Mitochondrial DNA and Distribution Modelling Evidenced the Lost Genetic Diversity and Wild-Residence of Star Tortoise, Geochelone elegans (Testudines: Testudinidae) in India
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
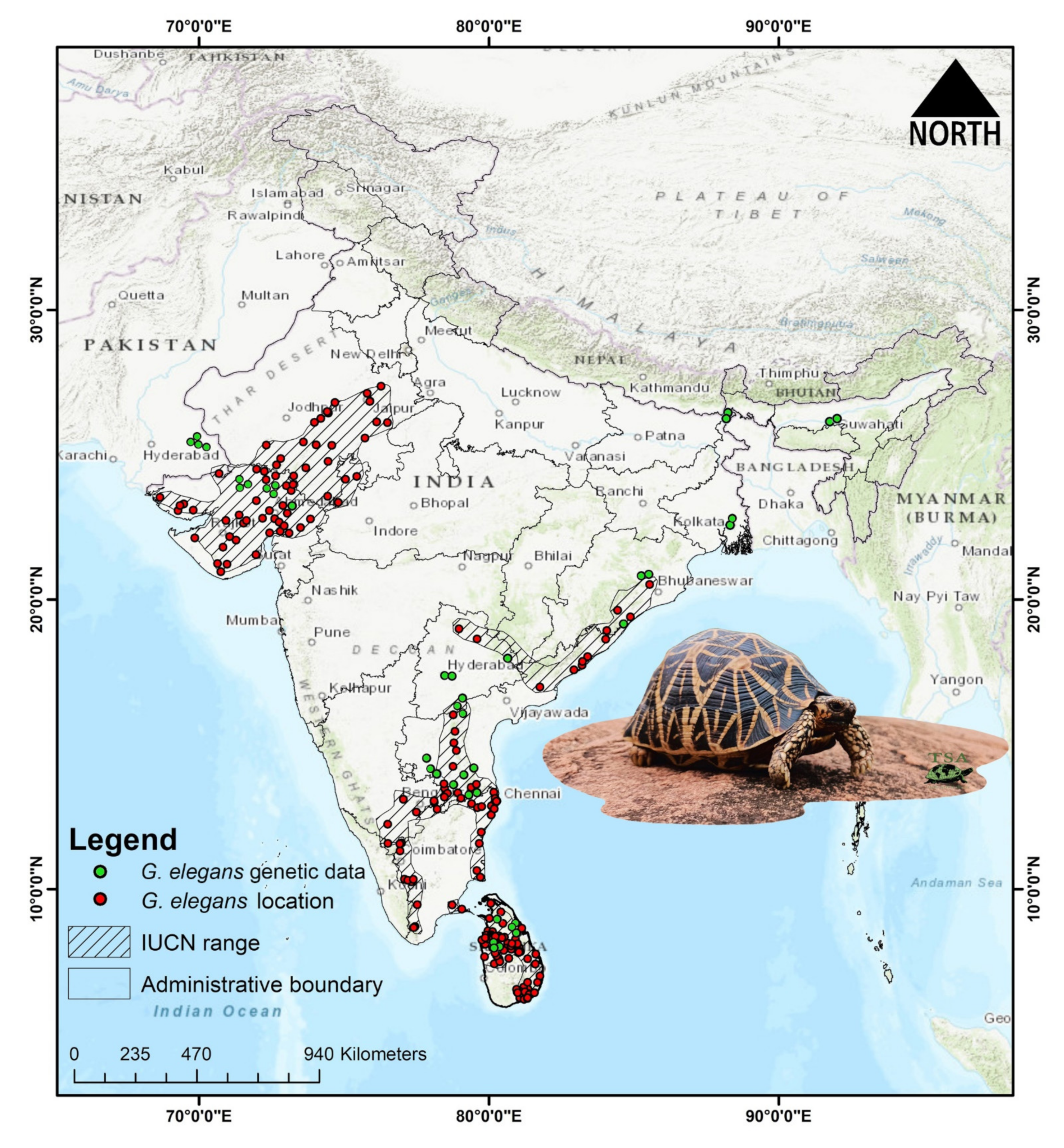
2.1. Sampling and Ethics Statement
2.2. Molecular Experiments
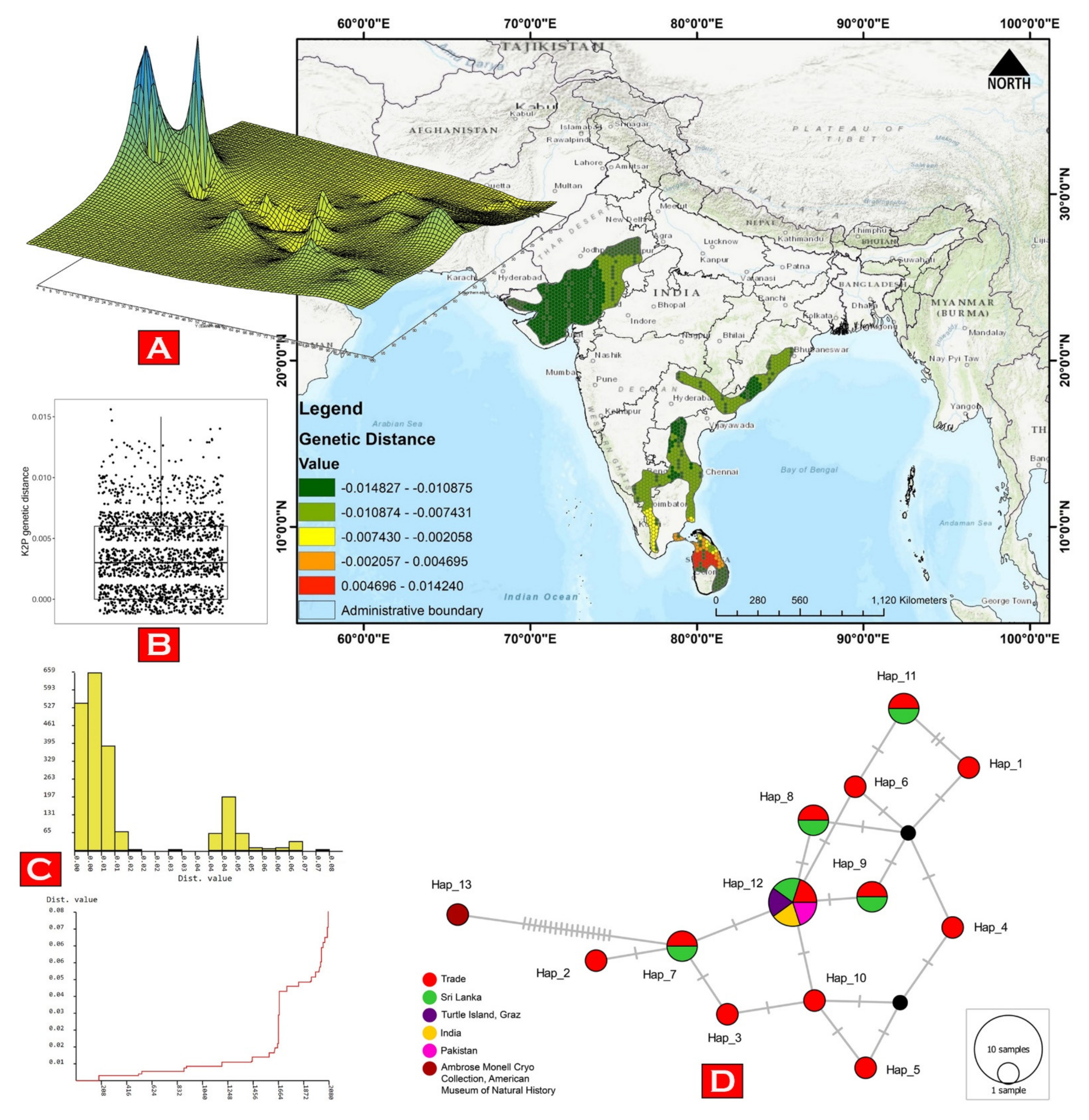
2.3. Genetic Analyses
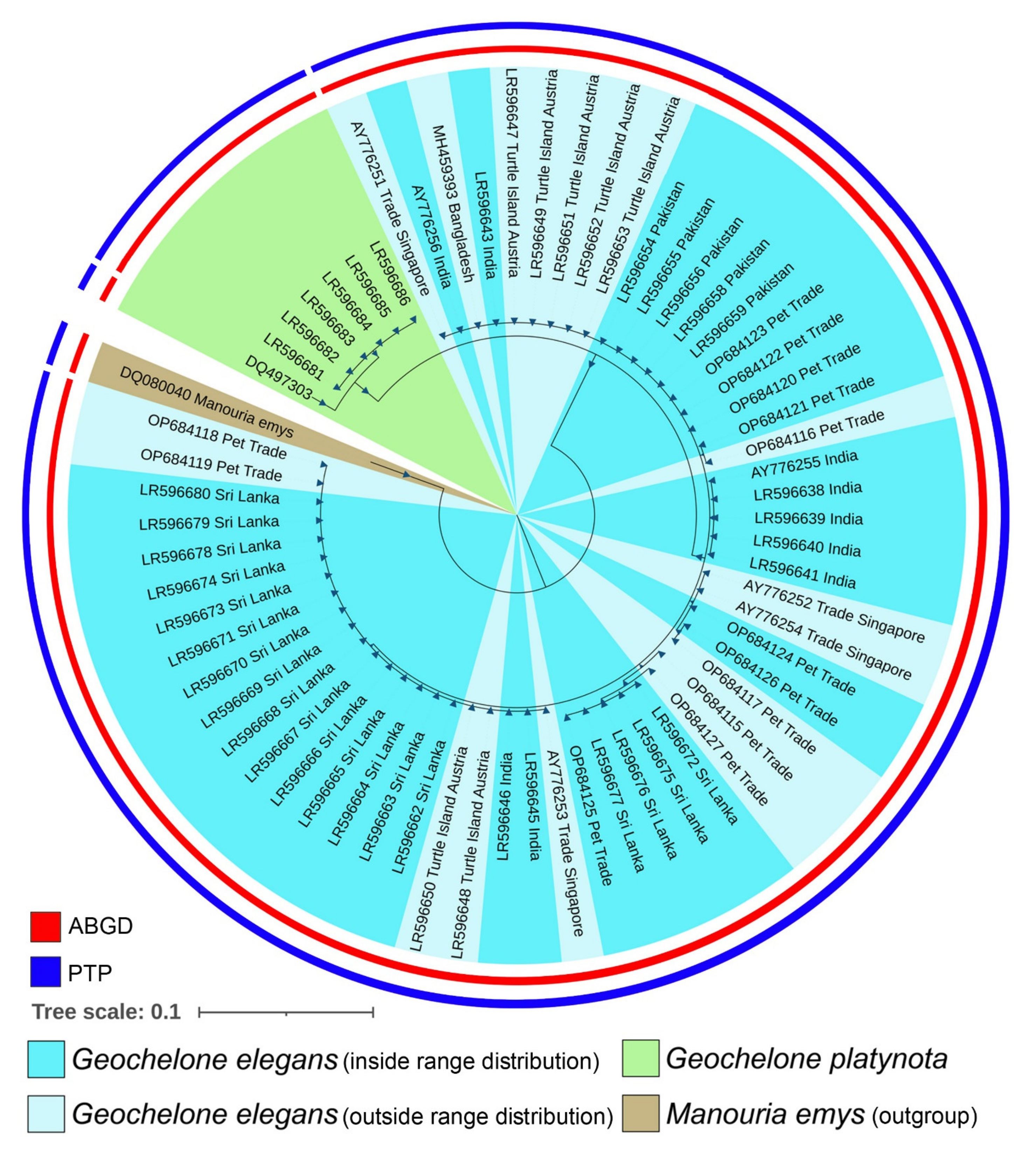
2.4. Phylogenetic Inference
2.5. Species Occurrence Data
2.6. Model Covariate Selection
