Towards a Framework for High-Performance Simulation of Livestock Disease Outbreak: A Case Study of Spread of African Swine Fever in Vietnam
Abstract
Simple Summary
Abstract
1. Introduction
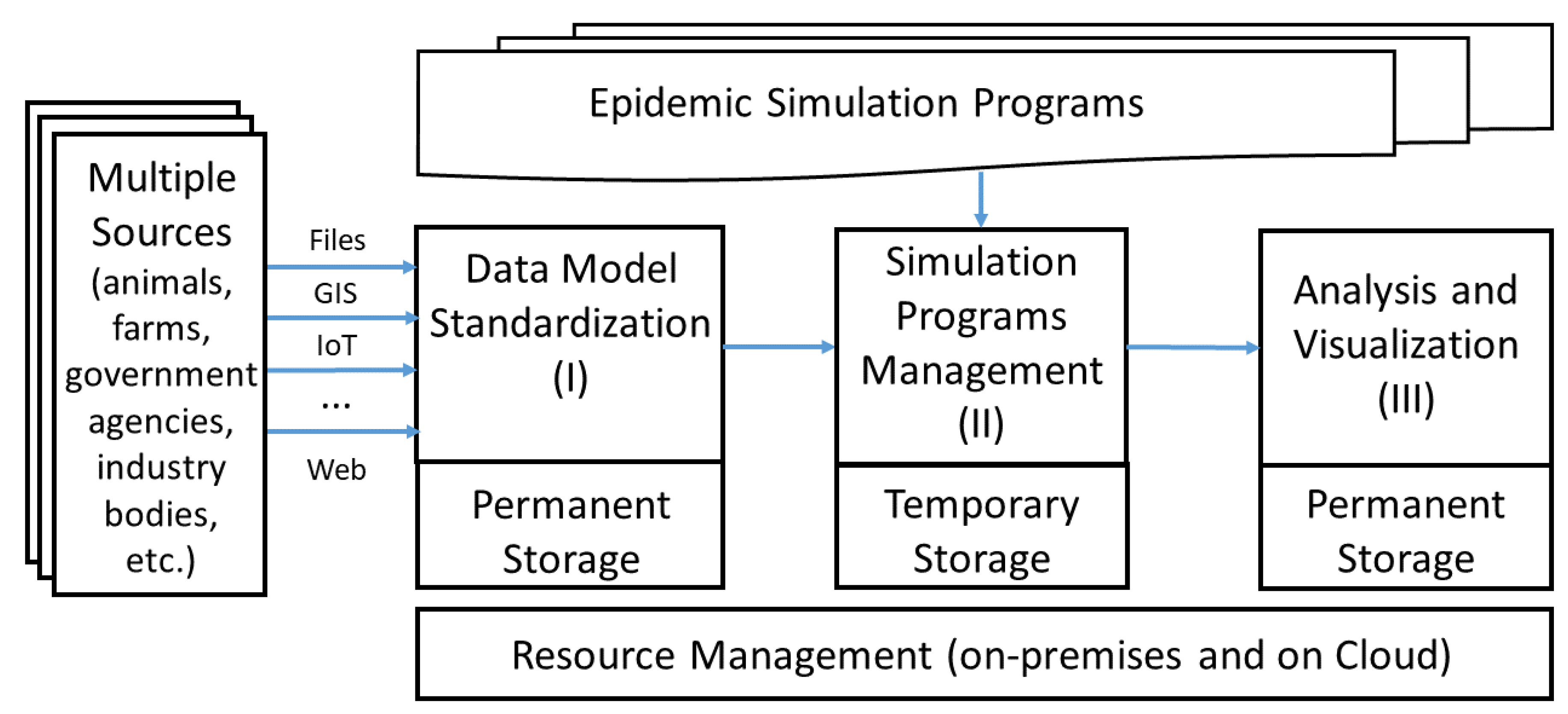
- Data Model Standardization is a module for transforming data related to livestock, veterinary epidemiology, etc., from many sources with different formats into uniform data models stored in tables of standardized database. These data models are designed according to the standards of each continent or region, for example, the SIGMA standard of European Food Safety Authority (EFSA), a standard for animal disease input data [28].
- Simulation Programs Management is a module for managing and distributing resources on premises as well as on the cloud for automatic and high-performance execution of simulations and centralized collection of outputs. This module also makes it possible to integrate different epidemic simulators on the same platform that manages and distributes resources for simulation computation, regardless of whether using a mathematical [16] or an agent-based model [29]. Moreover, a special new feature is that these programs can be allocated resources for operating and producing results at the same time.
