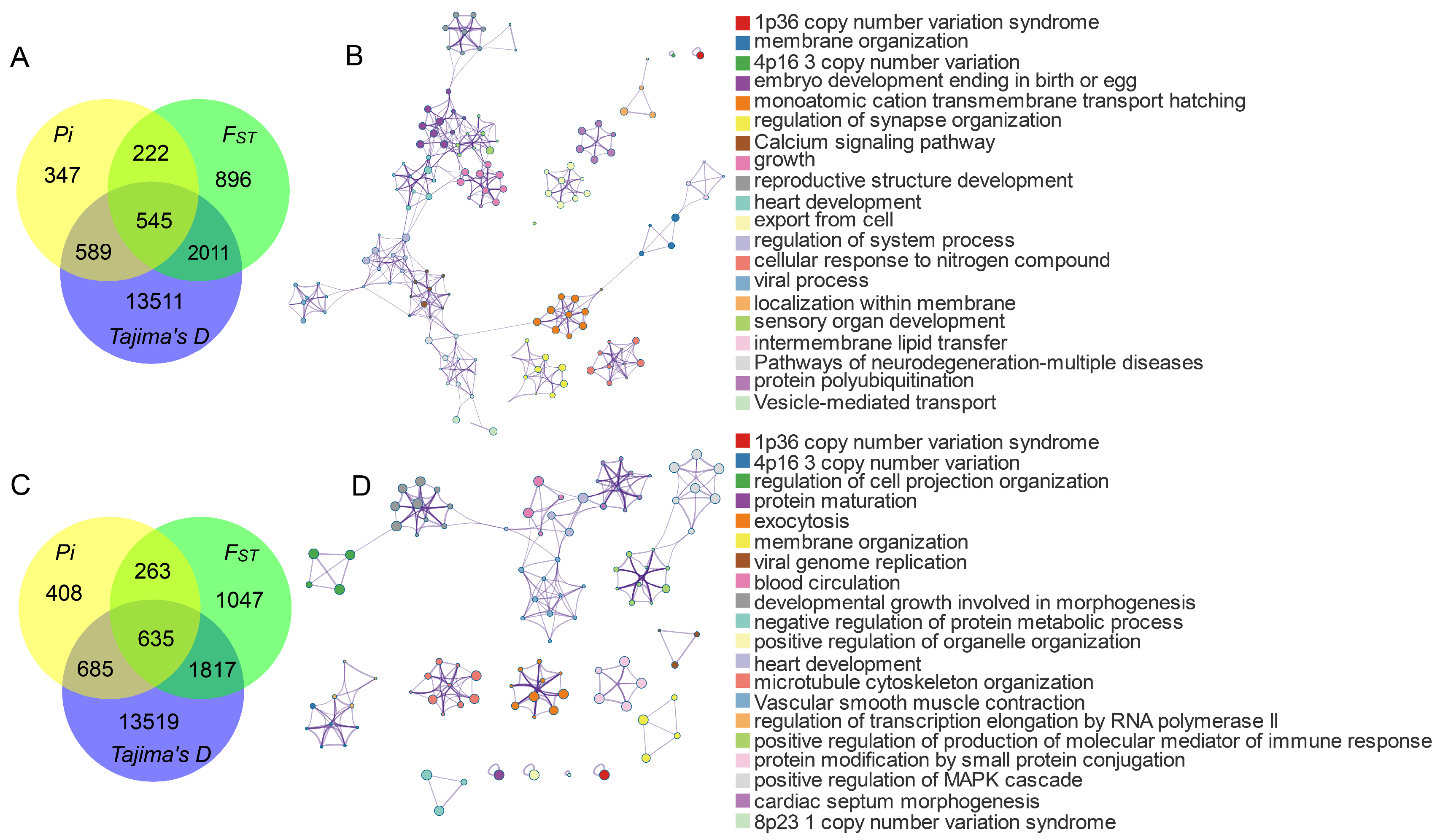
3.3. Genetic Signature of Positive Selection in Pinan Cattle
3.4. Consequences of Positively Selected SNPs on Protein
3.4.1. Effects of NDN c.581T > A on NDN Protein Structure and Function
3.4.2. Effects of PARVA c.893T > A on PARVA Protein Structure and Function
4. Discussion
4.1. Discussion on the Population Genetic Structure and Diversity Analysis
4.2. Signature of Selection Analysis
4.3. Positively Selected Loci NDN c.581T > A and PARVA c.893T > A in Pinan Cattle
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Li, F.; Chen, Y.; Wu, H.; Meng, Q.; Guan, L.L. Metatranscriptomic profiling reveals the effect of breed on active rumen eukaryotic composition in beef cattle with varied feed efficiency. Front. Microbiol. 2020, 11, 367. [Google Scholar] [CrossRef] [PubMed]
- Xue, Y.; Yan, J.; Cui, Y.; Zhao, H.; Zhang, Y.n.; Ma, C.; Zheng, H. The technical efficiency of beef calf production systems: Evidence from a survey in Hebei, China. Agriculture 2022, 12, 1604. [Google Scholar] [CrossRef]
- Wang, C.; Zhang, J.; Zhou, W. The effect of animal husbandry on economic growth: Evidence from 13 provinces of North China. Front. Environ. Sci. 2023, 10. [Google Scholar] [CrossRef]
- Liu, X.; Liu, X.Y.; Zhang, D.A.; Wang, J.Q.; Liu, Q.S.; Liang, S.; Song, W.W.; Huang, Y.Z. Progress in breeding and new variety approval of Pinan cattle. China Cattle Sci. 2023, 49, 54–56. [Google Scholar]
- Nong, Y. Pinan cattle grow fast. Fortune World 2016, 12, 67. [Google Scholar]
- Wang, J.Q.; Wang, Y.H.; Tan, S.J.; Ru, B.R.; Liu, X.; Li, Q.Z. Study on the performance of Pinan cattle growth, reproduction, slaughter and meat quality. China Cattle Sci. 2019, 45, 52–54. [Google Scholar]
- Yu, D.H.; Li, Q.Z.; Lu, T.Y.; Zhang, R.D.; Feng, W.C. Comparative study on fattening and slaughtering of Pinan cattle and Nanyang cattle. In Proceedings of the Progress in China’s Cattle Industry 2009, Nanyang, China, 10–11 November 2009. [Google Scholar]
- Feng, L. Reproductive performance of crossbreed bulls and cows of Pinan crossbreed. Contemp. Anim. Husb. 2011, 7, 39–41. [Google Scholar]
- Bo, D.; Feng, Y.; Bai, Y.; Li, J.; Wang, Y.; You, Z.; Shen, J.; Bai, Y. Whole-genome resequencing reveals genetic diversity and growth trait-related genes in Pinan cattle. Animals 2024, 14, 2163. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Biasini, M.; Bienert, S.; Waterhouse, A.; Arnold, K.; Studer, G.; Schmidt, T.; Kiefer, F.; Gallo Cassarino, T.; Bertoni, M.; Bordoli, L.; et al. SWISS-MODEL: Modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014, 42, 252–258. [Google Scholar] [CrossRef]