2.7. Model Building and Evaluation
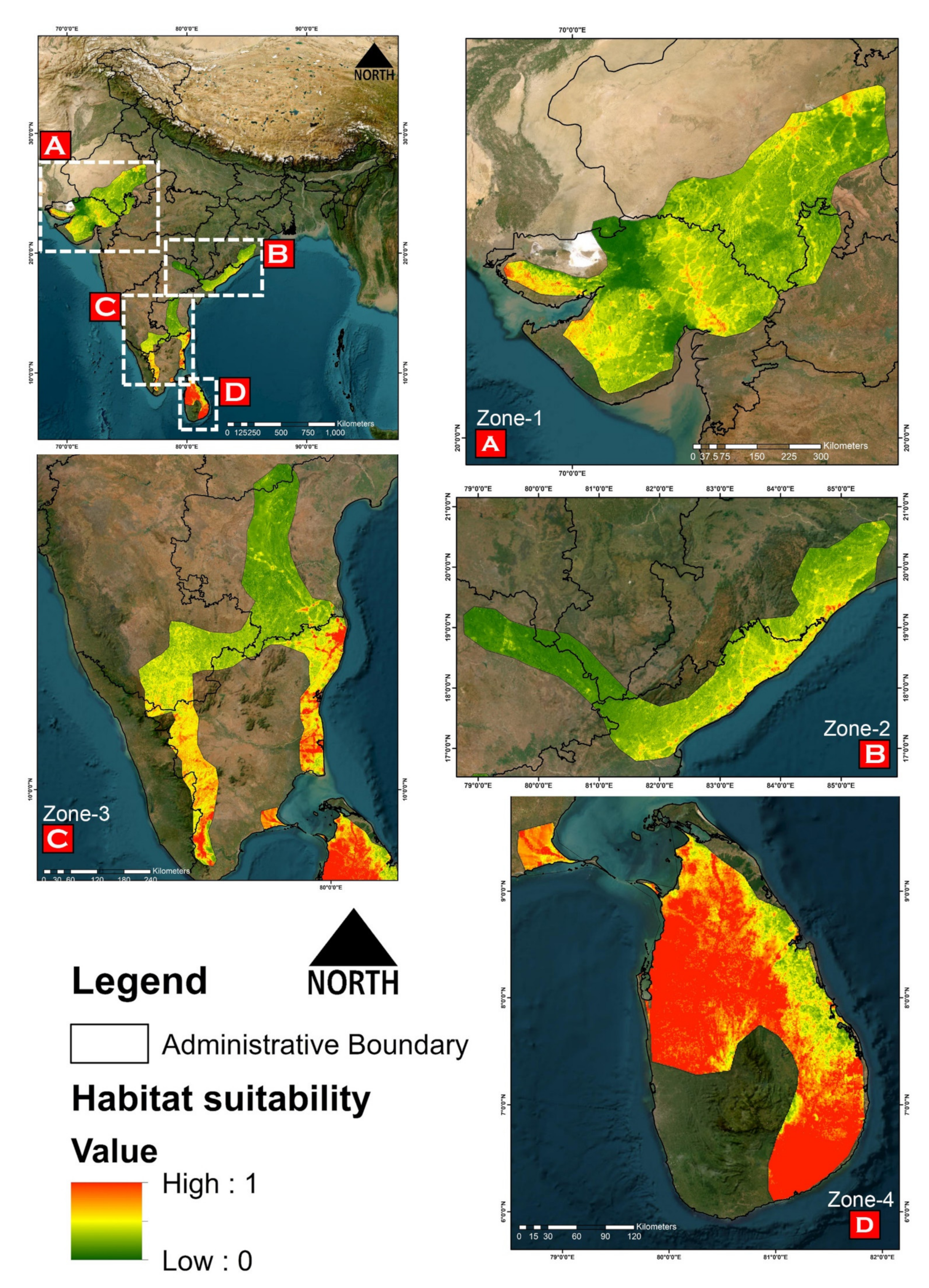
2.8. Habitat Quality Assessment
3. Results
3.1. Molecular Characterization
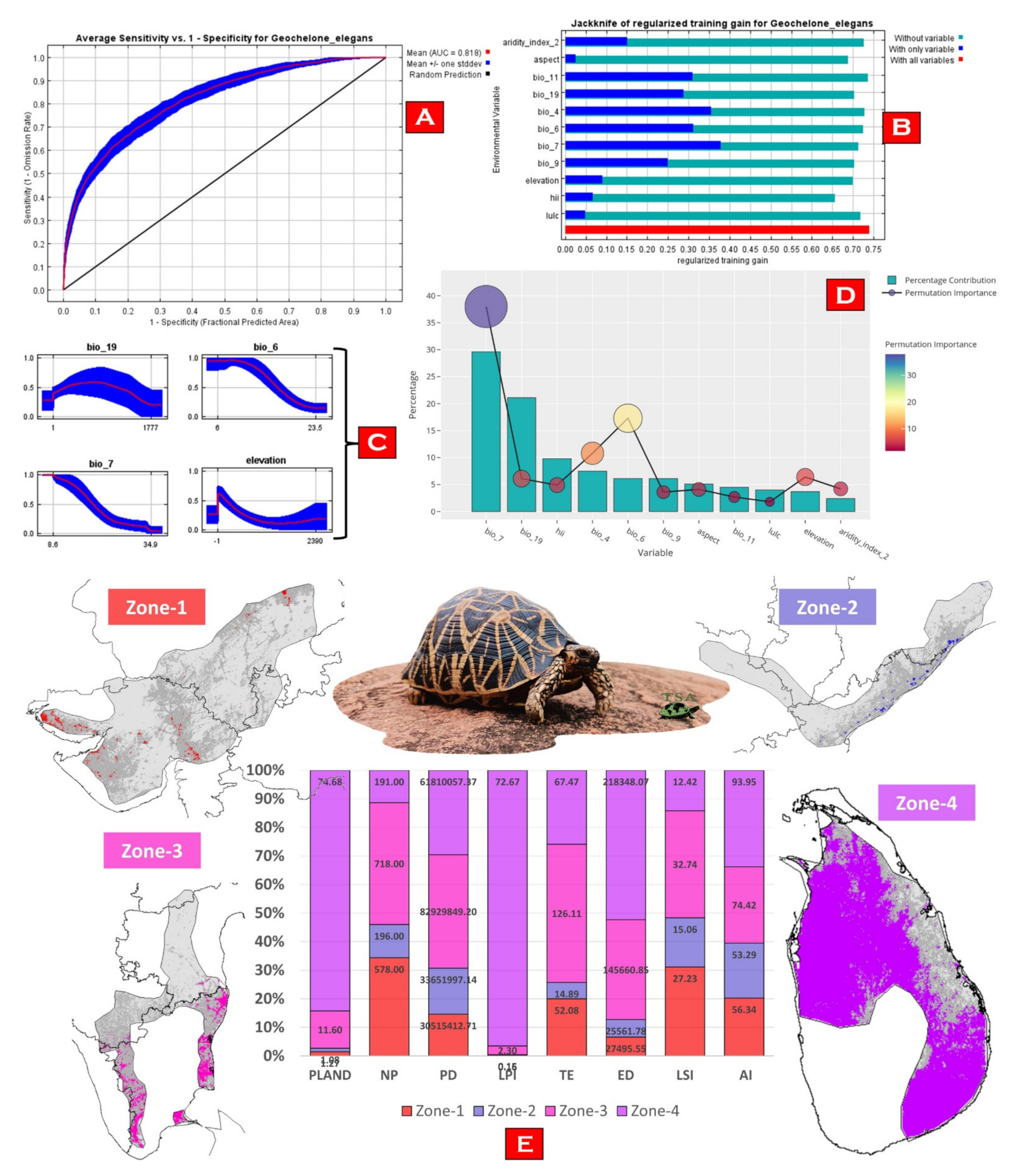
3.2. Model Performance and Habitat Suitability
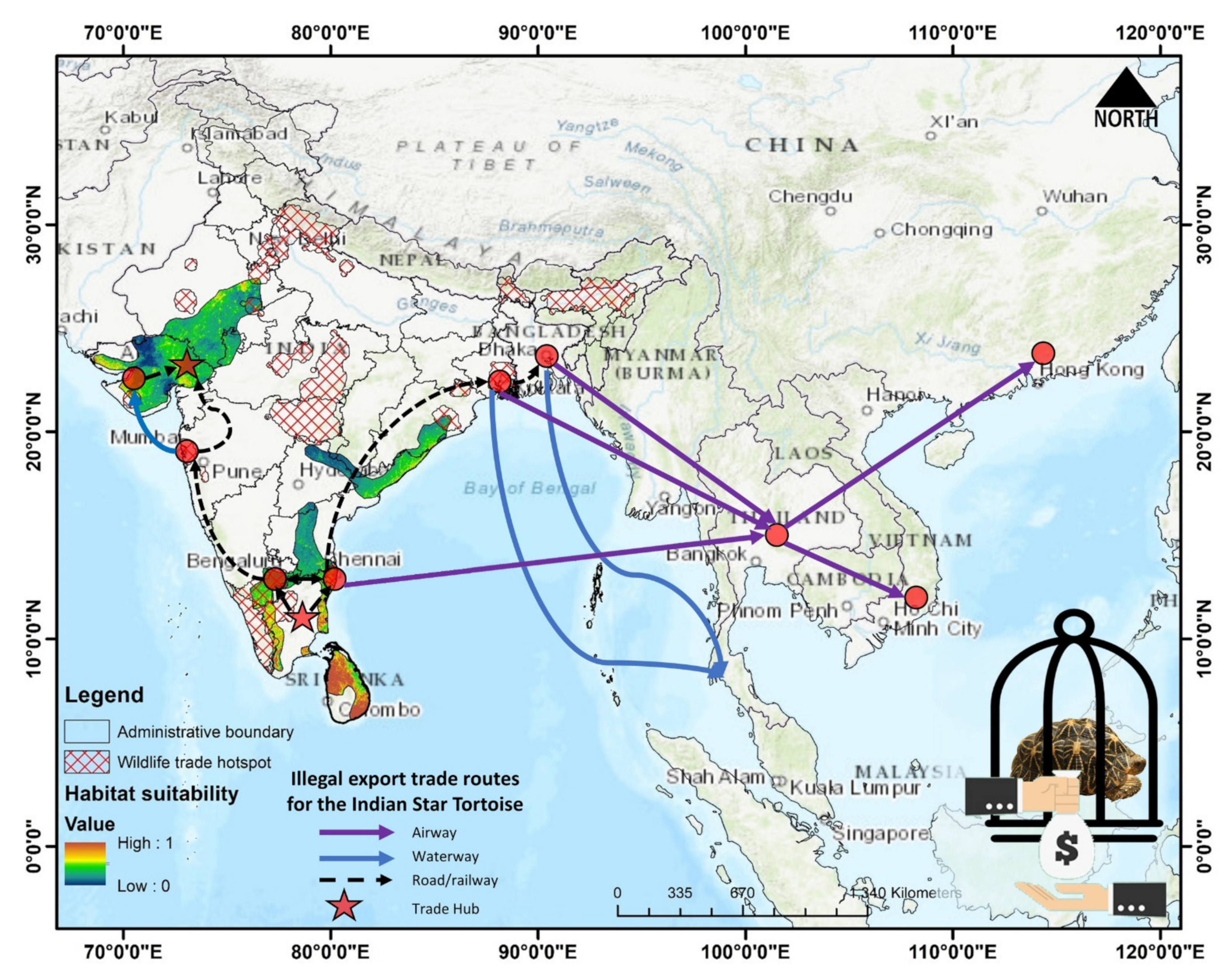
3.3. Habitat Quality Estimation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- De Silva, A.; Bauer, A. Origin of the name Geochelone elegans. Wildlanka 2016, 4, 22–28. [Google Scholar]
