A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Quantitative Real-Time PCR
2.3. Plasmid Construction
2.4. RNA Oligonucleotides and Transfection
2.5. Dual-Luciferase Reporter Assay
2.6. Western Blot
2.7. Fatty Acid Methyl Ester and Gas Chromatography (GC) Analysis
2.8. Mitochondrial Membrane Potential Detection
2.9. Electron Microscopy Analysis
2.10. Statistical Analysis
3. Results
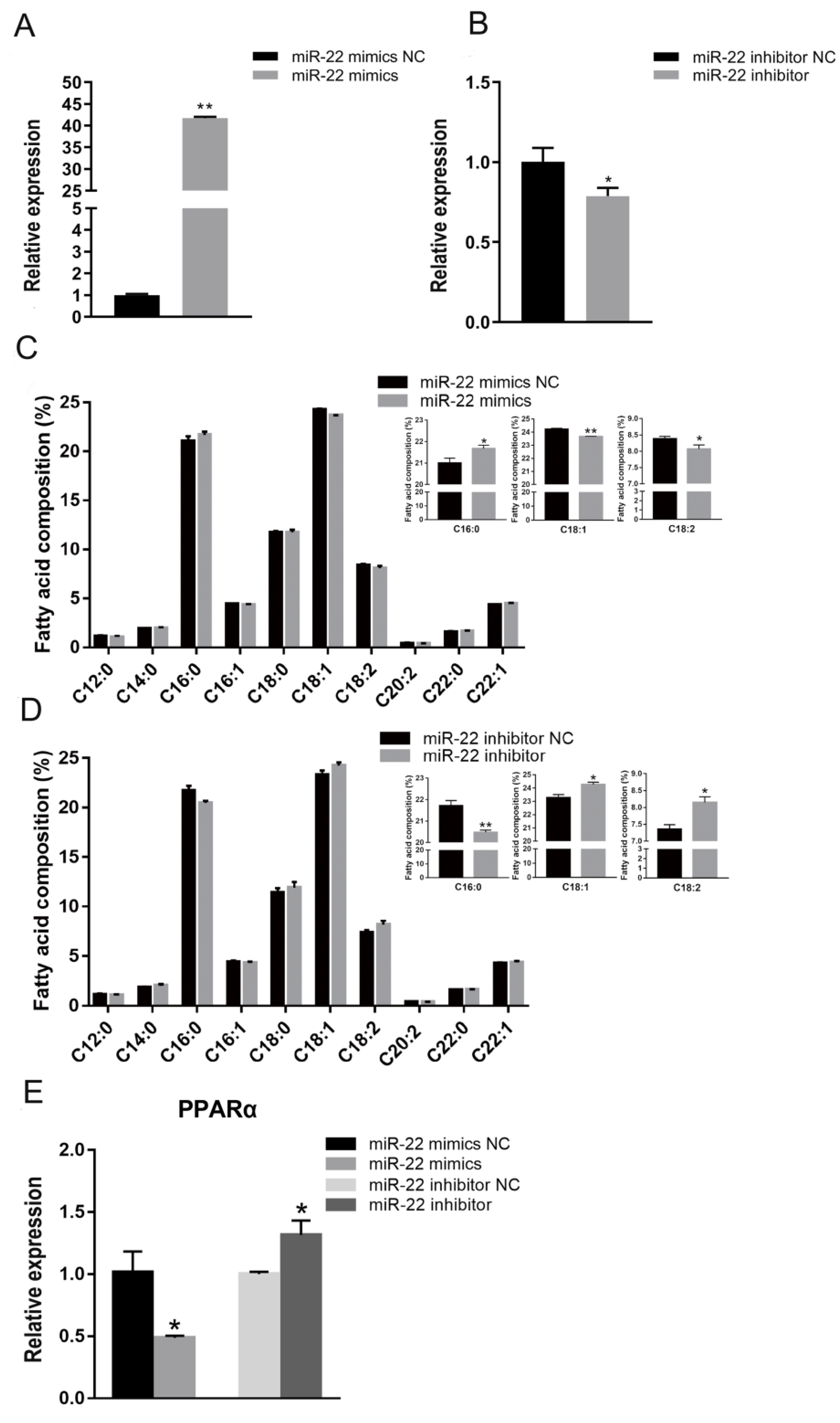
3.1. miR-22 Represses Fatty Acid Elongation in C2C12 Cells
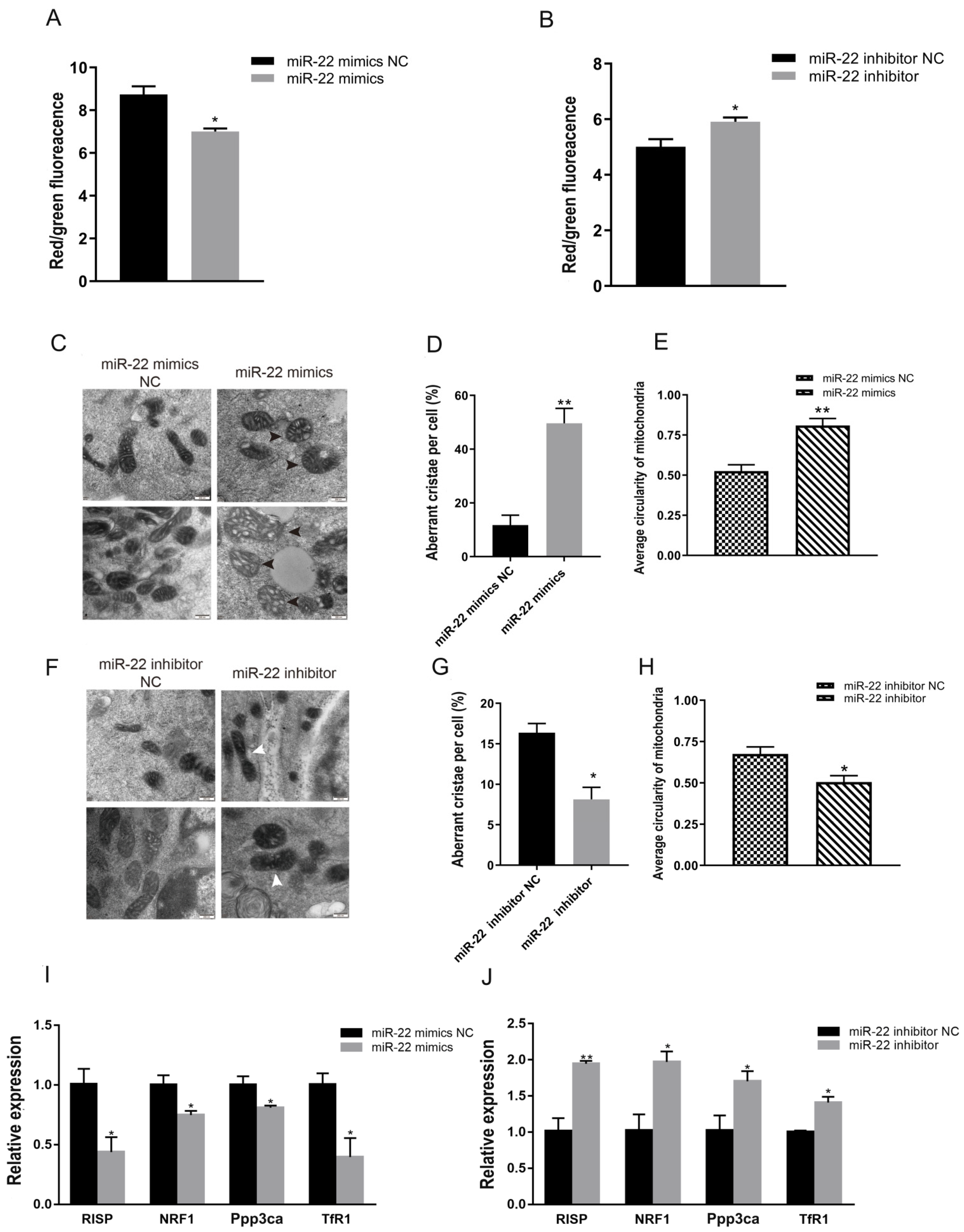
3.2. miR-22 Reduces Membrane Potential but Promotes Division and Abnormal Cristae of C2C12 Cell Mitochondria
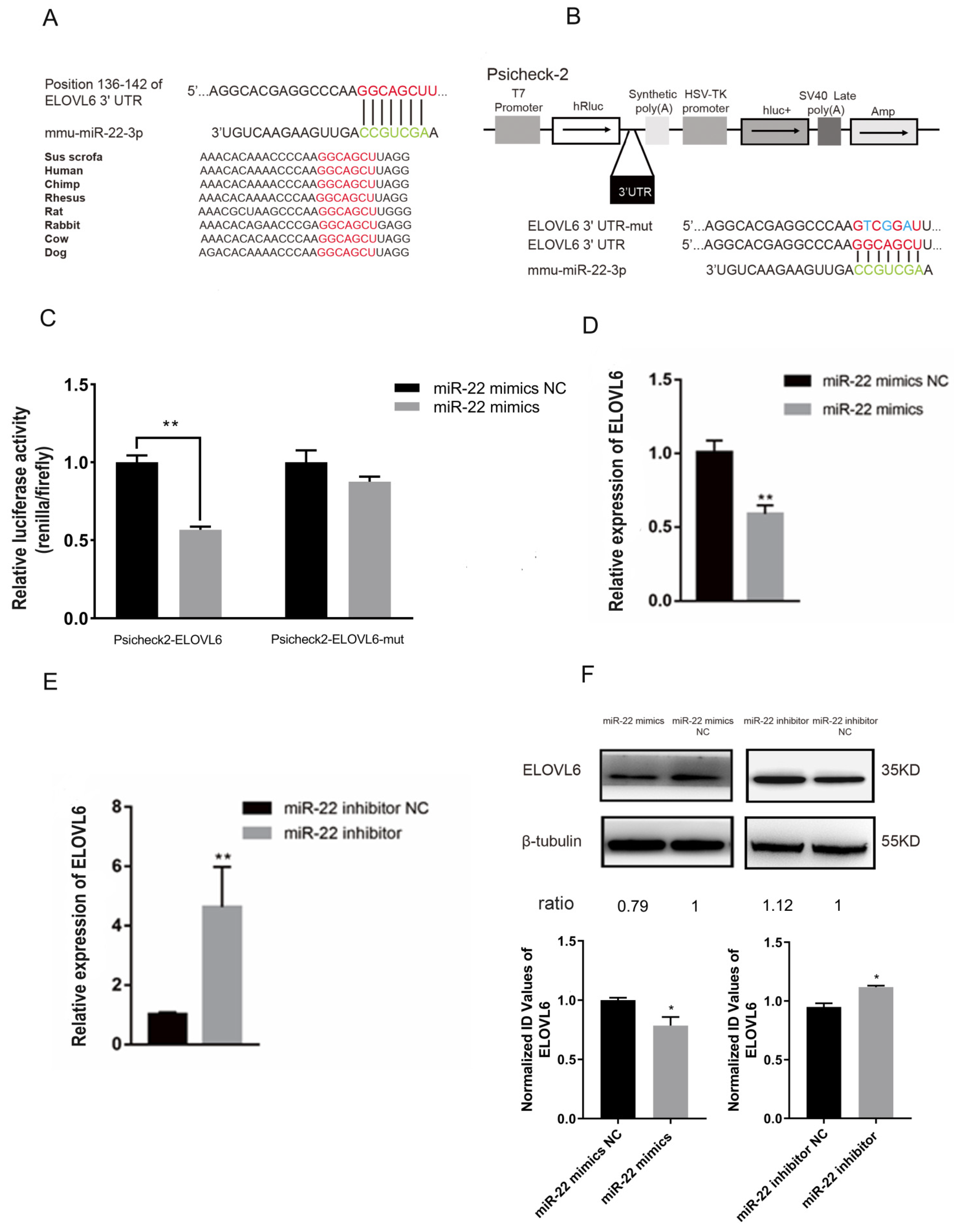
3.3. miR-22 Directly Targets the Elongase of Very Long Chain Fatty Acids 6 (ELOVL6) Gene in C2C12 Cells
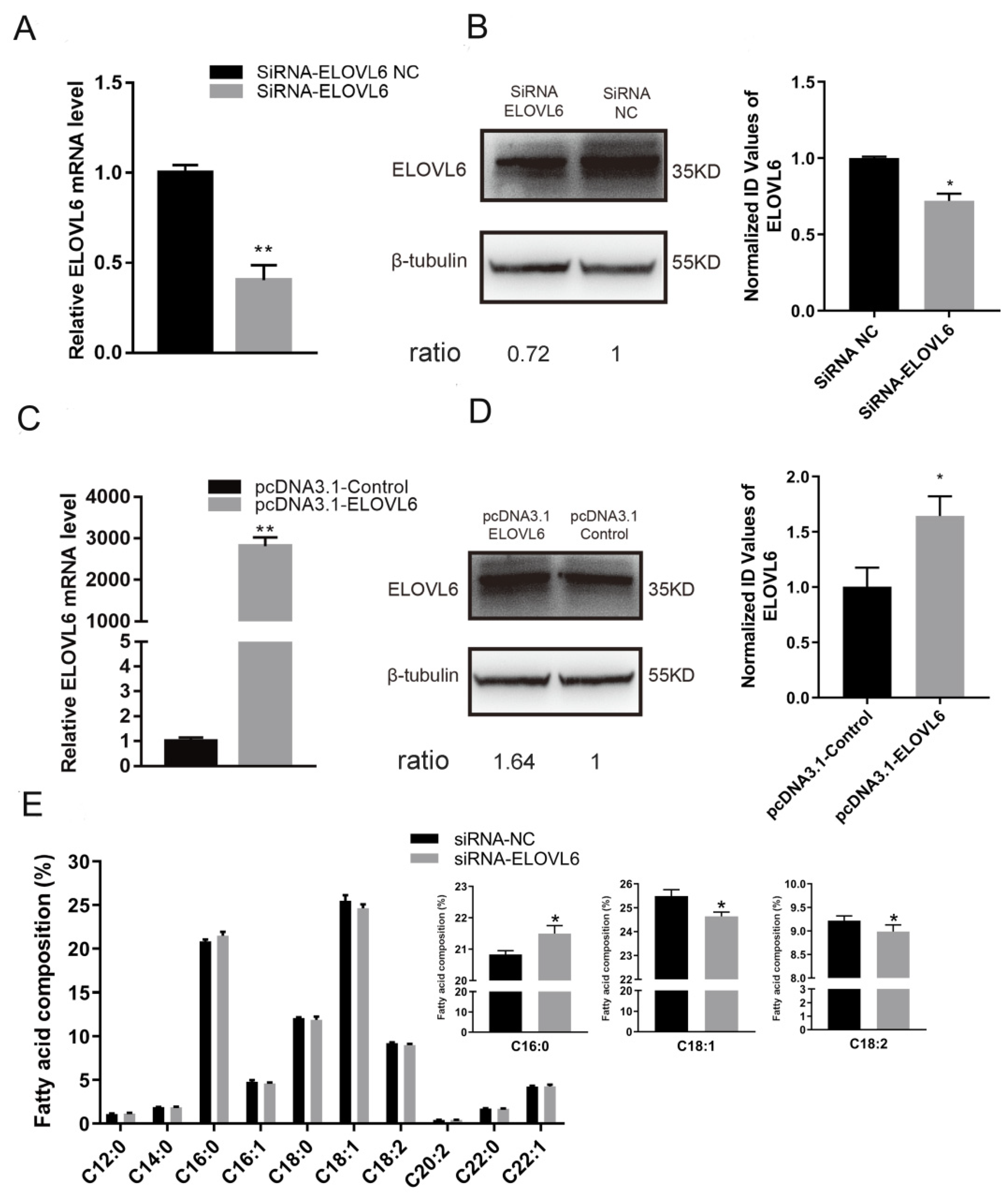
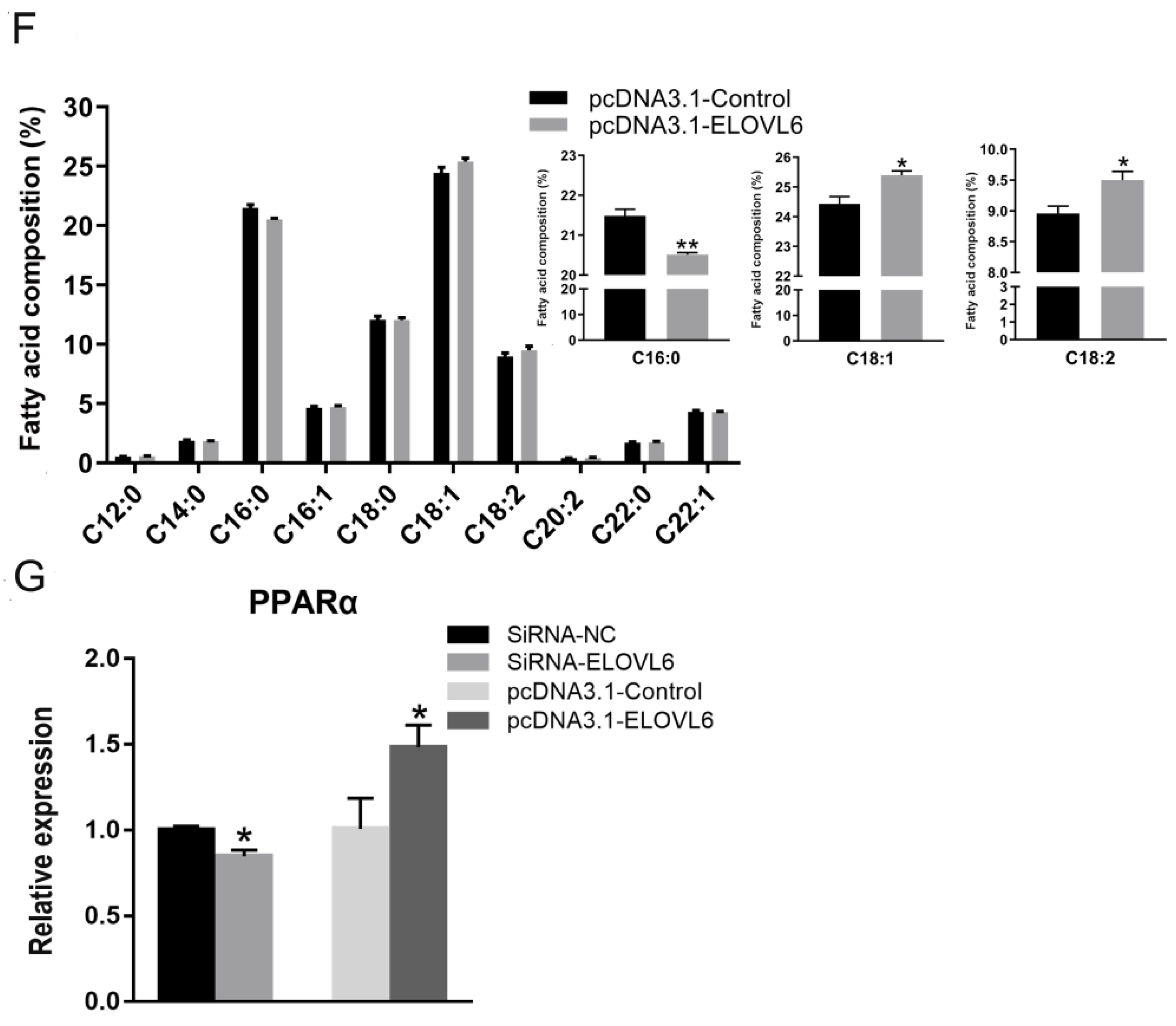
3.4. ELOVL6 Has a Positive Effect on Fatty Acid Elongation
3.5. ELOVL6 Promotes Cell Membrane Potential and Maintains Normal Mitochondrial Morphology of C2C12 Cells
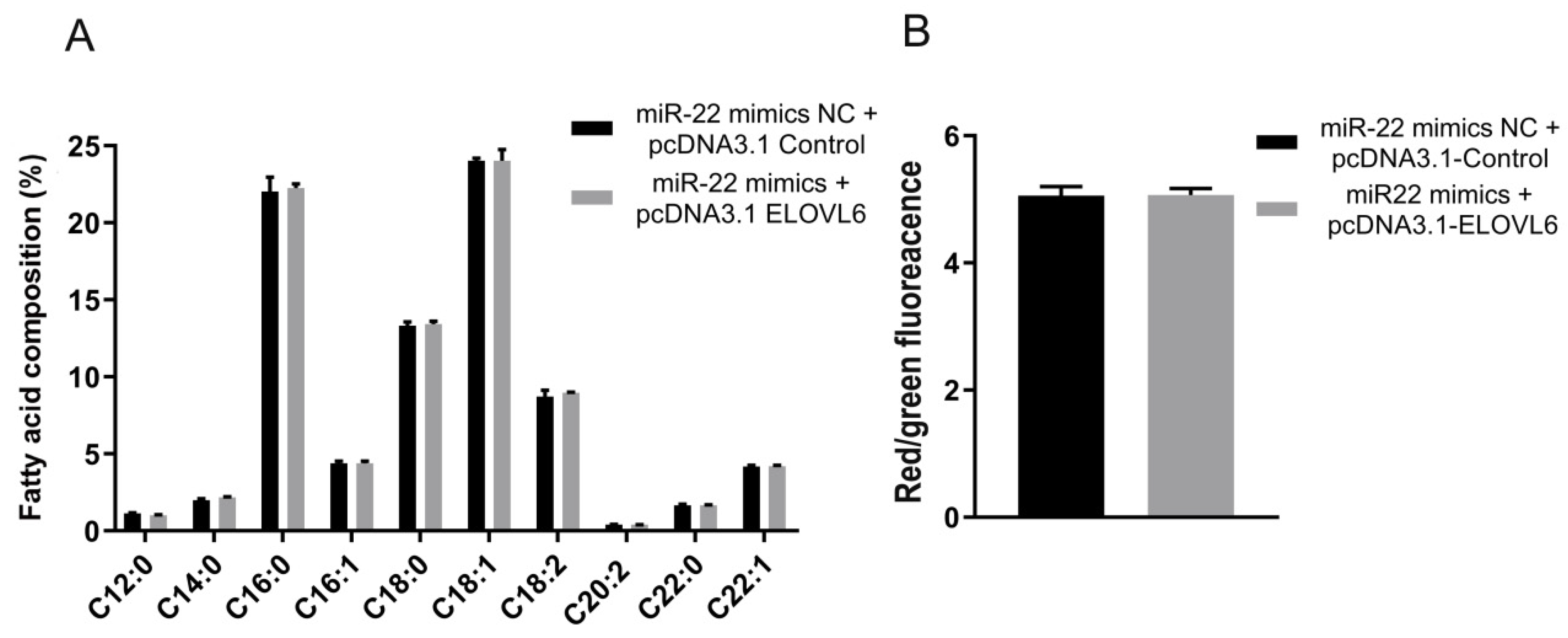
3.6. ELOVL6 Compensates for the Effects of miR-22 on Fatty Acid Composition and Mitochondrial Morphology in C2C12 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wood, J.; Enser, M.; Fisher, A.; Nute, G.; Sheard, P.; Richardson, R.; Hughes, S.; Whittington, F. Fat deposition, fatty acid composition and meat quality: A review. Meat Sci. 2008, 78, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Popp, J.; Wicke, M.; Klein, G.; Krischek, C. The relationship of pork longissimus muscle pH to mitochondrial respiratory activities, meat quality and muscle structure. Animal 2015, 9, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Cameron, N.; Enser, M.; Nute, G.; Whittington, F.; Penman, J.; Fisken, A.; Perry, A.; Wood, J. Genotype with nutrition interaction on fatty acid composition of intramuscular fat and the relationship with flavour of pig meat. Meat Sci. 2000, 55, 187–195. [Google Scholar] [CrossRef]
