Co-Infection by Leptospira montravelensis and Leptospira interrogans Serovar Pomona in Urine Samples of Donkeys and Pigs in Sardinia, Italy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection and MAT Analysis
2.2. DNA Extraction and Molecular Detection of Leptospira spp. by Multiplex qPCR
2.3. Cultural Assay
2.4. MLST Genotyping and Amplification of rrs, rpoB, and secY Genes
2.5. Whole-Genome Sequencing
3. Results
3.1. Sample Collection and MAT Results
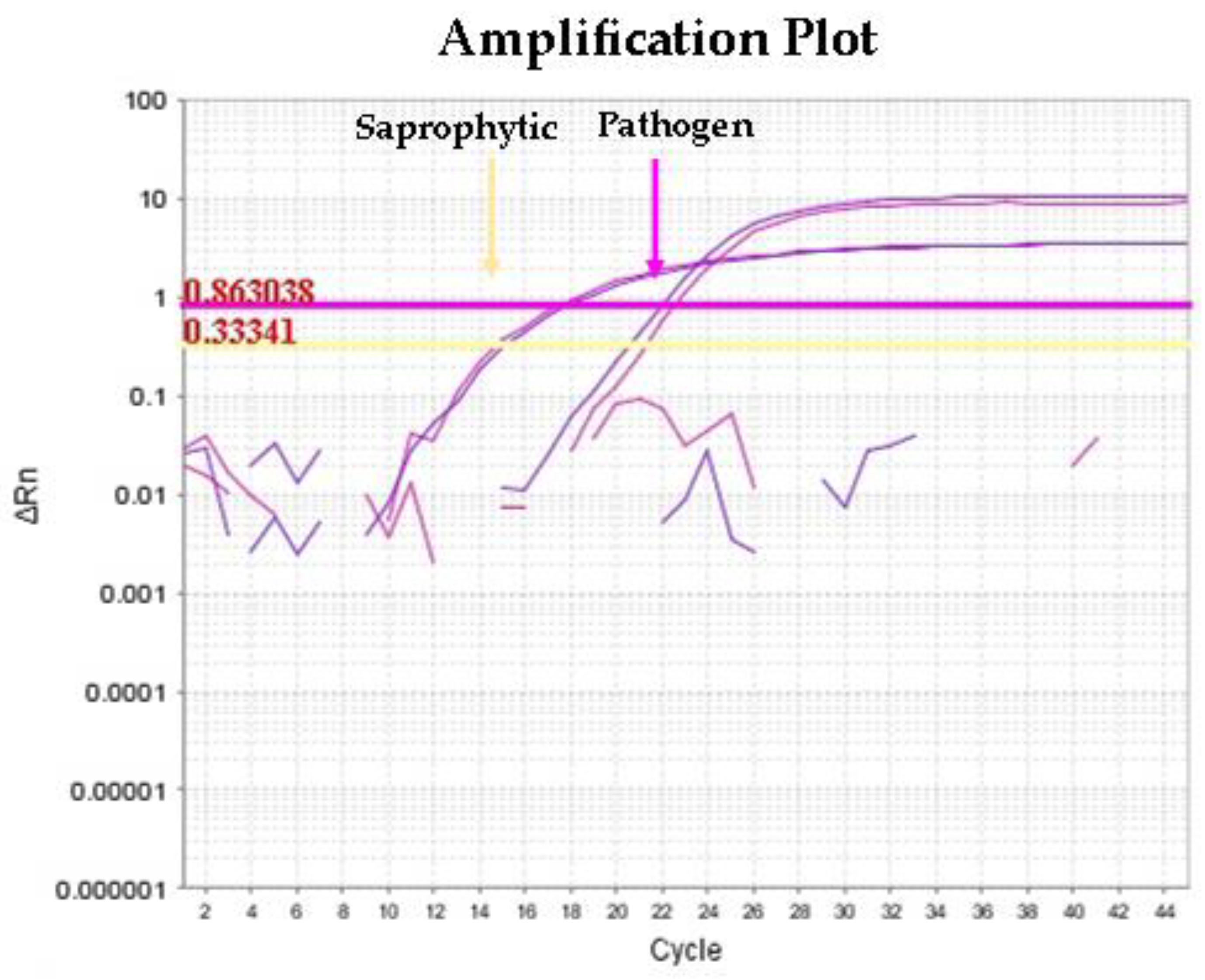
3.2. Molecular Detection of Leptospira spp. from Urine and Kidney Samples
3.3. Molecular Identification by PCR and MLST from Urine and Kidney Samples
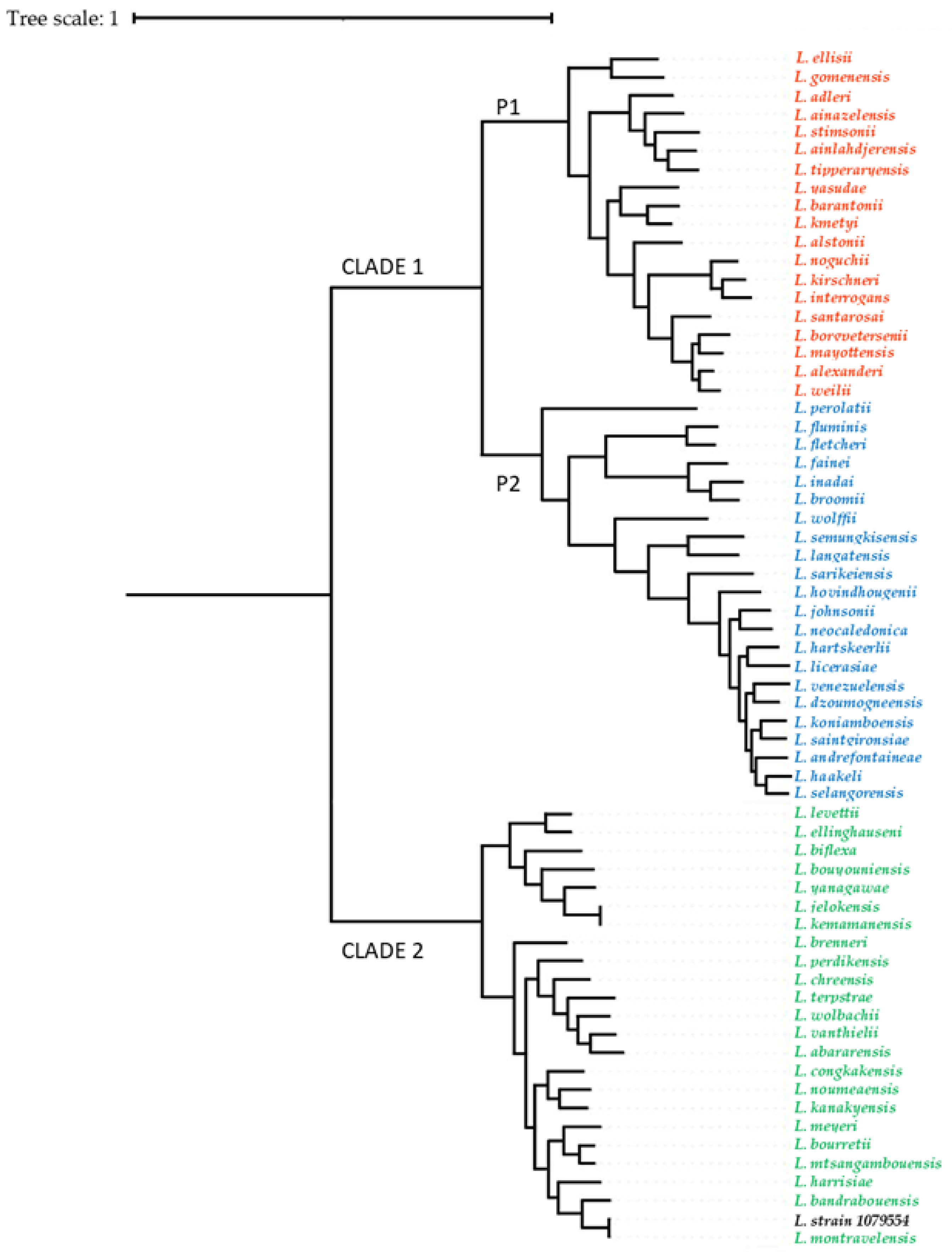
3.4. Whole-Genome Sequencing of Leptospira Strains and Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Soo, Z.M.P.; Khan, N.A.; Siddiqui, R. Leptospirosis: Increasing importance in developing countries. Acta Trop. 2020, 201. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Virulence of the zoonotic agent of leptospirosis: Still terra incognita? Nat. Rev. Microbiol. 2017, 15, 297–307. [Google Scholar] [CrossRef] [PubMed]
- WHO. Report of the Second Meeting of the Leptospirosis Burden Epidemiology Reference Group; WHO: Geneva, Switzerland, 2011. [Google Scholar]
- Adler, B.; de la Peña Moctezuma, A. Leptospira and leptospirosis. Vet. Microbiol. 2010, 140, 287–296. [Google Scholar] [CrossRef]
- Grippi, F.; Blanda, V.; Lo Presti, V.D.M.; Macaluso, G.; Galluzzo, P.; Bertasio, C.; Sciacca, C.; Arcuri, F.; D’agostino, R.; Ippolito, D.; et al. Leptospira interrogans Serogroup Pomona in a Dairy Cattle Farm in a Multi-Host Zootechnical System. Vet. Sci. 2022, 9, 83. [Google Scholar] [CrossRef]
- Bierque, E.; Thibeaux, R.; Girault, D.; Soupé-Gilbert, M.E.; Goarant, C. A systematic review of Leptospira in water and soil environments. PLoS ONE 2020, 15, e0227055. [Google Scholar] [CrossRef] [PubMed]
- Cilia, G.; Bertelloni, F.; Albini, S.; Fratini, F. Insight into the Epidemiology of Leptospirosis: A Review of Leptospira Isolations from “Unconventional” Hosts. Animals 2021, 11, 191. [Google Scholar] [CrossRef]
- Allan, K.J.; Biggs, H.M.; Halliday, J.E.B.; Kazwala, R.R.; Maro, V.P.; Cleaveland, S.; Crump, J.A. Epidemiology of Leptospirosis in Africa: A Systematic Review of a Neglected Zoonosis and a Paradigm for ‘One Health’ in Africa. PLoS Negl. Trop. Dis. 2015, 9, e0003899. [Google Scholar] [CrossRef]