- Mooers, B.H.M. Shortcuts for faster image creation in PyMOL. Protein Sci. 2020, 29, 268–276. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; et al. CDD: Specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009, 37, 205–210. [Google Scholar] [CrossRef]
- Vaser, R.; Adusumalli, S.; Leng, S.N.; Sikic, M.; Ng, P.C. SIFT missense predictions for genomes. Nat. Protoc. 2016, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Peters, S.O.; De Donato, M.; Hussain, T.; Rodulfo, H.; Babar, M.E.; Imumorin, I.G. Sequence variation of necdin gene in Bovidae. J. Anim. Sci. Technol. 2018, 60, 32. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Chen, W.; Li, D.; Gu, S.; Liu, X.; Dong, Y.; Jin, L.; Zhang, C.; Li, S. Conservation of imprinting and methylation of MKRN3, MAGEL2 and NDN genes in cattle. Animals 2021, 11, 1985. [Google Scholar] [CrossRef] [PubMed]
- Bo, D.D.; Li, M.Y.; Bai, Y.L.; Li, J.; Sheng, M.X.; Feng, Y.Q.; Bai, Y.Y. Study on the growth and development of Pinan crossbred cows. China Cattle Sci. 2024, 50, 24–28, 53. [Google Scholar]
- Liang, S.; Wang, J.Q.; Zhang, L. Accelerating cultivation of Pinan cattle and advancing approval of new breeds. China Anim. Ind. 2024, 14, 46–47. [Google Scholar]
- Zhang, S.; Yao, Z.; Li, X.; Zhang, Z.; Liu, X.; Yang, P.; Chen, N.; Xia, X.; Lyu, S.; Shi, Q.; et al. Assessing genomic diversity and signatures of selection in Pinan cattle using whole-genome sequencing data. BMC Genom. 2022, 23, 460. [Google Scholar] [CrossRef]
- Wei, X.; Zhu, Y.; Zhao, X.; Zhao, Y.; Jing, Y.; Liu, G.; Wang, S.; Li, H.; Ma, Y. Transcriptome profiling of mRNAs in muscle tissue of Pinan cattle and Nanyang cattle. Gene 2022, 825, 146435. [Google Scholar] [CrossRef]
- Wang, P.; Ou, G.; Li, G.; Li, H.; Zhao, T. Analysis of genetic diversity and structure of endangered Dengchuan cattle population using a single-nucleotide polymorphism chip. Anim. Biotechnol. 2024, 35, 2349625. [Google Scholar] [CrossRef]
- Suezawa, R.; Nikadori, H.; Sasaki, S. Genetic diversity and genomic inbreeding in Japanese Black cows in the islands of Okinawa Prefecture evaluated using single-nucleotide polymorphism array. Anim. Sci. J. 2021, 92, e13525. [Google Scholar] [CrossRef]
- Slatkin, M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat. Rev. Genet. 2008, 9, 477–485. [Google Scholar] [CrossRef]
- McVean, G.A. A genealogical interpretation of linkage disequilibrium. Genetics 2002, 162, 987–991. [Google Scholar] [CrossRef] [PubMed]
- Pang, P.Y.; Zhang, Z.J.; Liu, X.; Yao, Z.; Song, X.Y.; Wang, H.R.; Wang, J.Q.; Li, Q.Z.; Wang, E.Y.; Ru, B.R.; et al. Basic ideas and key measures for improving breeding of Pinan cattle. China Cattle Sci. 2022, 48, 83–85, 96. [Google Scholar]