- Uemoto, Y.; Abe, T.; Tameoka, N.; Hasebe, H.; Inoue, K.; Nakajima, H.; Shoji, N.; Kobayashi, M.; Kobayashi, E. Whole-genome association study for fatty acid composition of oleic acid in Japanese Black cattle. Anim. Genet. 2011, 42, 141–148. [Google Scholar] [CrossRef]
- Estany, J.; Ros-Freixedes, R.; Tor, M.; Pena, R. Triennial growth and development symposium: Genetics and breeding for intramuscular fat and oleic acid content in pigs. J. Anim. Sci. 2017, 95, 2261–2271. [Google Scholar] [CrossRef]
- Bekhit, A.E.D.A.; Hopkins, D.L.; Fahri, F.T.; Ponnampalam, E.N. Oxidative processes in muscle systems and fresh meat: Sources, markers, and remedies. Compr. Rev. Food Sci. Food Saf. 2013, 12, 565–597. [Google Scholar] [CrossRef]
- Yang, X.; Chen, J.; Jia, C.; Zhao, R. Gene expression of calpain 3 and PGC-1α is correlated with meat tenderness in the longissimus dorsi muscle of Sutai pigs. Livest. Sci. 2012, 147, 119–125. [Google Scholar] [CrossRef]
- Lee, J.S.; Kim, J.M.; Lim, K.S.; Hong, J.S.; Hong, K.C.; Lee, Y.S. Effects of polymorphisms in the porcine micro RNA MIR206/MIR133B cluster on muscle fiber and meat quality traits. Anim. Genet. 2013, 44, 101–106. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Q.; Wang, B.; Wu, W.; Wei, J.; Li, P.; Huang, R. miR-22 regulates C2C12 myoblast proliferation and differentiation by targeting TGFBR1. Eur. J. Cell Biol. 2018, 97, 257–268. [Google Scholar] [CrossRef]
- Gonçalves, T.M.; de Almeida Regitano, L.C.; Koltes, J.E.; Cesar, A.S.M.; da Silva Andrade, S.C.; Mourão, G.B.; Gasparin, G.; Moreira, G.C.M.; Fritz-Waters, E.; Reecy, J.M. Gene co-expression analysis indicates potential pathways and regulators of beef tenderness in nellore cattle. Front. Genet. 2018, 9, 441. [Google Scholar] [CrossRef] [Green Version]