- Dupouey, J.; Faucher, B.; Edouard, S.; Richet, H.; Kodjo, A.; Drancourt, M.; Davoust, B. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 77–83. [Google Scholar] [CrossRef]
- Magalhães, A.R.; Codeço, C.T.; Svenning, J.C.; Escobar, L.E.; Van de Vuurst, P.; Gonçalves-Souza, T. Neglected tropical diseases risk correlates with poverty and early ecosystem destruction. Infect. Dis. poverty 2023, 12, 32. [Google Scholar] [CrossRef]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global Morbidity and Mortality of Leptospirosis: A Systematic Review. PLoS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef]
- Agampodi, S.B.; Karunarathna, D.; Jayathilala, N.; Rathnayaka, H.; Agampodi, T.C.; Karunanayaka, L. Outbreak of leptospirosis after white-water rafting: Sign of a shift from rural to recreational leptospirosis in Sri Lanka? Epidemiol. Infect. 2014, 142, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Gomes de Araújo, H.; Limeira, C.H.; Viviane Ferreira de Aquino, V.; Longo Ribeiro Vilela, V.; José Alves, C.; Silvano dos Santos Higino, S.; Santos, C.d.S.A.B.; de Azevedo, S.S. Global Seropositivity of Swine Leptospirosis: Systematic Review and Meta-Analysis. Trop. Med. Infect. Dis. 2023, 8, 158. [Google Scholar] [CrossRef]
- Vincent, A.T.; Schiettekatte, O.; Goarant, C.; Neela, V.K.; Bernet, E.; Thibeaux, R.; Ismail, N.; Khalid, M.K.N.M.; Amran, F.; Masuzawa, T.; et al. Revisiting the taxonomy and evolution of pathogenicity of the genus Leptospira through the prism of genomics. PLoS Negl. Trop. Dis. 2019, 13, e0007270. [Google Scholar] [CrossRef]
- Thibeaux, R.; Girault, D.; Bierque, E.; Soupé-Gilbert, M.E.; Rettinger, A.; Douyère, A.; Meyer, M.; Iraola, G.; Picardeau, M.; Goarant, C. Biodiversity of environmental Leptospira: Improving identification and revisiting the diagnosis. Front. Microbiol. 2018, 9, 816. [Google Scholar] [CrossRef]
- Piredda, I.; Ponti, M.N.; Palmas, B.; Noworol, M.; Pedditzi, A.; Rebechesu, L.; Chisu, V. Molecular Typing of Pathogenic Leptospira Species Isolated from Wild Mammal Reservoirs in Sardinia. Animals 2021, 11, 1109. [Google Scholar] [CrossRef] [PubMed]
- Piredda, I.; Sechi, S.; Cocco, R.; Bertoldi, L.; Palmas, B.; Chisu, V. Isolation of Leptospira interrogans Serovar Canicola in a Vaccinated Dog without Clinical Symptoms. Pathogens 2022, 11, 406. [Google Scholar] [CrossRef] [PubMed]
