First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing
Abstract
Simple Summary
Abstract
1. Introduction
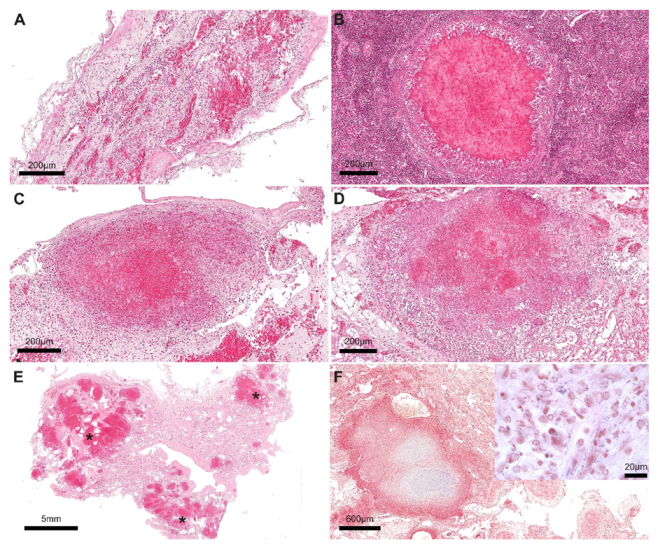
2. Case Description
3. Discussion
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Piveteau, P.; Depret, G.; Pivato, B.; Garmyn, D.; Hartmann, A. Changes in Gene Expression during Adaptationof Listeria monocytogenes to the Soil Environment. PLoS ONE 2011, 6, e24881. [Google Scholar] [CrossRef]
- Vivant, A.L.; Garmyn, D.; Piveteau, P. Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. 2013, 3, 87. [Google Scholar] [CrossRef] [PubMed]
- Raschle, S.; Stephan, R.; Stevens, M.J.A.; Cernela, N.; Zurfluh, K.; Muchaamba, F.; Nüesch-Inderbinen, M. Environmental dissemination of pathogenic Listeria monocytogenes in flowing surface waters in Switzerland. Sci. Rep. 2021, 11, 9066. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, A.; Cancellotti, F.M.; Turilli, C.; Randi, E. Serological investigations for some bacterial and viral pathogens in fallow deer (Cervus dama) and wild boar (Sus scrofa) of the San Rossore Preserve, Tuscany, Italy. J. Wildl. Dis. 1988, 24, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Sugimoto, T.; Sato, M.; Hirai, K. Incidence of Listeria monocytogenes in wild animals in Japan. J. Vet. Med. Sci. 2000, 62, 673–675. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dhama, K.; Verma, A.K.; Rajagunalan, S.; Kumar, A.; Tiwari, R.; Chakraborty, S.; Kumar, R. Listeria monocytogenes infection in poultry and its public health importance with special reference to food borne zoonoses. Pak. J. Biol. Sci. 2013, 16, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Frye, F.L. Fungal, actinomycete, bacterial, rickettsial, and viral diseases. In Biomedical and Surgical Aspects of Captive Reptile Husbandry; Krieger Publishing: Malabar, FL, USA, 1991; Volume 2, pp. 101–160. [Google Scholar]
- Girling, S.J.; Fraser, M.A. Listeria monocytogenes Septicaemia in an Inland Bearded Dragon, Pogona vitticeps. J. Herpeol. Med. Surg. 2004, 14, 6–9. [Google Scholar] [CrossRef]
- Vancraeynester, D.; Plasmans, F.; De Graef, E.; Hermans, K.; Decostere, A. Listeria monocytogenes associated myocardial perforation in a bearded dragon (Pogona vitticeps). Vlaams Diergeneeskundig Tijdschrift 2006, 75, 232–234. [Google Scholar]
- Matt, C.; Ramachandran, A.; Allison, R.W.; Wall, C.R.; Dieterly, A.M.; Brandao, J. Listeria monocytogenes in an inland bearded dragon (Pogona vitticeps). J. Exot. Pet. Med. 2019, 30, 76–81. [Google Scholar] [CrossRef]
- Bremer, P.J.; Osborne, C.M.; Kemp, R.A.; Smith, J.J. Survival of Listeria monocytogenes in sea water and effect of exposure on thermal resistence. J. Appl. Microbiol. 1998, 85, 545–553. [Google Scholar] [CrossRef]
- Beleveneva, I.A. Incidence and characteristics of Staphulococcus aureus and Listeria monocytogenes from the Japan and South China seas. Mar. Pollut. Boul. 2011, 62, 382–387. [Google Scholar] [CrossRef] [PubMed]
- Grattarola, C.; Giorda, F.; Iulini, B.; Pintore, M.D.; Pautasso, A.; Zoppi, S.; Goria, M.; Romano, A.; Peletto, S.; Varello, K.; et al. Meningoencephalitis and Listeria monocytogenes, Toxoplasma gondii and Brucella spp. coinfection in a dolphin in Italy. Dis. Aquat. Org. 2016, 118, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Sévellec, Y.; Torresi, M.; Félix, B.; Palma, F.; Centorotola, G.; Bilei, S.; Senese, M.; Terracciano, G.; Leblanc, J.C.; Pomilio, F.; et al. First Report on the Finding of Listeria monocytogenes ST121 Strain in a Dolphin Brain. Pathogens 2020, 9, 802. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, M.; Aguilar-Bultet, L.; Rupp, S.; Guildimann, C.; Stephan, R.; Schock, A.; Oevermann, A. Listeria monocytogenes sequence type 1 is predominant in ruminant rhombencephalitis. Sci. Rep. 2016, 6, 36419. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Chen, Y.; Gorski, L.; Ward, T.J.; Osborne, J.; Kathariou, S. Listeria monocytogenes Source Distribution Analysis Indicates Regional Heterogeneity and Ecological Niche Preference among Serotype 4b Clones. mBio 2018, 9, e00396-18. [Google Scholar] [CrossRef]
- Determinazione Dir. Regione Abruzzo n. DG/21/167 del 31.12.2014 “Applicazione nella Regione Abruzzo del Reg. CE 1069/2009 Recante: “Norme Sanitarie Relative ai Sottoprodotti di O.A. Non Destinati al Consumo Umano”. Available online: http://bura.regione.abruzzo.it/ (accessed on 6 September 2022).