- Schweisgut, J.; Schutt, C.; Wüst, S.; Wietelmann, A.; Ghesquière, B.; Carmeliet, P.; Dröse, S.; Korach, K.S.; Braun, T.; Boettger, T. Sex-specific, reciprocal regulation of ER α and miR-22 controls muscle lipid metabolism in male mice. EMBO J. 2017, 36, 1199–1214. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Senyilmaz, D.; Virtue, S.; Xu, X.; Tan, C.Y.; Griffin, J.L.; Miller, A.K.; Vidal-Puig, A.; Teleman, A.A. Regulation of mitochondrial morphology and function by stearoylation of TFR1. Nature 2015, 525, 124–128. [Google Scholar] [CrossRef]
- Ma, Z.; Li, H.; Zheng, H.; Jiang, K.; Yan, F.; Tian, Y.; Kang, X.; Wang, Y.; Liu, X. Hepatic ELOVL6 mRNA is regulated by the gga-miR-22-3p in egg-laying hen. Gene 2017, 623, 72–79. [Google Scholar] [CrossRef] [PubMed]
- Zulfakar, S.S.; White, J.D.; Ross, T.; Tamplin, M.L. Cultured C2C12 cell lines as a model for assessment of bacterial attachment to bovine primary muscle cells. Meat Sci. 2013, 94, 215–219. [Google Scholar] [CrossRef]
- Xiong, Q.; Chai, J.; Deng, C.; Jiang, S.; Liu, Y.; Huang, T.; Suo, X.; Zhang, N.; Li, X.; Yang, Q. Characterization of porcine SKIP gene in skeletal muscle development: Polymorphisms, association analysis, expression and regulation of cell growth in C2C12 cells. Meat Sci. 2012, 92, 490–497. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA using TRIzol (TRI reagent). Cold Spring Harb. Protoc. 2010, 2010, pdb.prot5439. [Google Scholar] [CrossRef]