- Piredda, I.; Bertoldi, L.; Benvenuto, G.; Palmas, B.; Pedditzi, A.; Pintore, P.; Chisu, V. First Isolation and Molecular Typing of Pathogenic and Intermediate Leptospira Species from Urine of Symptomatic Dogs. Vet. Sci. 2021, 8, 304. [Google Scholar] [CrossRef]
- Naudet, J.; Crespin, L.; Cappelle, J.; Kodjo, A.; Ayral, F. Circulating serogroups of Leptospira in swine from a 7-year study in France (2011–2017). Porc. Heal. Manag. 2022, 8, 15. [Google Scholar] [CrossRef]
- Miraglia, F.; Moreno, L.Z.; Morais, Z.M.; Langoni, H.; Shimabukuro, F.H.; Dellagostin, O.A.; Hartskeerl, R.; Vasconcellos, S.A.; Moreno, A.M. Characterization of Leptospira interrogans serovar Pomona isolated from swine in Brazil. J. Infect. Dev. Ctries. 2015, 9, 1054–1061. [Google Scholar] [CrossRef]
- Moreno, L.Z.; Kremer, F.S.; Jaeger, L.H.; Loureiro, A.P.; Miraglia, F.; Eslabao, M.R.; Dellagostin, O.A.; Lilenbaum, W.; Moreno, A.M. Genomic characterization and comparative analysis of Leptospira interrogans serogroup Australis isolated from swine. Pathog. Dis. 2017, 75, ftx119. [Google Scholar] [CrossRef]
- Sykes, J.E.; Haake, D.A.; Gamage, C.D.; Mills, W.Z.; Nally, J.E. A global one health perspective on leptospirosis in humans and animals. J. Am. Vet. Med. Assoc. 2022, 260, 1589–1596. [Google Scholar] [CrossRef]
- Piredda, I.; Ponti, M.N.; Piras, A.; Palmas, B.; Pintore, P.; Pedditzi, A.; Chisu, V. New Insights on Leptospira Infections in a Canine Population from North Sardinia, Italy: A Sero-Epidemiological Study. Biology 2021, 10, 507. [Google Scholar] [CrossRef]
- Stoddard, R.A. Detection of pathogenic Leptospira spp. through real-time PCR (qPCR) targeting the LipL32 gene. Methods Mol. Biol. 2013, 943, 257–266. [Google Scholar] [CrossRef]
- Barragan, V.; Chiriboga, J.; Miller, E.; Olivas, S.; Birdsell, D.; Hepp, C.; Hornstra, H.; Schupp, J.M.; Morales, M.; Gonzalez, M.; et al. High Leptospira Diversity in Animals and Humans Complicates the Search for Common Reservoirs of Human Disease in Rural Ecuador. PLoS Negl. Trop. Dis. 2016, 10, e0004990. [Google Scholar] [CrossRef]
- Bedir, O.; Kilic, A.; Atabek, E.; Kuskucu, A.M.; Turhan, V.; Basustaoglu, A.C. Simultaneous detection and differentiation of pathogenic and nonpathogenic Leptospira spp. by multiplex real-time PCR (TaqMan) assay. Polish J. Microbiol. 2010, 59, 167–173. [Google Scholar] [CrossRef]