- Analysis and Visualization is a module for handling data analysis on the output results stored centrally in the form of files or in the databases and is responsible for displaying the results in the form of tables, charts, histograms, or epidemiological maps using popular data representation programs in epidemiology, such as QGIS [30] or Epi Info [31], or in the web forms of cloud services.
2. Architecture
2.1. Data Model Standardization
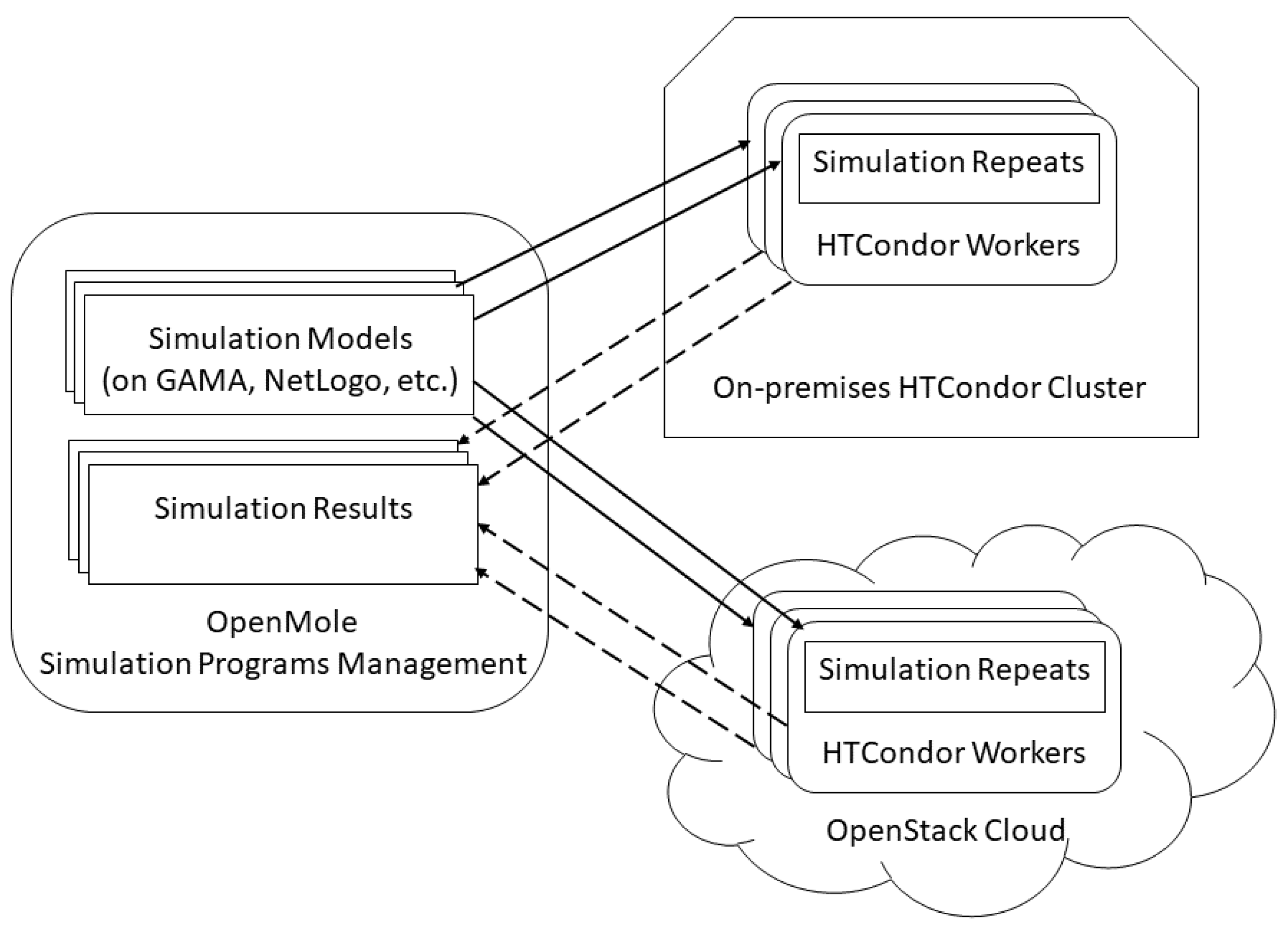
2.2. Simulation Programs Management
- -
- The population module: This module can describe the entire population or describe individual livestock (especially in agent-based models). The population needs to be generated at the beginning of each simulation run and assigned macro attributes such as quantity and composition or micro attributes such as age, gender, or medical history, etc.