- Wang, H.; Shi, L.; Liang, T.; Wang, B.; Wu, W.; Su, G.; Wei, J.; Li, P.; Huang, R. MiR-696 regulates C2C12 cell proliferation and differentiation by targeting CNTFRα. Int. J. Biol. Sci. 2017, 13, 413. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Petenuci, M.E.; Dos Santos, V.J.; Gualda, I.P.; Lopes, A.P.; Schneider, V.V.A.; dos Santos, O.O.; Visentainer, J.V. Fatty acid composition and nutritional profiles of Brycon spp. from central Amazonia by different methods of quantification. J. Food Sci. Technol. 2019, 56, 1551–1558. [Google Scholar] [CrossRef]
- Kislinger, T.; Gramolini, A.O.; Pan, Y.; Rahman, K.; MacLennan, D.H.; Emili, A. Proteome Dynamics during C2C12 Myoblast Differentiation* S. Mol. Cell. Proteom. 2005, 4, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Chen, L.; Zhang, S.; Zhang, Y.; Wang, J.; Zhu, L. MicroRNA-23a reduces slow myosin heavy chain isoforms composition through myocyte enhancer factor 2C (MEF2C) and potentially influences meat quality. Meat Sci. 2016, 116, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.-P.; Chen, J.; Seok, H.Y.; Zhang, Z.; Kataoka, M.; Hu, X.; Wang, D.-Z. MicroRNA-22 regulates cardiac hypertrophy and remodeling in response to stress. Circ. Res. 2013, 112, 1234–1243. [Google Scholar] [CrossRef] [PubMed]
- Koufaris, C.; Valbuena, G.; Pomyen, Y.; Tredwell, G.; Nevedomskaya, E.; Lau, C.-H.; Yang, T.; Benito, A.; Ellis, J.; Keun, H. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene 2016, 35, 2766–2776. [Google Scholar] [CrossRef] [PubMed]
- Weiss, K.; Mihály, J.; Liebisch, G.; Marosvölgyi, T.; Schmitz, G.; Decsi, T.; Rühl, R. Effect of synthetic ligands of PPAR α, β/δ, γ, RAR, RXR and LXR on the fatty acid composition of phospholipids in mice. Lipids 2011, 46, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xue, W.; Jin, B.; Zhang, X.; Ma, F.; Xu, X. Candidate gene expression affects intramuscular fat content and fatty acid composition in pigs. J. Appl. Genet. 2013, 54, 113–118. [Google Scholar] [CrossRef]