- ISO11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria Spp.—Part 1: Detection Method. ISO: Geneva, Switzerland, 2017.
- Portmann, A.C.; Fournier, C.; Gimonet, J.; Ngom-Bru, C.; Barretto, C.; Baert, L. A Validation Approach of an End-To-End Whole Genome Sequencing Workflow for Source Tracking of Listeria monocytogenes and Salmonella enterica. Front. Microbiol. 2018, 9, 446. [Google Scholar] [CrossRef]
- Cito, F.; Di Pasquale, A.; Cammà, C.; Cito, P. The Italian Information System for the Collection and Analysis of Complete Genome Sequence of Pathogens Isolated from Animal, Food and Environment. Int. J. Infect. Dis. 2018, 73, 296–297. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Timme, R.E.; Lafon, P.C.; Balkey, M.; Adams, J.K.; Wagner, D.; Carleton, H.; Strain, E.; Hoffmann, M.; Sabol, A.; Rand, H.; et al. Gen-FS coordinated proficiency test data for genomic foodborne pathogen surveillance, 2017 and 2018 exercises. Sci. Data 2020, 7, 402. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A complete suite for gene-by-gene schema creation and strain identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef] [PubMed]
- Vázquez-Boland, J.A.; Kuhn, M.; Berche, P.; Chakraborty, T.; Domínguez-Bernal, G.; Goebel, W.; Gonzalez-Zorn, B.; Wehland, J.; Kreft, J. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 2001, 14, 584–640. [Google Scholar] [CrossRef] [PubMed]
- Orsi, R.H.; Borowsky, M.L.; Lauer, P.; Young, S.K.; Nusbaum, C.; Galagan, J.E.; Birren, B.W.; Ivy, R.A.; Sun, Q.; Graves, L.M.; et al. Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genom. 2008, 9, 539. [Google Scholar] [CrossRef] [PubMed]
- Parsons, C.; Niedermeyer, J.; Gould, N.; Brown, P.; Strules, J.; Parsons, A.W.; Bernardo Mesa-Cruz, J.; Kelly, M.J.; Hooker, M.J.; Chamberlain, M.J.; et al. Listeria monocytogenes at the human-wildlife interface: Black bears (Ursus americanus) as potential vehicles for Listeria. Microb Biotechnol. 2020, 13, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gorba, C.; Moura, A.; Leclercq, A.; Gómez-Martín, Á.; Gomis, J.; Jiménez-Trigos, E.; Mocé, M.L.; Lecuit, M.; Quereda, J.J. Listeria spp. Isolated from Tonsils of Wild Deer and Boars: Genomic Characterization. Appl. Environ. Microbiol. 2021, 87, e02651-20. [Google Scholar] [CrossRef] [PubMed]
- Filipello, V.; Mughini-Gras, L.; Gallina, S.; Vitale, N.; Mannelli, A.; Pontello, M.; Decastelli, L.; Allard, M.W.; Brown, E.W.; Lomonaco, S. Attribution of Listeria monocytogenes human infections to food and animal sources in Northern Italy. Food Microbiol. 2020, 89, 103433. [Google Scholar] [CrossRef]
- Guidi, F.; Chiaverini, A.; Repetto, A.; Lorenzetti, C.; Centorotola, G.; Bazzucchi, V.; Palombo, B.; Gattuso, A.; Pomilio, F.; Blasi, G. Hyper-Virulent Listeria monocytogenes Strains Associated With Respiratory Infections in Central Italy. Front. Cell. Infect. Microbiol. 2021, 11, 765540. [Google Scholar] [CrossRef]
- Gouin, E.; Mengaud, J.; Cossart, P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect. Immun. 1994, 62, 3550–3553. [Google Scholar] [CrossRef]
- Šteingolde, Ž.; Meistere, I.; Avsejenko, J.; Ķibilds, J.; Bergšpica, I.; Streikiša, M.; Gradovska, S.; Alksne, L.; Roussel, S.; Terentjeva, M.; et al. Characterization and Genetic Diversity of Listeria monocytogenes Isolated from Cattle Abortions in Latvia, 2013–2018. Vet. Sci. 2021, 8, 195. [Google Scholar] [CrossRef]
- Cossart, P.; Toledo-Arana, A. Listeria monocytogenes, a unique model in infection biology: An overview. Microbes Infect. 2008, 10, 1041–1050. [Google Scholar] [CrossRef]