- D’Cruze, N.; Mookerjee, A.; Vyas, R.; Macdonald, D.W.; de Silva, A. Geochelone elegans (Schoepff 1795)-Indian Star Tortoise, Star Tortoise. In Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group; Rhodin, A.G.J., Iverson, J.B., van Dijk, P.P., Stanford, C.B., Goode, E.V., Buhlmann, K.A., Pritchard, P.C.H., Mittermeier, R.A., Eds.; Chelonian Research Foundation: Arlington, VT, USA, 2018; Volume 5, p. 106.1–106.13. [Google Scholar]
- D’Cruze, N.; Singh, B.; Morrison, T.; Schmidt-Burbach, J.; Macdonald, D.W.; Mookerjee, A. A star attraction: The illegal trade in Indian Star Tortoises. Nat. Conserv. 2015, 13, 1–19. [Google Scholar] [CrossRef]
- Mendiratta, U.; Sheel, V.; Singh, S. Enforcement seizures reveal large-scale illegal trade in India’s tortoises and freshwater turtles. Biol. Conserv. 2017, 207, 100–105. [Google Scholar] [CrossRef]
- Choudhury, B.C.; de Silva, A.; Shepherd, C. Geochelone elegans—In The IUCN Red List of Threatened Species 2020: e.T39430A123815345. Available online: https://dx.doi.org/10.2305/IUCN.UK.2020-2.RLTS.T39430A123815345.en (accessed on 25 November 2022).