- Park, S.Y.; Kim, M.H.; Ahn, J.H.; Lee, S.J.; Lee, J.H.; Eum, W.S.; Choi, S.Y.; Kwon, H.Y. The stimulatory effect of essential fatty acids on glucose uptake involves both Akt and AMPK activation in C2C12 skeletal muscle cells. Korean J. Physiol. Pharmacol. Off. J. Korean Physiol. Soc. Korean Soc. Pharmacol. 2014, 18, 255. [Google Scholar] [CrossRef]
- Ros-Freixedes, R.; Reixach, J.; Tor, M.; Estany, J. Expected genetic response for oleic acid content in pork. J. Anim. Sci. 2012, 90, 4230–4238. [Google Scholar] [CrossRef]
- Aaslyng, M.D.; Schäfer, A. The effect of free fatty acids on the odour of pork investigated by sensory profiling and GC-O-MS. Eur. Food Res. Technol. 2008, 226, 937–948. [Google Scholar] [CrossRef]
- Kim, H.-Y.; Kim, Y.-J.; Park, G.-B. Effect of probiotic supplemention on the performance and quality characteristics of meat from finishing pigs. Food Sci. Anim. Resour. 2007, 27, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Matarneh, S.K.; Silva, S.L.; Gerrard, D.E. New insights in muscle biology that alter meat quality. Annu. Rev. Anim. Biosci. 2021, 9, 355–377. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Ryan, M.T.; Schlame, M.; Zhao, M.; Gu, Z.; Klingenberg, M.; Pfanner, N.; Greenberg, M.L. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 2000, 275, 22387–22394. [Google Scholar] [CrossRef]
- Du, J.-K.; Cong, B.-H.; Yu, Q.; Wang, H.; Wang, L.; Wang, C.-N.; Tang, X.-L.; Lu, J.-Q.; Zhu, X.-Y.; Ni, X. Upregulation of microRNA-22 contributes to myocardial ischemia-reperfusion injury by interfering with the mitochondrial function. Free Radic. Biol. Med. 2016, 96, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ginsburg, K.S.; Hall, D.D.; Zimmermann, M.; Stein, I.S.; Zhang, M.; Tandan, S.; Hill, J.A.; Horne, M.C.; Bers, D. Targeting of protein phosphatases PP2A and PP2B to the C-terminus of the L-type calcium channel Cav1. 2. Biochemistry 2010, 49, 10298–10307. [Google Scholar] [CrossRef] [PubMed]