- Woollard, J.; Schmitz, C.B.; Freeman, A.E.; Tuggle, C.K. Rapid communication: HinfI polymorphism at the bovine PIT1 locus. J. Anim. Sci. 1994, 72, 3267. [Google Scholar] [CrossRef][Green Version]
- Di Stasio, L.; Sartore, S.; Albera, A. Lack of association of GH1 and POU1F1 gene variants with meat production traits in Piemontese cattle. Anim. Genet. 2002, 33, 61–64. [Google Scholar] [CrossRef]
- Qiu, G.Y.; Cheng, H.; Pan, C.Y.; Wang, J.Q.; Niu, H. Genetic variations of HinfⅠ, AluⅠ and PstⅠ loci within POU1F1 gene exon 6 and its association with growth traits in Jiaxian Red Cattle. J. Northwest Sci-Tech Univ. Agric. For. (Nat. Sci. Ed.) 2009, 37, 43–48. [Google Scholar]
- Liu, B.; Cheng, H.; Lan, X.Y.; Lei, C.C.; Zhang, Z.Q.; Zhang, R.F. Correlation of polymorphisms of POU1F1 gene and growth traits in Qinchuan cattle and its hybrid cattle. Sci. Agric. Sin. 2005, 38, 2520–2525. [Google Scholar]
- Hirose, K.; Shimoda, N.; Kikuchi, Y. Expression patterns of lgr4 and lgr6 during zebrafish development. GEP 2011, 11, 378–383. [Google Scholar] [CrossRef]
- Doherty, L.; Wan, M.; Peterson, A.; Youngstrom, D.W.; King, J.S.; Kalajzic, I.; Hankenson, K.D.; Sanjay, A. Wnt-associated adult stem cell marker Lgr6 is required for osteogenesis and fracture healing. Bone 2023, 169, 116681. [Google Scholar] [CrossRef]
- Lehoczky, J.A.; Tabin, C.J. Lgr6 marks nail stem cells and is required for digit tip regeneration. Proc. Natl. Acad. Sci. USA 2015, 112, 13249–13254. [Google Scholar] [CrossRef]
- Nurgulsim, K.; Rajwali, K.; Abbas, R.S.H.; Amel, A.-A.; Mouna, J.; Ijaz, A.; Mumtaz, A.U.; Hassan, A.E.-A.A.; Hamayun, K.; Zan, L. Bioinformatics and genetic variants analysis of FGF10 gene promoter with their association at carcass quality and body measurement traits in Qinchuan beef cattle. Anim. Biotechnol. 2023, 34, 1950–1959. [Google Scholar] [CrossRef]
- Chen, L.; Pan, L.; Zeng, Y.; Zhu, X.; You, L. CPNE1 regulates myogenesis through the PERK-eIF2α pathway mediated by endoplasmic reticulum stress. Cell Tissue Res. 2023, 391, 545–560. [Google Scholar] [CrossRef] [PubMed]
- Hernandez Cordero, A.I.; Gonzales, N.M.; Parker, C.C.; Sokolof, G.; Vandenbergh, D.J.; Cheng, R.; Abney, M.; Sko, A.; Douglas, A.; Palmer, A.A.; et al. Genome-wide Associations Reveal Human-Mouse Genetic Convergence and Modifiers of Myogenesis, CPNE1 and STC2. Am. J. Hum. Genet. 2019, 105, 1222–1236. [Google Scholar] [CrossRef]