- De Silva, A. The Biology and Status of the Star Tortoise in Sri Lanka; Ministry of Environment and Natural Resources, AMP Print Shop: Gampola, Sri Lanka, 2003; 100p.
- Fyfe, J.D. Star Tortoises: The Natural History, Captive Care and Breeding; Living Art Publishing: Ada, OK, USA, 2007. [Google Scholar]
- Gaur, A.; Reddy, A.; Annapoorni, S.; Satyarebala, B.; Shivaji, S. The origin of Indian Star tortoises (Geochelone elegans) based on nuclear and mitochondrial DNA analysis: A story of rescue and repatriation. Conserv. Genet. 2006, 7, 231–240. [Google Scholar] [CrossRef]
- Vamberger, M.; Spitzweg, C.; de Silva, A.; Masroor, R.; Praschag, P.; Fritz, U. Already too late? Massive trade in Indian star tortoises (Geochelone elegans) might have wiped out its phylogeographic differentiation. Amphib.-Reptil. 2020, 41, 133–138. [Google Scholar] [CrossRef]
- Gray, T.N.; Marx, N.; Khem, V.; Lague, D.; Nijman, V.; Gauntlett, S. Holistic management of live animals confiscated from illegal wildlife trade. J. Appl. Ecol. 2017, 54, 726–730. [Google Scholar] [CrossRef]
- Robinson, J.E.; Sinovas, P. Challenges of analyzing the global trade in CITES-listed wildlife. Conserv. Biol. 2018, 32, 1203–1206. [Google Scholar] [CrossRef]
- International Union for Conservation of Nature (IUCN). Guidelines for the Management of Confiscated, Live Organisms; IUCN: Gland, Switzerland, 2019. [Google Scholar]
- International Union for Conservation of Nature (IUCN). IUCN Guidelines for the Placement of Confiscated Animals. In Proceedings of the 51st Meeting of The IUCN Council, Gland, Switzerland, 8–11 February 2010. [Google Scholar]
- TRAFFIC. Wildlife Trade in Southeast Asia. CoP 13 Briefing TRAFFIC International. 2004. Available online: http://www.trafficj.org/cop13/pdf/cop13briefing_SoutheastAsia.pdf (accessed on 25 November 2022).
- Le, M.D.; McCormack, T.E.M.; Hoang, H.V.; Duong, H.T.; Nguyen, T.Q.; Ziegler, T.; Nguyen, H.D.; Ngo, H.T. Threats from wildlife trade: The importance of genetic data in safeguarding the endangered Four-eyed Turtle (Sacalia quadriocellata). Nat. Conserv. 2020, 41, 91–111. [Google Scholar] [CrossRef]
- Mothes, C.C.; Howell, H.J.; Searcy, C.A. Habitat suitability models for the imperiled wood turtle (Glyptemys insculpta) raise concerns for the species’ persistence under future climate change. Glob. Ecol. Conserv. 2020, 24, e01247. [Google Scholar] [CrossRef]
- Willey, L.L.; Jones, M.T.; Sievert, P.R.; Akre, T.S.B.; Marchand, M.; deMaynadier, P.; Yorks, D.; Mays, J.; Dragon, J.; Erb, L.; et al. Distribution models combined with standardized surveys reveal widespread habitat loss in a threatened turtle species. Biol. Conserv. 2022, 266, 109437. [Google Scholar] [CrossRef]
- Escoriza, D.; Ben Hassine, J. Niche diversification of Mediterranean and southwestern Asian tortoises. PeerJ 2022, 10, e13702. [Google Scholar] [CrossRef]
- Gibbs, J.P.; Hunter, E.A.; Shoemaker, K.T.; Tapia, W.H. Frtoise reintroduction to Española Island, Galapagos. PLoS ONE 2014, 9, e110742. [Google Scholar] [CrossRef]
- Munro, N.T.; McIntyre, S.; Macdonald, B.; Cunningham, S.A.; Gordon, I.J.; Cunningham, R.B.; Manning, A.D. Returning a lost process by reintroducing a locally extinct digging marsupial. PeerJ 2019, 7, e6622. [Google Scholar] [CrossRef]
- Percie du Sert, N.; Hurst, V.; Ahluwalia, A.; Alam, S.; Avey, M.T.; Baker, M.; Browne, W.J.; Clark, A.; Cuthill, I.C.; Dirnagl, U.; et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020, 18, e3000410. [Google Scholar]
- Verma, S.K.; Singh, L. Novel universal primers establish identity of an enormous number of animal species for forensic application. Mol. Ecol. Notes. 2002, 3, 28–31. [Google Scholar] [CrossRef]
- Miller, M.P. Alleles In Space (AIS): Computer software for the joint analysis of individual spatial and genetic information. J. Hered. 2005, 96, 722–724. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Puillandre, N.; Lambert, A.; Brouillet, S.; Achaz, G. ABGD, Automatic Barcode Gap Discovery for primary species delimitation. Mol. Ecol. 2012, 21, 1864–1877. [Google Scholar] [CrossRef]
- Zhang, J.; Kapli, P.; Pavlidis, P.; Stamatakis, A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics 2013, 29, 2869–2876. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Clement, M.; Snell, Q.; Walker, P.; Posada, D.; Crandall, K. TCS: A computer program to estimate gene genealogies. Mol. Ecol. 2000, 9, 1657–1659. [Google Scholar] [CrossRef] [PubMed]