- Sharikabad, M.N.; Østbye, K.M.; Brørs, O. Increased [Mg2+] oreduces Ca2+ influx and disruption of mitochondrial membrane potential during reoxygenation. Am. J. Physiol. Heart Circ. Physiol. 2001, 281, H2113–H2123. [Google Scholar] [CrossRef]
- You, L.-h.; Li, Z.; Duan, X.-L.; Zhao, B.-L.; Chang, Y.-Z.; Shi, Z.-h. Mitochondrial ferritin suppresses MPTP-induced cell damage by regulating iron metabolism and attenuating oxidative stress. Brain Res. 2016, 1642, 33–42. [Google Scholar] [CrossRef]
- Barbieri, E.; Battistelli, M.; Casadei, L.; Vallorani, L.; Piccoli, G.; Guescini, M.; Gioacchini, A.M.; Polidori, E.; Zeppa, S.; Ceccaroli, P. Morphofunctional and biochemical approaches for studying mitochondrial changes during myoblasts differentiation. J. Aging Res. 2011, 2011, 845379. [Google Scholar] [CrossRef]
- Wai, T.; Langer, T. Mitochondrial dynamics and metabolic regulation. Trends Endocrinol. Metab. 2016, 27, 105–117. [Google Scholar] [CrossRef]
- Hughes, B.G.; Hekimi, S. A mild impairment of mitochondrial electron transport has sex-specific effects on lifespan and aging in mice. PLoS ONE 2011, 6, e26116. [Google Scholar] [CrossRef]
- Scarpulla, R.C. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol. Rev. 2008, 88, 611–638. [Google Scholar] [CrossRef] [Green Version]
- Scarpulla, R.C. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 2006, 97, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How muscle structure and composition influence meat and flesh quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Chen, X.; Huang, Z.; Chen, D.; Zheng, P.; He, J.; Chen, H.; Yu, J.; Luo, Y.; Yu, B. miR-22-3p regulates muscle fiber-type conversion through inhibiting AMPK/SIRT1/PGC-1α pathway. Anim. Biotechnol. 2021, 32, 254–261. [Google Scholar] [CrossRef] [PubMed]