- Zuerner, R.L. Laboratory maintenance of pathogenic Leptospira. Curr. Protoc. Microbiol. 2005, 12, 12E.1.1–12E.1.13. [Google Scholar] [CrossRef]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.G.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A Single Multilocus Sequence Typing (MLST) Scheme for Seven Pathogenic Leptospira Species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef]
- Scola, B.L.; Bui, L.T.M.; Baranton, G.; Khamis, A.; Raoult, D. Partial rpoB gene sequencing for identification of Leptospira species. FEMS Microbiol. Lett. 2006, 263, 142–147. [Google Scholar] [CrossRef]
- Piredda, I.; Bertoldi, L.; Chisu, V. Genome Sequence of a Leptospira montravelensis Strain Isolated from Donkey Urine during a Leptospirosis Outbreak in Sardinia Island, Italy. Microbiol. Resour. Announc. 2023, 12, e01009-22. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. Elife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Tiedje, J.M. Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 2567–2572. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Piredda, I.; Scarpa, F.; Sanna, D.; Casu, M.; Ponti, M.N.; Chisu, V. Draft Genome Sequences of Four Different Strains Belonging to Leptospira interrogans Serovar Pomona Isolated from Mammals in the Island of Sardinia, Italy. Microbiol. Resour. Announc. 2021, 10, e00698-21. [Google Scholar] [CrossRef]
- Piredda, I.; Palmas, B.; Noworol, M.; Tola, S.; Longheu, C.; Bertasio, C.; Scaltriti, E.; Denurra, D.; Cherchi, M.; Picardeau, M.; et al. Isolation of Leptospira interrogans from a Bottlenose Dolphin (Tursiops truncatus) in the Mediterranean Sea. J. Wildl. Dis. 2020, 56, 727–729. [Google Scholar] [CrossRef]
- Bertasio, C.; Papetti, A.; Scaltriti, E.; Tagliabue, S.; D’incau, M.; Boniotti, M.B. Serological survey and molecular typing reveal new Leptospira serogroup pomona strains among pigs of northern Italy. Pathogens 2020, 9, 332. [Google Scholar] [CrossRef]
- Arent, Z.; Frizzell, C.; Gilmore, C.; Allen, A.; Ellis, W.A. Leptospira interrogans serovars Bratislava and Muenchen animal infections: Implications for epidemiology and control. Vet. Microbiol. 2016, 190, 19–26. [Google Scholar] [CrossRef]