- Leigh, J.W.; Bryant, D. Popart: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Nylander, J.A.A. MrModeltest v2, Program Distributed by the Author; Evolutionary Biology Centre, Uppsala University: Uppsala, Sweden, 2004. [Google Scholar]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Brown, J.L. SDM toolbox: A python-based GIS toolkit for landscape genetic, biogeographic and species distribution model analyses. Methods Ecol. Evol. 2014, 5, 694–700. [Google Scholar] [CrossRef]
- Peterson, A.T.; Soberón, J.; Pearson, R.G.; Anderson, R.P.; Martínez-Meyer, E.; Nakamura, M.; Araújo, M.B. Ecological Niches and Geographic Distributions; Princeton University Press: Princeton, NJ, USA, 2011; 328p. [Google Scholar]
- Fick, S.E.; Hijmans, R.J. Worldclim 2: New 1-km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 2017, 37, 4302–4315. [Google Scholar] [CrossRef]
- Zomer, R.J.; Xu, J.; Trabucco, A. Version 3 of the global aridity index and potential evapotranspiration database. Sci. Data. 2022, 9, 409. [Google Scholar] [CrossRef]
- Socioeconomic Data and Applications Center (SEDAD); Wildlife Conservation Society (WCS); Columbia University Center for International Earth Science Information Network (CIESIN). Last of the Wild Project, Version 2, 2005 (LWP-2): Global Human Footprint Dataset (Geographic); NASA Socioeconomic Data and Applications Center (SEDAC): Palisades, NY, USA, 2005. [CrossRef]
- Peacock, H. Assessment of the Protected Areas Network in Madagascar for Lemur Conservation; University of Calgary: Calgary, AB, Canada, 2011. [Google Scholar]
- Erinjery, J.J.; Singh, M.; Kent, R. Diet-dependent habitat shifts at different life stages of two sympatric primate species. J. Biosci. 2019, 44, 43. [Google Scholar] [CrossRef]
- Phillips, S.J.; Anderson, R.P.; Dudík, M.; Schapire, R.E.; Blair, M.E. Opening the black box: An open-source release of Maxent. Ecography 2017, 40, 887–893. [Google Scholar] [CrossRef]
- Elith, J.; Phillips, S.J.; Hastie, T.; Dudık, M.; Chee, Y.E.; Yates, C.J. A statistical explanation of MaxEnt for ecologists. Divers. Distrib 2011, 17, 43–57. [Google Scholar] [CrossRef]
- Phillips, S.J.; Dudık, M. Modeling of species distributions with MaxEnt: New extensions and a comprehensive evaluation. Ecography 2008, 31, 161–175. [Google Scholar] [CrossRef]
- Halvorsen, R.; Mazzoni, S.; Dirksen, J.W.; Næsset, E.; Gobakken, T.; Ohlson, M. How important are choice of model selection method and spatial autocorrelation of presence data for distribution modelling by MaxEnt? Ecol. Modell. 2016, 328, 108–118. [Google Scholar] [CrossRef]
- Kamilar, J.M.; Tecot, S.R. Anthropogenic and climatic effects on the distribution of eulemur species: An ecological niche modelling approach. Int. J. Primatol. 2016, 37, 47–68. [Google Scholar] [CrossRef]
- Johnson, S.E.; Delmore, K.E.; Brown, K.A.; Wyman, T.M.; Louis, E.E. Niche divergence in a brown lemur (Eulemur spp.) hybrid zone: Using ecological niche models to test models of stability. Int. J. Primatol. 2016, 37, 69–88. [Google Scholar] [CrossRef]
- McGarigal, K. FRAGSTATS Help; University of Massachusetts: Amherst, MA, USA, 2015. [Google Scholar]
- Mukherjee, T.; Sharma, L.K.; Saha, G.K.; Thakur, M.; Chandra, K. Past, present and future: Combining habitat suitability and future landcover simulation for long-term conservation management of Indian rhino. Sci. Rep. 2020, 10, 606. [Google Scholar] [CrossRef]
- Mukherjee, T.; Sharma, V.; Sharma, L.K.; Thakur, M.; Joshi, B.D.; Sharief, A.; Thapa, A.; Dutta, R.; Dolker, S.; Tripathy, B.; et al. Landscape-level habitat management plan through geometric reserve design for critically endangered Hangul (Cervus hanglu hanglu). Sci. Total Environ. 2021, 777, 146031. [Google Scholar] [CrossRef]
- Butchart, S.H.; Walpole, M.; Collen, B.; van Strien, A.; Scharlemann, J.P.; Almond, R.E.; Baillie, J.E.; Bomhard, B.; Brown, C.; Bruno, J.; et al. Global biodiversity: Indicators of recent declines. Science 2010, 328, 1164–1168. [Google Scholar] [CrossRef]
- IPBES. Global Assessment Report on Biodiversity and Ecosystem Services; Brondizio, E.S., Ed.; Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services: Bonn, Germany, 2019. [Google Scholar]
- Tilman, D.; Clark, M.; Williams, D.R.; Kimmel, K.; Polasky, S.; Packer, C. Future threats to biodiversity and pathways to their prevention. Nature 2017, 546, 73–81. [Google Scholar] [CrossRef]
- Conde, D.A.; Staerk, J.; Colchero, F.; da Silva, R.; Schöley, J.; Baden, H.M.; Jouvet, L.; Fa, J.E.; Syed, H.; Jongejans, E.; et al. Data gaps and opportunities for comparative and conservation biology. Proc. Natl. Acad. Sci. USA 2019, 116, 9658–9664. [Google Scholar] [CrossRef] [PubMed]