- Sunaga, H.; Matsui, H.; Ueno, M.; Maeno, T.; Iso, T.; Syamsunarno, M.R.A.; Anjo, S.; Matsuzaka, T.; Shimano, H.; Yokoyama, T. Deranged fatty acid composition causes pulmonary fibrosis in Elovl6-deficient mice. Nat. Commun. 2013, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Shimano, H. Elovl6: A new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. 2009, 87, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Azar, S.; Udi, S.; Drori, A.; Hadar, R.; Nemirovski, A.; Vemuri, K.V.; Miller, M.; Sherill-Rofe, D.; Arad, Y.; Gur-Wahnon, D. Reversal of diet-induced hepatic steatosis by peripheral CB1 receptor blockade in mice is p53/miRNA-22/SIRT1/PPARα dependent. Mol. Metab. 2020, 42, 101087. [Google Scholar] [CrossRef]
- Magnani, N.D.; Marchini, T.; Vanasco, V.; Tasat, D.R.; Alvarez, S.; Evelson, P. Reactive oxygen species produced by NADPH oxidase and mitochondrial dysfunction in lung after an acute exposure to residual oil fly ashes. Toxicol. Appl. Pharmacol. 2013, 270, 31–38. [Google Scholar] [CrossRef]
- Whitelaw, P.F.; Hesketh, J.E. Expression of c-myc and c-fos in rat skeletal muscle. Evidence for increased levels of c-myc mRNA during hypertrophy. Biochem. J. 1992, 281, 143–147. [Google Scholar] [CrossRef]
- Caracciolo, D.; Di Martino, M.T.; Amodio, N.; Morelli, E.; Montesano, M.; Botta, C.; Scionti, F.; Talarico, D.; Altomare, E.; Cantafio, M.E.G. miR-22 suppresses DNA ligase III addiction in multiple myeloma. Leukemia 2019, 33, 487–498. [Google Scholar] [CrossRef]
- Kang, D.; Hamasaki, N. Maintenance of mitochondrial DNA integrity: Repair and degradation. Curr. Genet. 2002, 41, 311–322. [Google Scholar] [CrossRef]


| Gene Name | Sequence (5′-3′) | Accession Number |
|---|---|---|