- Zamora, J.; Frías, S.R.y.M. Swine leptospirosis. First isolation in Chile of Leptospira interrogans serovar tarassovi. Zentralbl. Veterinarmed. B 1988, 35, 105–108. [Google Scholar] [CrossRef]
- Mazzotta, E.; Bellinati, L.; Bertasio, C.; Boniotti, M.B.; Lucchese, L.; Ceglie, L.; Martignago, F.; Leopardi, S.; Natale, A. Synanthropic and Wild Animals as Sentinels of Zoonotic Agents: A Study of Leptospira Genotypes Circulating in Northeastern Italy. Int. J. Environ. Res. Public Health 2023, 20, 3783. [Google Scholar] [CrossRef]
- Ellis, W.A. Animal leptospirosis. Curr. Top. Microbiol. Immunol. 2015, 387, 99–137. [Google Scholar] [CrossRef]
- Strutzberg-Minder, K.; Tschentscher, A.; Beyerbach, M.; Homuth, M.; Kreienbrock, L. Passive surveillance of Leptospira infection in swine in Germany. Porc. Health Manag. 2018, 4, 10. [Google Scholar] [CrossRef]
- Ellis, W.A.; McParland, P.J.; Bryson, D.G.; Thiermann, A.B.; Montgomery, J. Isolation of leptospires from the genital tract and kidneys of aborted sows. Vet. Rec. 1986, 118, 294–295. [Google Scholar] [CrossRef]
- Ellis, W.A.; McParland, P.J.; Bryson, D.G.; Cassells, J.A. Boars as carriers of leptospires of the Australis serogroup on farms with an abortion problem. Vet. Rec. 1986, 118, 563. [Google Scholar] [CrossRef]
- Vitale, M.; Agnello, S.; Chetta, M.; Amato, B.; Vitale, G.; Bella, C.D.; Vicari, D.; Presti, V.D.M.L. Human leptospirosis cases in Palermo Italy. The role of rodents and climate. J. Infect. Public Health 2018, 11, 209–214. [Google Scholar] [CrossRef]
- Myers, D.M. Serological studies and isolations of serotype hardjo and Leptospira biflexa strains from horses of Argentina. J. Clin. Microbiol. 1976, 3, 548–555. [Google Scholar] [CrossRef]
- Pui, C.F.; Bilung, L.M.; Apun, K.; Su’ut, L. Diversity of Leptospira spp. in Rats and Environment from Urban Areas of Sarawak, Malaysia. J. Trop. Med. 2017, 2017, 3760674. [Google Scholar] [CrossRef]
- Dhayabaran, V.; Chidambaram, D.; Krishnaswamy, P.R. Identification of compounds for improved growth of Leptospira in culture and isolation. Diagn. Microbiol. Infect. Dis. 2020, 96, 114923. [Google Scholar] [CrossRef]
- Ahmed, N.; Manjulata Devi, S.; de los Á Valverde, M.; Vijayachari, P.; Machang’u, R.S.; Ellis, W.A.; Hartskeerl, R.A. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Ann. Clin. Microbiol. Antimicrob. 2006, 5, 28. [Google Scholar] [CrossRef]