- Cox, N.; Young, B.E.; Bowles, P.; Fernandez, M.; Marin, J.; Rapacciuolo, G.; Böhm, M.; Brooks, T.M.; Hedges, S.B.; Hilton-Taylor, C.; et al. A global reptile assessment highlights shared conservation needs of tetrapods. Nature 2022, 605, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Roll, U.; Feldman, A.; Novosolov, M.; Allison, A.; Bauer, A.M.; Bernard, R.; Böhm, M.; Castro-Herrera, F.; Chirio, L.; Collen, B.; et al. The global distribution of tetrapods reveals a need for targeted reptile conservation. Nat. Ecol. Evol. 2017, 1, 1677–1682. [Google Scholar] [CrossRef] [PubMed]
- Stanford, C.B.; Iverson, J.B.; Rhodin, A.G.J.; van Dijk, P.P.; Mittermeier, R.A.; Kuchling, G.; Berry, K.H.; Bertolero, A.; Bjorndal, K.A.; Blanck, T.E.G.; et al. Turtles and Tortoises Are in Trouble. Curr. Biol. 2020, 30, R721–R735. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.E.; St John, F.A.V.; Griffiths, R.A.; Roberts, D.L. Captive reptile mortality rates in the home and implications for the wildlife trade. PLoS ONE 2016, 11, e0157519. [Google Scholar] [CrossRef]
- Teixeira, J.C.; Huber, C.D. The inflated significance of neutral genetic diversity in conservation genetics. Proc. Natl. Acad. Sci. USA 2021, 118, e2015096118. [Google Scholar] [CrossRef]
- Poulakakis, N.; Edwards, D.L.; Chiari, Y.; Garrick, R.C.; Russello, M.A.; Benavides, E.; Watkins-Colwell, G.J.; Glaberman, S.; Tapia, W.; Gibbs, J.P.; et al. Description of a New Galapagos Giant Tortoise Species (Chelonoidis; Testudines: Testudinidae) from Cerro Fatal on Santa Cruz Island. PLoS ONE 2015, 10, e0138779. [Google Scholar] [CrossRef]
- Russello, M.A.; Glaberman, S.; Gibbs, J.P.; Marquez, C.; Powell, J.R.; Caccone, A. A cryptic taxon of Galápagos tortoise in conservation peril. Biol. Lett. 2005, 1, 287–290. [Google Scholar] [CrossRef]
- Caccone, A.; Gentile, G.; Gibbs, J.P.; Frirts, T.H.; Snell, H.L.; Betts, J.; Powell, J.R. Phylogeography and history of giant Galápagos tortoises. Evolution 2002, 56, 2052–2066. [Google Scholar]
- Van der Kuyl, A.C.; Ballasina, D.L.; Zorgdrager, F. Mitochondrial haplotype diversity in the tortoise species Testudo graeca from North Africa and the Middle East. BMC Evol. Biol. 2005, 5, 29. [Google Scholar] [CrossRef]
- Loire, E.; Chiari, Y.; Bernard, A.; Cahais, V.; Romiguier, J.; Nabholz, B.; Lourenço, J.M.; Galtier, N. Population genomics of the endangered giant Galápagos tortoise. Genome Biol. 2013, 14, R136. [Google Scholar] [CrossRef]
- Poulakakis, N.; Russello, M.; Geist, D.; Caccone, A. Unravelling the peculiarities of island life: Vicariance, dispersal and the diversification of the extinct and extant giant Galápagos tortoises. Mol. Ecol. 2012, 21, 160–173. [Google Scholar] [CrossRef]
- Caccone, A.; Amato, G.; Gratry, O.C.; Behler, J.; Powell, J.R. A molecular phylogeny of four endangered Madagascar tortoises based on MtDNA sequences. Mol. Phylogenet. Evol. 1999, 12, 1–9. [Google Scholar] [CrossRef]
- Van der Kuyl, A.C.; Ph Ballasina, D.L.; Dekker, J.T.; Maas, J.; Willemsen, R.E.; Goudsmit, J. Phylogenetic relationships among the species of the genus Testudo (Testudines: Testudinidae) inferred from mitochondrial 12S rRNA gene sequences. Mol. Phylogenet. Evol. 2002, 22, 174–183. [Google Scholar] [CrossRef]
- Fritz, U.; Bininda-Emonds, O.R. When genes meet nomenclature: Tortoise phylogeny and the shifting generic concepts of Testudo and Geochelone. Zoology 2007, 110, 298–307. [Google Scholar] [CrossRef]
- Parham, J.F.; Macey, J.R.; Papenfuss, T.J.; Feldman, C.R.; Türkozan, O.; Polymeni, R.; Boore, J. The phylogeny of Mediterranean tortoises and their close relatives based on complete mitochondrial genome sequences from museum specimens. Mol. Phylogenet. Evol. 2006, 38, 50–64. [Google Scholar] [CrossRef]
- Zhang, X.; Unmack, P.J.; Kuchling, G.; Wang, Y.; Georges, A. Resolution of the enigmatic phylogenetic relationship of the critically endangered Western Swamp Tortoise Pseudemydura umbrina (Pleurodira: Chelidae) using a complete mitochondrial genome. Mol. Phylogenet. Evol. 2017, 115, 58–61. [Google Scholar] [CrossRef]
- Quesada, V.; Freitas-Rodríguez, S.; Miller, J.; Pérez-Silva, J.G.; Jiang, Z.F.; Tapia, W.; Santiago-Fernández, O.; Campos-Iglesias, D.; Kuderna, L.F.K.; Quinzin, M.; et al. Giant tortoise genomes provide insights into longevity and age-related disease. Nat. Ecol. Evol. 2019, 3, 87–95. [Google Scholar] [CrossRef]
- Poulakakis, N.; Glaberman, S.; Russello, M.; Beheregaray, L.B.; Ciofi, C.; Powell, J.R.; Caccone, A. Historical DNA analysis reveals living descendants of an extinct species of Galápagos tortoise. Proc. Natl. Acad. Sci. USA 2008, 105, 15464–15469. [Google Scholar] [CrossRef]
- Kehlmaier, C.; Barlow, A.; Hastings, A.K.; Vamberger, M.; Paijmans, J.L.; Steadman, D.W.; Albury, N.A.; Franz, R.; Hofreiter, M.; Fritz, U. Tropical ancient DNA reveals relationships of the extinct Bahamian giant tortoise Chelonoidis alburyorum. Proc. Biol. Sci. 2017, 284, 20162235. [Google Scholar] [CrossRef]