- Shi, D.; Anwar, T.M.; Pan, H.; Chai, W.; Xu, S.; Yue, M. Genomic Determinants of Pathogenicity and Antimicrobial Resistance for 60 Global Listeria monocytogenes Isolates Responsible for Invasive Infections. Front. Cell. Infect. Microbiol. 2021, 11, 718840. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Zhao, D.; Xie, X.; Yao, H.; Wang, Y.; Kong, S.; Chen, X.; Pan, Z.; Jiao, X.; Yin, Y. inlF Enhances Listeria monocytogenes Early-Stage Infection by Inhibiting the Inflammatory Response. Front. Cell. Infect. Microbiol. 2022, 11, 1202. [Google Scholar] [CrossRef]
- Guerreiro, D.N.; Arcari, T.; O’Byrne, C.P. The σB-Mediated General Stress Response of Listeria monocytogenes: Life and Death Decision Making in a Pathogen. Front. Microbiol. 2020, 11, 1505. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wu, L.; Zhu, M.; Yang, Z.; Pilar, G.; Bao, H.; Zhou, Y.; Wang, R.; Zhang, H. Non-coding RNA regulates phage sensitivity in Listeria monocytogenes. PLoS ONE 2021, 16, e0260768. [Google Scholar] [CrossRef] [PubMed]
- Kastner, R.; Dussurget, O.; Archambaud, C.; Kernbauer, E.; Soulat, D.; Cossart, P.; Decker, T. LipA, a tyrosine and lipid phosphatase involved in the virulence of Listeria monocytogenes. Infect. Immun. 2011, 79, 2489–2498. [Google Scholar] [CrossRef] [PubMed]
- Cortes, B.W.; Naditz, A.L.; Anast, J.M.; Schmitz-Esser, S. Transcriptome Sequencing of Listeria monocytogenes Reveals Major Gene Expression Changes in Response to Lactic Acid Stress Exposure but a Less Pronounced Response to Oxidative Stress. Front. Microbiol. 2020, 10, 3110. [Google Scholar] [CrossRef]
- Kurpas, M.; Osek, J.; Moura, A.; Leclercq, A.; Lecuit, M.; Wieczorek, K. Genomic Characterization of Listeria monocytogenes Isolated from Ready-to-Eat Meat and Meat Processing Environments in Poland. Front. Microbiol. 2020, 11, 1412. [Google Scholar] [CrossRef] [PubMed]
- Hanes, R.M.; Huang, Z. Investigation of Antimicrobial Resistance Genes in Listeria monocytogenes from 2010 through to 2021. Int. J. Environ. Res. Public. Health. 2022, 19, 5506. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Renzo, L.; De Angelis, M.E.; Torresi, M.; Di Lollo, V.; Di Teodoro, G.; Averaimo, D.; Defourny, S.V.P.; Di Giacinto, F.; Profico, C.; Olivieri, V.; et al. First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing. Animals 2022, 12, 2364. https://doi.org/10.3390/ani12182364
Di Renzo L, De Angelis ME, Torresi M, Di Lollo V, Di Teodoro G, Averaimo D, Defourny SVP, Di Giacinto F, Profico C, Olivieri V, et al. First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing. Animals. 2022; 12(18):2364. https://doi.org/10.3390/ani12182364
Chicago/Turabian StyleDi Renzo, Ludovica, Maria Elisabetta De Angelis, Marina Torresi, Valeria Di Lollo, Giovanni Di Teodoro, Daniela Averaimo, Sabrina Vanessa Patrizia Defourny, Federica Di Giacinto, Chiara Profico, Vincenzo Olivieri, and et al. 2022. "First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing" Animals 12, no. 18: 2364. https://doi.org/10.3390/ani12182364
APA StyleDi Renzo, L., De Angelis, M. E., Torresi, M., Di Lollo, V., Di Teodoro, G., Averaimo, D., Defourny, S. V. P., Di Giacinto, F., Profico, C., Olivieri, V., Pomilio, F., Cammà, C., Ferri, N., & Di Francesco, G. (2022). First Report of Septicaemic Listeriosis in a Loggerhead Sea Turtle (Caretta caretta) Stranded along the Adriatic Coast: Strain Detection and Sequencing. Animals, 12(18), 2364. https://doi.org/10.3390/ani12182364