- Bae, J.H.; Hong, M.; Jeong, H.J.; Kim, H.; Lee, S.J.; Ryu, D.; Bae, G.U.; Cho, S.C.; Lee, Y.S.; Krauss, R.S.; et al. Satellite cell-specific ablation of Cdon impairs integrin activation, FGF signalling, and muscle regeneration. J. Cachexia Sarcopenia 2020, 11, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Echevarría-Andino, M.L.; Franks, N.E.; Schrader, H.E.; Hong, M.; Krauss, R.S.; Allen, B.L. CDON contributes to Hedgehog-dependent patterning and growth of the developing limb. Dev. Biol. 2023, 493, 1–11. [Google Scholar] [CrossRef]
- Pruller, J.; Figeac, N.; Zammit, P.S. DVL1 and DVL3 require nuclear localisation to regulate proliferation in human myoblasts. Sci. Rep. 2022, 12, 8388. [Google Scholar] [CrossRef]
- Mawaribuchi, S.; Musashijima, M.; Wada, M.; Izutsu, Y.; Kurakata, E.; Park, M.K.; Takamatsu, N.; Ito, M. Molecular evolution of two distinct dmrt1 promoters for germ and somatic cells in vertebrate gonads. Mol. Biol. Evol. 2017, 34, 724–733. [Google Scholar] [CrossRef]
- Yoshimoto, S.; Ikeda, N.; Izutsu, Y.; Shiba, T.; Takamatsu, N.; Ito, M. Opposite roles of DMRT1 and its W-linked paralogue, DM-W, in sexual dimorphism of Xenopus laevis: Implications of a ZZ/ZW-type sex-determining system. Development 2010, 137, 2519–2526. [Google Scholar] [CrossRef]
- Masuyama, H.; Yamada, M.; Kamei, Y.; Fujiwara-Ishikawa, T.; Todo, T.; Nagahama, Y.; Matsuda, M. Dmrt1 mutation causes a male-to-female sex reversal after the sex determination by Dmy in the medaka. Chromosome Res. 2012, 20, 163–176. [Google Scholar] [CrossRef] [PubMed]
- Irie, N.; Lee, S.-M.; Lorenzi, V.; Xu, H.; Chen, J.; Inoue, M.; Kobayashi, T.; Sancho-Serra, C.; Drousioti, E.; Dietmann, S.; et al. DMRT1 regulates human germline commitment. Nat. Cell Biol. 2023, 25, 1439–1452. [Google Scholar] [CrossRef]
- Wei, Y.; Yang, D.; Du, X.; Yu, X.; Zhang, M.; Tang, F.; Ma, F.; Li, N.; Bai, C.; Li, G.; et al. Interaction between DMRT1 and PLZF protein regulates self-renewal and proliferation in male germline stem cells. Mol. Cell. Biochem. 2021, 476, 1123–1134. [Google Scholar] [CrossRef]
- Jorgensen, E.M.; Ruman, J.I.; Doherty, L.; Taylor, H.S. A novel mutation of HOXA13 in a family with hand-foot-genital syndrome and the role of polyalanine expansions in the spectrum of Müllerian fusion anomalies. Fertil. Steril. 2010, 94, 1235–1238. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Taylor, H.S. The role of hox genes in female reproductive tract development, adult function, and fertility. Cold Spring Harb. Perspect. Med. 2015, 6, a023002. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Modi, D. Role of HOXA10 in pathologies of the endometrium. Rev. Endocr. Metab. Disord. 2025, 26, 81–96. [Google Scholar]
- Yu, M.; Tang, J.; Huang, Y.; Guo, C.; Du, P.; Li, N.; Quan, Q. HOXA10 regulates the synthesis of cholesterol in endometrial stromal cells. Front. Endocrinol. 2022, 13, 852671. [Google Scholar]