- -
- The contact module: There are many ways that diseases can be transmitted in livestock, either by direct contact (animal-to-animal and droplet spread) or by indirect contact (airborne transmission, contaminated objects, food and drinking water, etc.). At the farm level, indirect transmission can also be caused by trade, grazing practices, or seasonal movements of livestock. Contacts also have properties such as duration or intensity.
- -
- The disease module: The task of this module is to deal with everything related to diseases. An agent-based model, for example, has individual-level information that may be divided into three parts: first, animal disease and health state; second, contact-causing state changes; and third, contact-independent state change (e.g., movement control, vaccination strategy, etc.). A disease might alter animals’ everyday habit, for example, making them lie still or stop eating. Animal death that is managed by the population module may result from the disease.
2.3. Analysis and Visualization
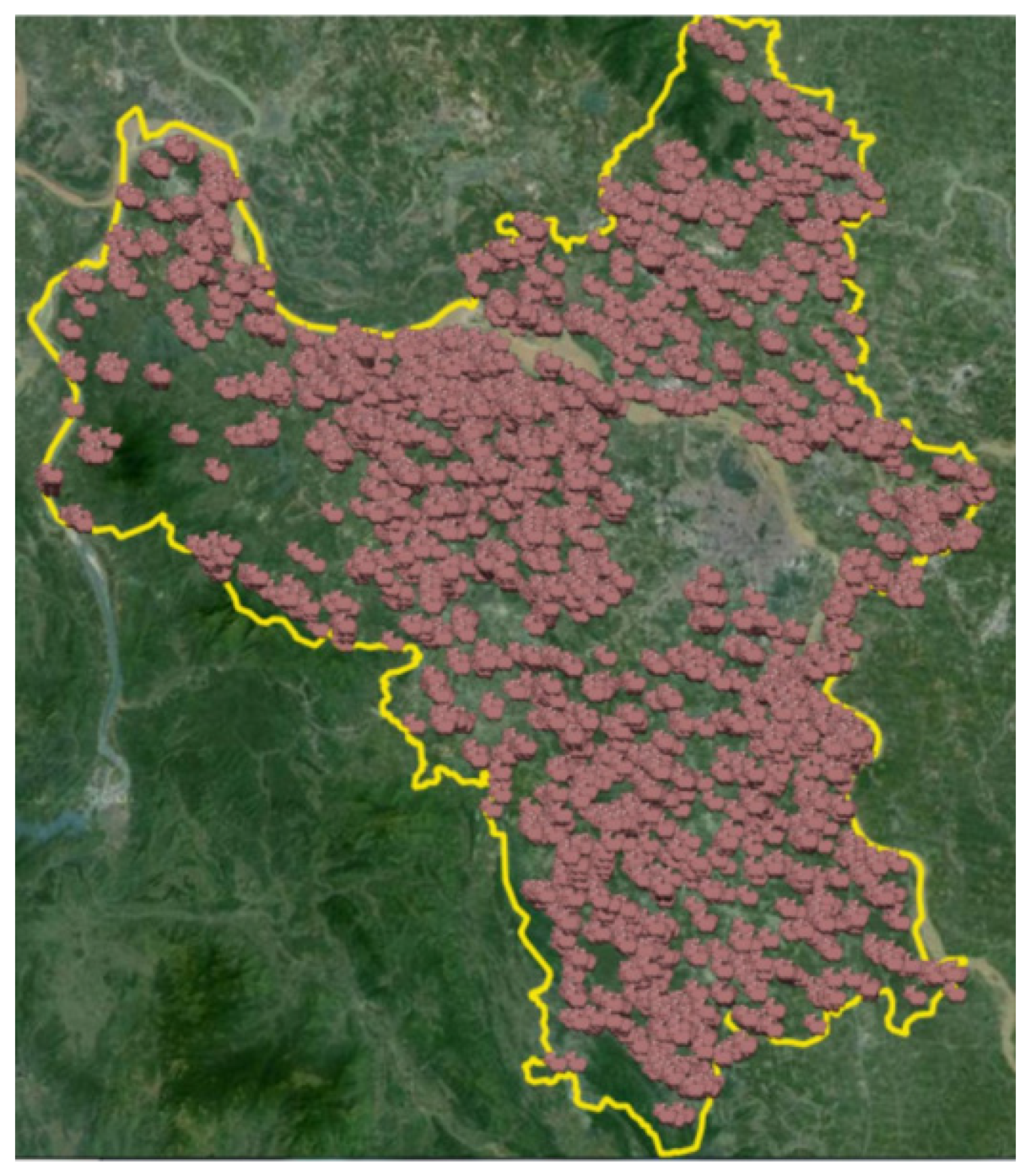
3. ASF Case Study and Simulation Model
4. Evaluation
4.1. Settings of Model Parameters
4.2. System Setup
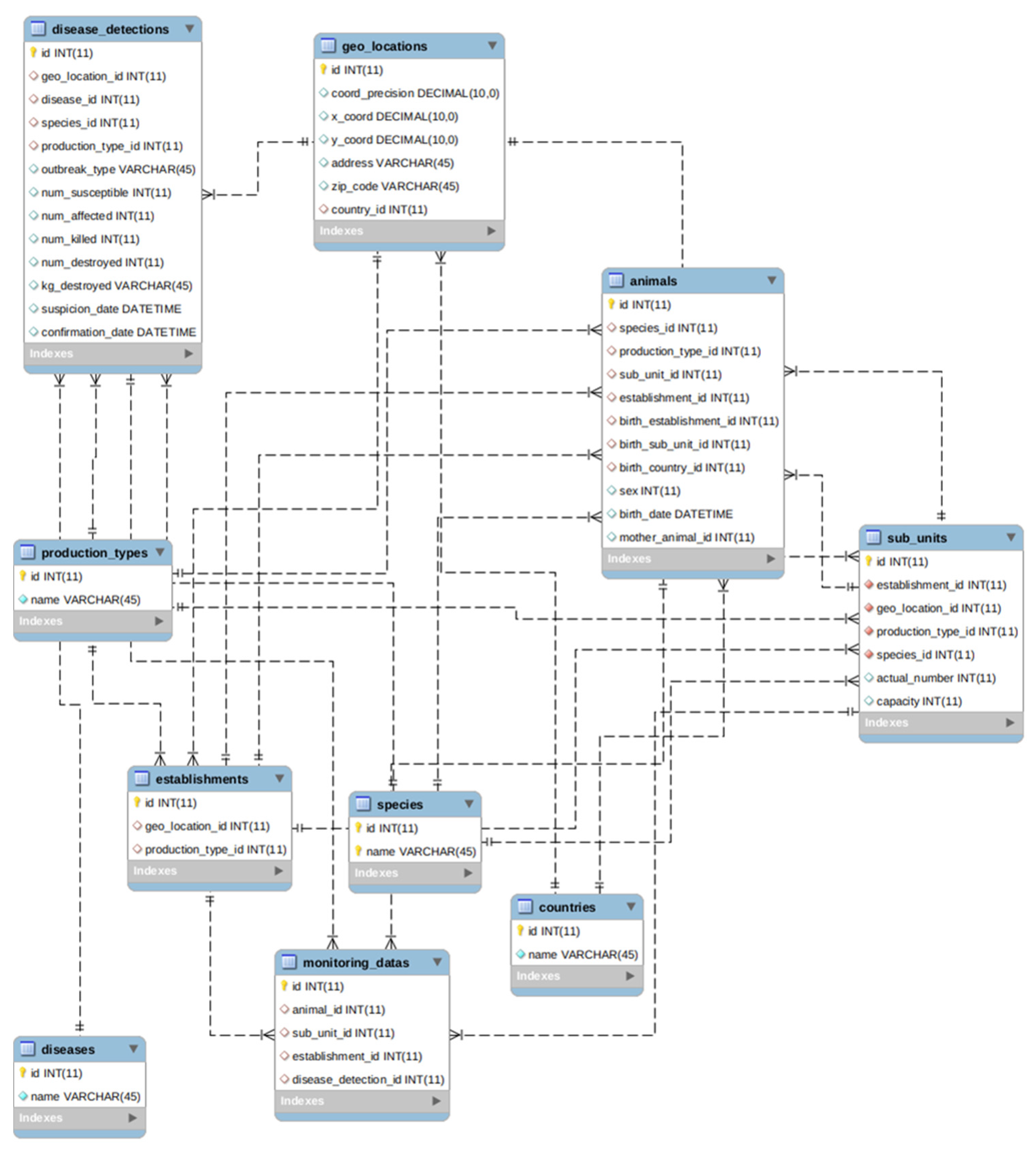
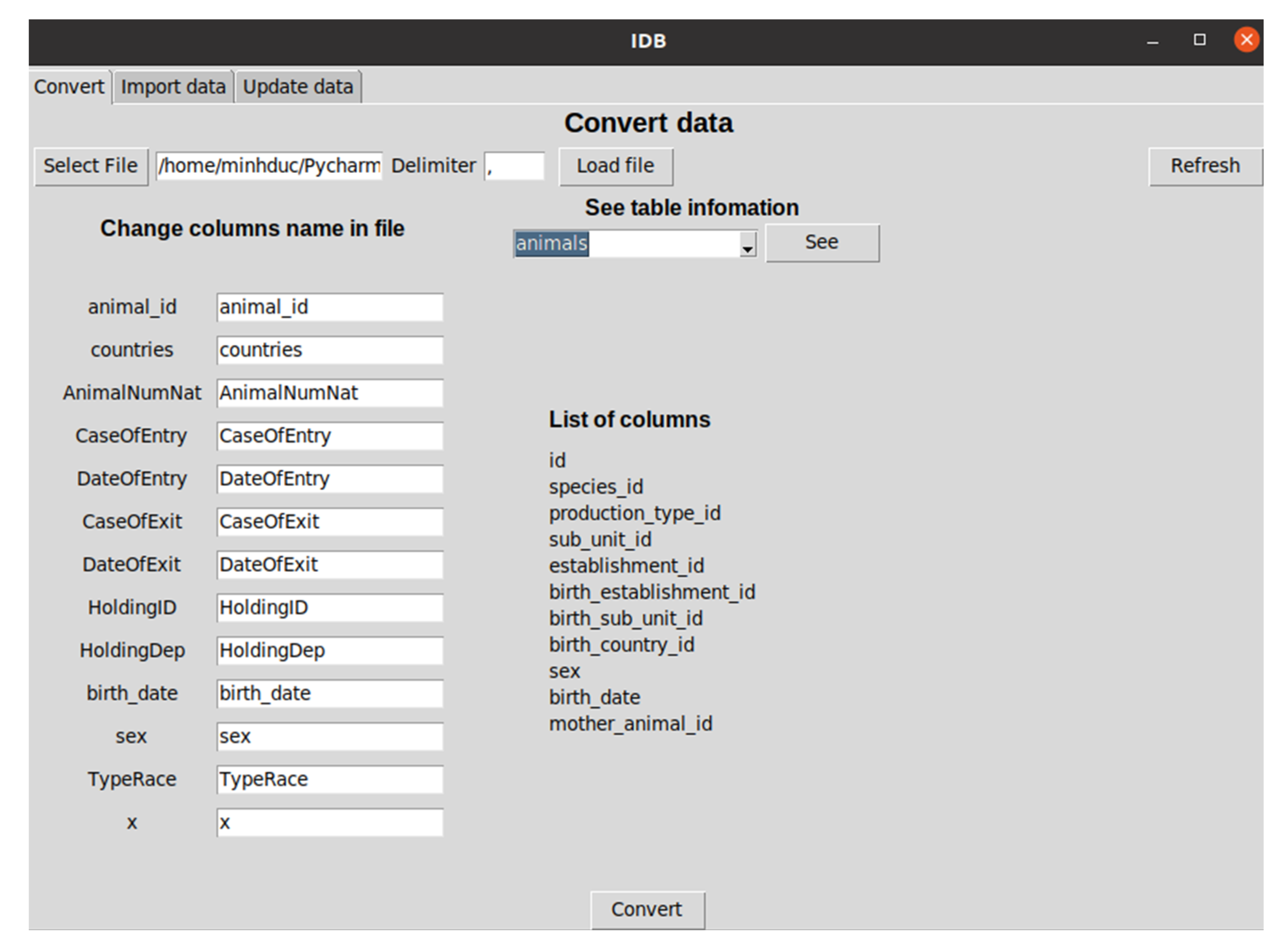
4.3. Standardization of Animal Disease Data Model
- Establishments: Any premises, buildings or any habitat or location where animals or germinal products are kept temporarily or permanently, excluding houses in which pets are kept as well as veterinary offices or clinics;
- Sub_units: Animals management group as part of an establishment such as flocks, pen, herds, houses, sheds, etc;
- Animals: Any terrestrial human-kept and registered animal that has a single identity number;
- Geo_locations: Positioning in the greatest possible resolution of the unit of interest, i.e., an establishment or a single animal;
- Disease_detections: Information string on probable epidemic reports, as documented in government veterinary agencies’ information system where accessible or in other public systems (e.g., EFSA DCF, and WAHIS);
- Production_types: Type of the establishment’s finished product or objective for which animals are maintained and/or raised;
- Species: The name, genus, species, and breed of the sub-unit of interest. This is especially significant in instances where the individual animals have no animal identification;
- Diseases: Disease to be reported;
- Countries: The ISO code of the country of birth or farm of the kept animal;
- Monitoring_data: Data from the surveillance or monitoring of kept animals, farms, diseases, and others.