| GAPDH | F: ATCACTGCCACCCAGAAGACT | NM_008084.2 |
| R: CATGCCAGTGAGCTTCCCGTT | ||
| ELOVL6 | F: GAAAAGCAGTTCAACGAGAACG | NM_130450.2 |
| R: AGATGCCGACCACCAAAGATA | ||
| PPARa | F: AGAGCCCCATCTGTCCTCTC | NM_011144.6 |
| R: ACTGGTAGTCTGCAAAACCAAA | ||
| RISP | F: CTTCTGTCCGTTTTTCCC | NM_025710.2 |
| R: GGTTTGCCTCTCCATTTA | ||
| NRF1 | F: GCCGTCGGAGCACTTACT | NM_001164226.1 |
| R: CTGTTCCAAGGTCACCACC | ||
| Ppp3ca | F: TGTACACGGTGGTTTGTCTCCAG | NM_001293622.1 |
| R: GGCCCATAAGCAGGTGGTTC | ||
| TfR1 | F: GCAGCTATTGCACTAGTC | NM_011638.4 |
| R: TGACTGCACTATGGTCAC | ||
| U6 | F: GTGCTCGCTTCGGCAGCACATAT | NR_003027.2 |
| R: AAAATATGGAACGCTTCACGAA | ||
| miR-22 | F: CAGGAAGCTGCCAGTTGAA | NR_030711.1 |
| R: TCAACTGGTGTCGTGGAGTC | ||
| RT-loop-miR-22 | CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGACAGTTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Hu, M.; Shen, Z.; Zhou, X.; Yang, S.; He, K.; Li, X.; Yan, F.; Zhao, A. A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells. Animals 2022, 12, 2274. https://doi.org/10.3390/ani12172274
Wang H, Hu M, Shen Z, Zhou X, Yang S, He K, Li X, Yan F, Zhao A. A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells. Animals. 2022; 12(17):2274. https://doi.org/10.3390/ani12172274
Chicago/Turabian StyleWang, Han, Moran Hu, Zhonghao Shen, Xiaolong Zhou, Songbai Yang, Ke He, Xiangchen Li, Feifei Yan, and Ayong Zhao. 2022. "A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells" Animals 12, no. 17: 2274. https://doi.org/10.3390/ani12172274
APA StyleWang, H., Hu, M., Shen, Z., Zhou, X., Yang, S., He, K., Li, X., Yan, F., & Zhao, A. (2022). A Specific microRNA Targets an Elongase of Very Long Chain Fatty Acids to Regulate Fatty Acid Composition and Mitochondrial Morphology of Skeletal Muscle Cells. Animals, 12(17), 2274. https://doi.org/10.3390/ani12172274