- Connell, K.A.; Guess, M.K.; Chen, H.; Andikyan, V.; Bercik, R.; Taylor, H.S. HOXA11 is critical for development and maintenance of uterosacral ligaments and deficient in pelvic prolapse. J. Clin. Investig. 2008, 118, 1050–1055. [Google Scholar]
- Ma, Y.; Guess, M.; Datar, A.; Hennessey, A.; Cardenas, I.; Johnson, J.; Connell, K.A. Knockdown of Hoxa11 in vivo in the uterosacral ligament and uterus of mice results in altered collagen and matrix metalloproteinase activity. Biol. Reprod. 2012, 86, 100. [Google Scholar] [CrossRef]
- Kobielak, A.; Pasolli, H.A.; Fuchs, E. Mammalian formin-1 participates in adherens junctions and polymerization of linear actin cables. Nat. Cell Biol. 2004, 6, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Mruk, D.D.; Tang, E.I.; Lee, W.M.; Wong, C.K.; Cheng, C.Y. Formin 1 regulates microtubule and F-Actin organization to support spermatid transport during spermatogenesis in the rat testis. Endocrinology 2016, 157, 2894–2908. [Google Scholar] [CrossRef]
- Buzanskas, M.E.; Grossi, D.d.A.; Ventura, R.V.; Schenkel, F.S.; Chud, T.C.S.; Stafuzza, N.B.; Rola, L.D.; Meirelles, S.L.C.; Mokry, F.B.; Mudadu, M.d.A.; et al. Candidate genes for male and female reproductive traits in Canchim beef cattle. J. Anim. Sci. Biotechnol. 2017, 8, 67. [Google Scholar] [CrossRef]
- Padua, M.B.; Fox, S.C.; Jiang, T.; Morse, D.A.; Tevosian, S.G. Simultaneous gene deletion of gata4 and gata6 leads to early disruption of follicular development and germ cell loss in the murine ovary. Biol. Reprod. 2014, 91, 24. [Google Scholar] [CrossRef]
- Mehanovic, S.; Pierre, K.J.; Viger, R.S.; Tremblay, J.J. Chicken ovalbumin upstream promoter transcription factor type II interacts and functionally cooperates with GATA4 to regulate anti-Müllerian hormone receptor type 2 transcription in mouse MA-10 Leydig cells. Andrology 2022, 10, 1411–1425. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.F.; Taniguchi, H.; Viger, R.S. The effect of human GATA4 gene mutations on the activity of target gonadal promoters. J. Mol. Endocrinol. 2009, 42, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Zhang, T.; Liu, N.; Wang, C.; Guo, Z.; Pan, C.; Zhu, H.; Lan, X. Investigation of copy number variations (CNVs) of the goat PPP3CA gene and their effect on litter size and semen quality. Animals 2022, 12, 445. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, W.; Du, J.; Liu, H.; Wu, J.; Wang, C.; Tang, M.; Liu, Y.; Ju, Y.; Qu, W.; et al. Aluminum promotes B1 cells to produce IL-10 and impairs adaptive immune system. Environ. Pollut. 2025, 368, 125791. [Google Scholar] [CrossRef]