- Benacer, D.; Who, P.Y.; Zain, S.N.M.; Amran, F.; Thong, K.L. Pathogenic and saprophytic Leptospira species in water and soils from selected urban sites in peninsular Malaysia. Microbes Environ. 2013, 28, 135–140. [Google Scholar] [CrossRef]
- Meganathan, Y.; Vishwakarma, A.; Ramya, M. Biofilm formation and social interaction of Leptospira in natural and artificial environments. Res. Microbiol. 2022, 173, 103981. [Google Scholar] [CrossRef]
- Ristow, P.; Bourhy, P.; Kerneis, S.; Schmitt, C.; Prevost, M.C.; Lilenbaum, W.; Picardeau, M. Biofilm formation by saprophytic and pathogenic leptospires. Microbiology 2008, 154, 1309–1317. [Google Scholar] [CrossRef] [PubMed]
- Santecchia, I.; Bonhomme, D.; Papadopoulos, S.; Escoll, P.; Giraud-Gatineau, A.; Moya-Nilges, M.; Vernel-Pauillac, F.; Boneca, I.G.; Werts, C. Alive Pathogenic and Saprophytic Leptospires Enter and Exit Human and Mouse Macrophages With No Intracellular Replication. Front. Cell. Infect. Microbiol. 2022, 12, 936931. [Google Scholar] [CrossRef]
- Santos, A.A.N.; dos Santos Ribeiro, P.; da França, G.V.; Souza, F.N.; Ramos, E.A.G.; Figueira, C.P.; Reis, M.G.; Costa, F.; Ristow, P. Leptospira interrogans biofilm formation in Rattus norvegicus (Norway rats) natural reservoirs. PLoS Negl. Trop. Dis. 2021, 15, e0009736. [Google Scholar] [CrossRef]
- Cataldo, C.; Bellenghi, M.; Masella, R.; Busani, L. One Health challenges and actions: Integration of gender considerations to reduce risks at the human-animal-environmental interface. One Health 2023, 16, 100530. [Google Scholar] [CrossRef] [PubMed]
| Species | Sex | Titer Found |
|---|---|---|
| Pig 1 | female | 1:1600 |
| Pig 2 | female | 1:1600 |
| Pig 3 | female | 1:3200 |
| Pig 4 | female | 1:3200 |
| Pig 5 | female | 1:1600 |
| Pig 6 | female | 1:800 |
| Pig 7 | female | 1:3200 |
| Pig 8 | female | 1:1600 |
| Pig 9 | female | 1:1600 |
| Pig 10 | male | 1:3200 |
| Pig 11 | male | 1:1600 |
| Donkey 1 | male | 1:800 |
| Donkey 2 | female | 1:1600 |
| Donkey 3 | female | 1:800 |
| Samples | Matrix | qPCR1 (Ct Value) | qPCR2 (Ct Value) | Culture |
|---|---|---|---|---|
| Pig 1 | kidney | Pos (35.8) | Neg | Neg |
| Pig 2 | kidney | Pos (38.5) | Neg | Neg |
| Pig 3 | kidney | Pos (24.0) | Neg | Pos |
| Pig 4 | kidney | Pos (25.5) | Neg | Pos |
| Pig 5 | kidney | Pos (32.2) | Neg | Pos |