- Austin, J.J.; Arnold, E.N. Ancient mitochondrial DNA and morphology elucidate an extinct island radiation of Indian Ocean giant tortoises (Cylindraspis). Proc. Biol. Sci. 2001, 268, 2515–2523. [Google Scholar] [CrossRef] [PubMed]
- Palkovacs, E.P.; Gerlach, J.; Caccone, A. The evolutionary origin of Indian Ocean tortoises (Dipsochelys). Mol. Phylogenet. Evol. 2002, 24, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Le, M.; Raxworthy, C.J.; McCord, W.P.; Mertz, L. A molecular phylogeny of tortoises (Testudines: Testudinidae) based on mitochondrial and nuclear genes. Mol. Phylogenet. Evol. 2006, 40, 517–531. [Google Scholar] [CrossRef] [PubMed]
- Reid, B.N.; Le, M.; McCord, W.P.; Iverson, J.B.; Georges, A.; Bergmann, T.; Amato, G.; Desalle, R.; Naro-Maciel, E. Comparing and combining distance-based and character-based approaches for barcoding turtles. Mol. Ecol. Resour. 2011, 11, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Thomson, R.C.; Spinks, P.Q.; Shaffer, H.B. A global phylogeny of turtles reveals a burst of climate-associated diversification on continental margins. Proc. Natl. Acad. Sci. USA 2021, 118, e2012215118. [Google Scholar] [CrossRef]
- Chang, C.H.; Dai, W.Y.; Chen, T.Y.; Lee, A.H.; Hou, H.Y.; Liu, S.H.; Jang-Liaw, N.H. DNA barcoding reveals CITES-listed species among Taiwanese government-seized chelonian specimens. Genome 2018, 61, 615–624. [Google Scholar] [CrossRef]
- Das, S.; Rakib, T.M.; Islam, M.S.; Rahaman, M.M.; Das, T.; Hossain, M.A.; Sarker, S.; Raidal, S.R. The first complete mitogenome of Indian star tortoise (Geochelone elegans). Mitochondrial DNA B Resour. 2018, 3, 1112–1113. [Google Scholar] [CrossRef]
- Neumann, D.; Borisenko, A.V.; Coddington, J.A.; Häuser, C.L.; Butler, C.R.; Casino, A.; Vogel, J.C.; Haszprunar, G.; Giere, P. Global biodiversity research tied up by juridical interpretations of access and benefit sharing. Org. Divers. Evol. 2018, 18, 1–18. [Google Scholar] [CrossRef]
- Prathapan, K.D.; Pethiyagoda, R.; Bawa, K.S.; Raven, P.H.; Rajan, D. And 172 Co-Signatories from 35 Countries. When the cure kills—CBD limits biodiversity research. National laws fearing biopiracy squelch taxonomy studies. Science 2018, 360, 1405–1406. [Google Scholar] [CrossRef]
- D’Cruze, N.; Macdonald, D.W. A review of global trends in CITES live wildlife confiscations. Nat. Conserv. 2016, 15, 47–63. [Google Scholar] [CrossRef]
- Marshall, B.M.; Strine, C.; Hughes, A.C. Thousands of reptile species threatened by under-regulated global trade. Nat. Commun. 2020, 11, 4738. [Google Scholar] [CrossRef]
- Rhodin, A.G.J.; Stanford, C.B.; van Dijk, P.P.; Eisemberg, C.; Luiselli, L.; Mittermeier, R.A.; Hudson, R.; Horne, B.D.; Goode, E.; Kuchling, G.; et al. Global conservation status of turtles and tortoises (order Testudines). Chelonian Conserv. Biol. 2018, 17, 135–161. [Google Scholar] [CrossRef]
- Gumbs, R.; Gray, C.L.; Böhm, M.; Hoffmann, M.; Grenyer, R.; Jetz, W.; Meiri, S.; Roll, U.; Owen, N.R.; Rosindell, J. Global priorities for conservation of reptilian phylogenetic diversity in the face of human impacts. Nat. Commun. 2020, 11, 2616. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kundu, S.; Mukherjee, T.; Kim, A.R.; Lee, S.-R.; Mukherjee, A.; Jung, W.-K.; Kim, H.-W. Mitochondrial DNA and Distribution Modelling Evidenced the Lost Genetic Diversity and Wild-Residence of Star Tortoise, Geochelone elegans (Testudines: Testudinidae) in India. Animals 2023, 13, 150. https://doi.org/10.3390/ani13010150
Kundu S, Mukherjee T, Kim AR, Lee S-R, Mukherjee A, Jung W-K, Kim H-W. Mitochondrial DNA and Distribution Modelling Evidenced the Lost Genetic Diversity and Wild-Residence of Star Tortoise, Geochelone elegans (Testudines: Testudinidae) in India. Animals. 2023; 13(1):150. https://doi.org/10.3390/ani13010150
Chicago/Turabian StyleKundu, Shantanu, Tanoy Mukherjee, Ah Ran Kim, Soo-Rin Lee, Abhishek Mukherjee, Won-Kyo Jung, and Hyun-Woo Kim. 2023. "Mitochondrial DNA and Distribution Modelling Evidenced the Lost Genetic Diversity and Wild-Residence of Star Tortoise, Geochelone elegans (Testudines: Testudinidae) in India" Animals 13, no. 1: 150. https://doi.org/10.3390/ani13010150
APA StyleKundu, S., Mukherjee, T., Kim, A. R., Lee, S.-R., Mukherjee, A., Jung, W.-K., & Kim, H.-W. (2023). Mitochondrial DNA and Distribution Modelling Evidenced the Lost Genetic Diversity and Wild-Residence of Star Tortoise, Geochelone elegans (Testudines: Testudinidae) in India. Animals, 13(1), 150. https://doi.org/10.3390/ani13010150