- El-Shemi, A.G.; Alqurashi, A.; Abdulrahman, J.A.; Alzahrani, H.D.; Almwalad, K.S.; Felfilan, H.H.; Alomiri, W.S.; Aloufi, J.A.; Madkhali, G.H.; Maqliyah, S.A.; et al. IL-10-Directed cancer immunotherapy: Preclinical advances, clinical insights, and future perspectives. Cancers 2025, 17, 1012. [Google Scholar] [CrossRef]
- Li, Q.; Zhu, J.; Zhang, Y.; Pan, Y.; Li, Z.; Wang, M.; Gao, Y.; Feng, D.; He, X.; Zhang, C. Association of WHSC1/NSD2 and T-cell infiltration with prostate cancer metastasis and prognosis. Sci. Rep. 2023, 13, 21629. [Google Scholar] [CrossRef]
- Moon, S.W.; Son, H.J.; Mo, H.Y.; Choi, E.J.; Yoo, N.J.; Lee, S.H. Mutation and expression alterations of histone methylation-related NSD2, KDM2B and SETMAR genes in colon cancers. Pathol. Res. Pract. 2021, 219, 153354. [Google Scholar] [CrossRef]
- Zhou, S.; Liu, S.; Tian, G.; Zhao, L.; Wang, H.; Li, Y.; Shen, Y.; Han, L. KLK5 is associated with the radioresistance, aggression, and progression of cervical cancer. Gynecol. Oncol. 2022, 166, 138–147. [Google Scholar] [CrossRef]
- Garcia-Vidal, E.; Calba, I.; Riveira-Muñoz, E.; García, E.; Clotet, B.; Serra-Mitjà, P.; Cabrera, C.; Ballana, E.; Badia, R. Nucleotide-Binding oligomerization domain 1 (NOD1) agonists prevent SARS-CoV-2 Infection in human lung epithelial cells through harnessing the innate immune response. Int. J. Mol. Sci. 2024, 25, 5318. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhao, J.; Zhou, M.; Wei, K.; Jiang, P.; Xu, L.; Chang, C.; Shan, Y.; Xu, L.; Shi, Y.; et al. Role of signaling lymphocytic activation molecule family of receptors in the pathogenesis of rheumatoid arthritis: Insights and application. Front. Pharmacol. 2023, 14, 1306584. [Google Scholar] [CrossRef]
- Nilsen, K.E.; Zhang, B.; Skjesol, A.; Ryan, L.; Vagle, H.; Bøe, M.H.; Orning, P.; Kim, H.; Bakke, S.S.; Elamurugan, K.; et al. Peptide derived from SLAMF1 prevents TLR4-mediated inflammation in vitro and in vivo. Life Sci. Alliance 2023, 6, e202302164. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Dong, X.; Liu, J.; Liu, T.; Chen, X.; Bian, X.; Li, M.; Liu, Y. TRIM67 promotes non-small cell lung cancer development by positively regulating the notch pathway through DLK1 ubiquitination. J. Cancer 2024, 15, 1870–1879. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Miao, K.; Gao, L.; He, L. TRIM67 interacts with ENAH to regulate the apoptosis and autophagy of lung cancer cells. Gen. Physiol. Biophys. 2024, 43, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Couto, N.; Elzanowska, J.; Maia, J.; Batista, S.; Pereira, C.E.; Beck, H.C.; Carvalho, A.S.; Strano Moraes, M.C.; Carvalho, C.; Oliveira, M.; et al. IgG(+) extracellular vesicles measure therapeutic response in advanced pancreatic cancer. Cells 2022, 11, 2800. [Google Scholar] [CrossRef]