| urine | Pos (23.8) | Pos (15.1) | Pos | |
| Pig 6 | kidney | Pos (33.4) | Neg | Neg |
| Pig 7 | kidney | Neg | Neg | Neg |
| Pig 8 | kidney | Pos (22.7) | Neg | Pos |
| Pig 9 | kidney | Neg | Neg | Neg |
| urine | Pos (36.0) | Pos (26.8) | Neg | |
| Pig 10 | kidney | Neg | Neg | Neg |
| urine | Pos (35.8) | Pos (21.5) | Neg | |
| Pig 11 | kidney | Pos (35.0) | Neg | Neg |
| Donkey 1 | urine | Pos (37.4) | Neg | Neg |
| Donkey 2 | urine | Pos (29.0) | Neg | Neg |
| Donkey 3 | urine | Pos (29.2) | Neg | Pos |
| Samples | Matrix | rrs Gene | rpoB Gene | secY Gene | MLST |
|---|---|---|---|---|---|
| Pig 3 | kidney | L. interrogans | L. interrogans | L. interrogans | ST = 140 |
| Pig 4 | kidney | L. interrogans | L. interrogans | L. interrogans | ST = 140 |
| Pig 5 | kidney | L. interrogans | L. interrogans | L. interrogans | ST = 140 |
| urine | L. interrogans | L. biflexa | L. biflexa | Not determined | |
| Pig 8 | kidney | L. interrogans | L. interrogans | L. interrogans | ST = 140 |
| Donkey 3 | urine | L. interrogans | L. biflexa | L. biflexa | Not determined |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piredda, I.; Bertoldi, L.; Pedditzi, A.; Pintore, P.; Palmas, B.; Chisu, V. Co-Infection by Leptospira montravelensis and Leptospira interrogans Serovar Pomona in Urine Samples of Donkeys and Pigs in Sardinia, Italy. Animals 2023, 13, 1803. https://doi.org/10.3390/ani13111803
Piredda I, Bertoldi L, Pedditzi A, Pintore P, Palmas B, Chisu V. Co-Infection by Leptospira montravelensis and Leptospira interrogans Serovar Pomona in Urine Samples of Donkeys and Pigs in Sardinia, Italy. Animals. 2023; 13(11):1803. https://doi.org/10.3390/ani13111803
Chicago/Turabian StylePiredda, Ivana, Loris Bertoldi, Aureliana Pedditzi, Pierangela Pintore, Bruna Palmas, and Valentina Chisu. 2023. "Co-Infection by Leptospira montravelensis and Leptospira interrogans Serovar Pomona in Urine Samples of Donkeys and Pigs in Sardinia, Italy" Animals 13, no. 11: 1803. https://doi.org/10.3390/ani13111803
APA StylePiredda, I., Bertoldi, L., Pedditzi, A., Pintore, P., Palmas, B., & Chisu, V. (2023). Co-Infection by Leptospira montravelensis and Leptospira interrogans Serovar Pomona in Urine Samples of Donkeys and Pigs in Sardinia, Italy. Animals, 13(11), 1803. https://doi.org/10.3390/ani13111803