- Zhang, Y.; Tang, J.; Zheng, Y.; Guo, W.; Guo, Y.; Chang, M.; Wang, H.; Li, Y.; Chang, Z.; Xu, Y.; et al. Evolutionary and expression analysis of the pig MAGE gene family. Animals 2024, 14, 2095. [Google Scholar] [CrossRef]
- Legate, K.R.; Montañez, E.; Kudlacek, O.; Füssler, R. ILK, PINCH and parvin: The tIPP of integrin signalling. Nat. Rev. Mol. Cell Biol. 2006, 7, 20–31. [Google Scholar] [CrossRef]
- Onetti, Y.; Kälin, R.E.; Pitter, B.; Hou, M.; Arribas, V.; Glass, R.; Montanez, E. Deletion of endothelial α-parvin inhibits tumour angiogenesis, reduces tumour growth and induces tumour cell apoptosis. Angiogenesis 2022, 25, 155–158. [Google Scholar] [CrossRef]


| Missense Mutation | Chromosome | ΔAF1 1 | ΔAF2 1 | Amino Acid Variation |
|---|---|---|---|---|
| DMRT1 c.101G > C | Chr8 | 0.6212 | 0.8905 | G/A |
| DMRT1 c.634T > A | Chr8 | 0.5791 | 0.7759 | Y/N |
| DMRT1 c.881C > G | Chr8 | 0.5970 | 0.8905 | T/S |
| FMN1 c.58G > C | Chr10 | 0.6045 | 0.8869 | E/Q |
| FMN1 c.745C > G | Chr10 | 0.6119 | 0.8923 | L/V |
| CPNE1 c.2T > C | Chr13 | 0.7778 | 0.6667 | M/T |
| PARVA c.893T > A 2 | Chr15 | 0.6045 | 0.8905 | V/E |
| LGR6 c.887C > G | Chr16 | 0.5417 | 0.6689 | S/W |
| LGR6 c.571G > C | Chr16 | 0.5625 | 0.8837 | A/P |
| NDN c.581T > A 2 | Chr21 | 0.6045 | 0.8901 | L/H |
| PPP3CB c.12G > C | Chr28 | 0.5561 | 0.8259 | E/D |
| TRIM67 c.30G > C | Chr28 | 0.5896 | 0.8901 | C/W |
| TRIM67 c.154G > C | Chr28 | 0.5896 | 0.8901 | A/P |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bo, D.; Wang, Y.; Bai, Y.; Li, J.; Shen, J.; Wei, J.; Bai, Y. Global Whole-Genome Resequencing of Beef Cattle Reveals Characteristic Traits Related Genes in Pinan Cattle. Animals 2025, 15, 1626. https://doi.org/10.3390/ani15111626
Bo D, Wang Y, Bai Y, Li J, Shen J, Wei J, Bai Y. Global Whole-Genome Resequencing of Beef Cattle Reveals Characteristic Traits Related Genes in Pinan Cattle. Animals. 2025; 15(11):1626. https://doi.org/10.3390/ani15111626
Chicago/Turabian StyleBo, Dongdong, Yuanyuan Wang, Yilin Bai, Jing Li, Jiameng Shen, Jinxiao Wei, and Yueyu Bai. 2025. "Global Whole-Genome Resequencing of Beef Cattle Reveals Characteristic Traits Related Genes in Pinan Cattle" Animals 15, no. 11: 1626. https://doi.org/10.3390/ani15111626
APA StyleBo, D., Wang, Y., Bai, Y., Li, J., Shen, J., Wei, J., & Bai, Y. (2025). Global Whole-Genome Resequencing of Beef Cattle Reveals Characteristic Traits Related Genes in Pinan Cattle. Animals, 15(11), 1626. https://doi.org/10.3390/ani15111626