4.4. Performance Analysis
4.4.1. Cost-Effective Assessment
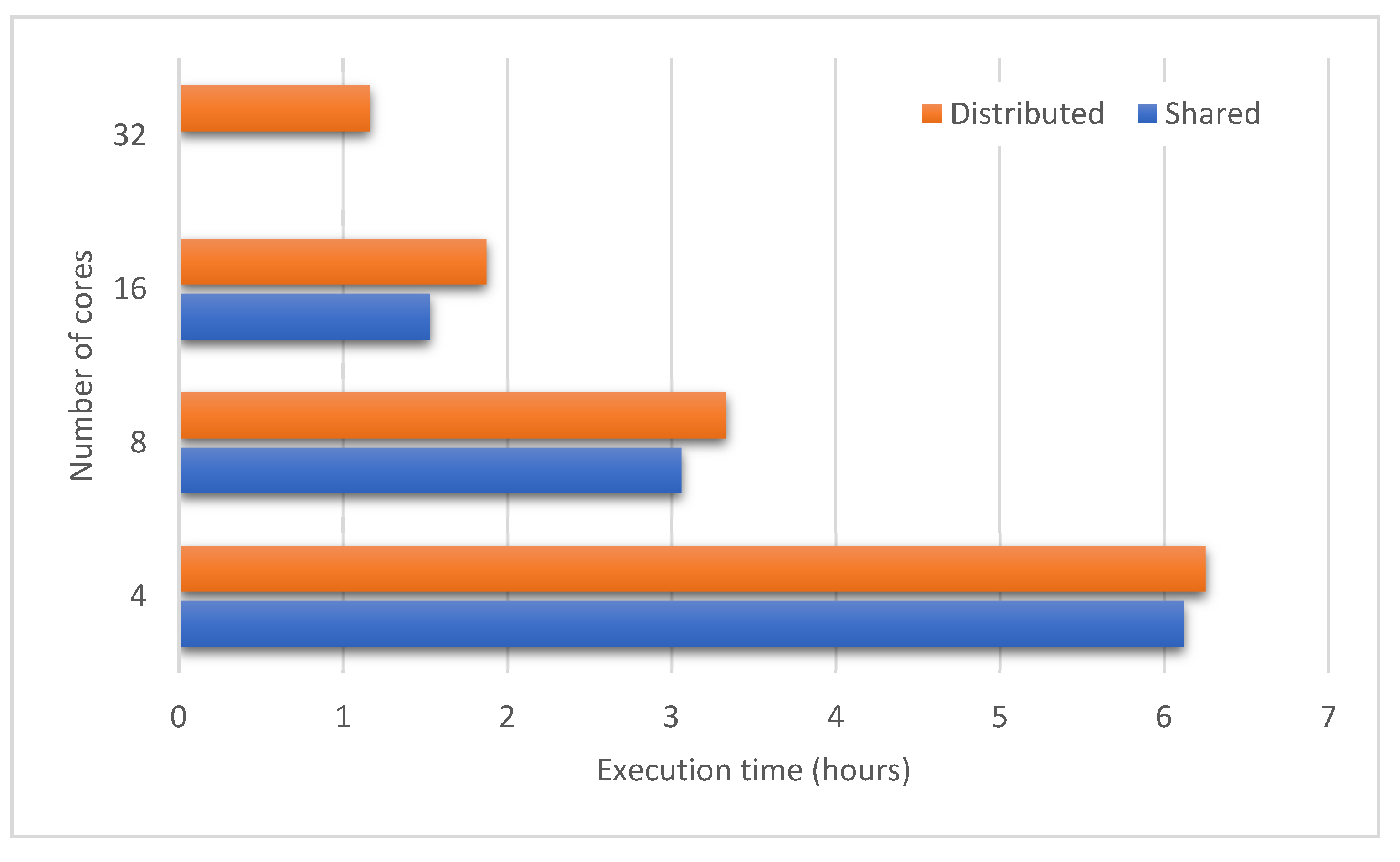
4.4.2. Scalability
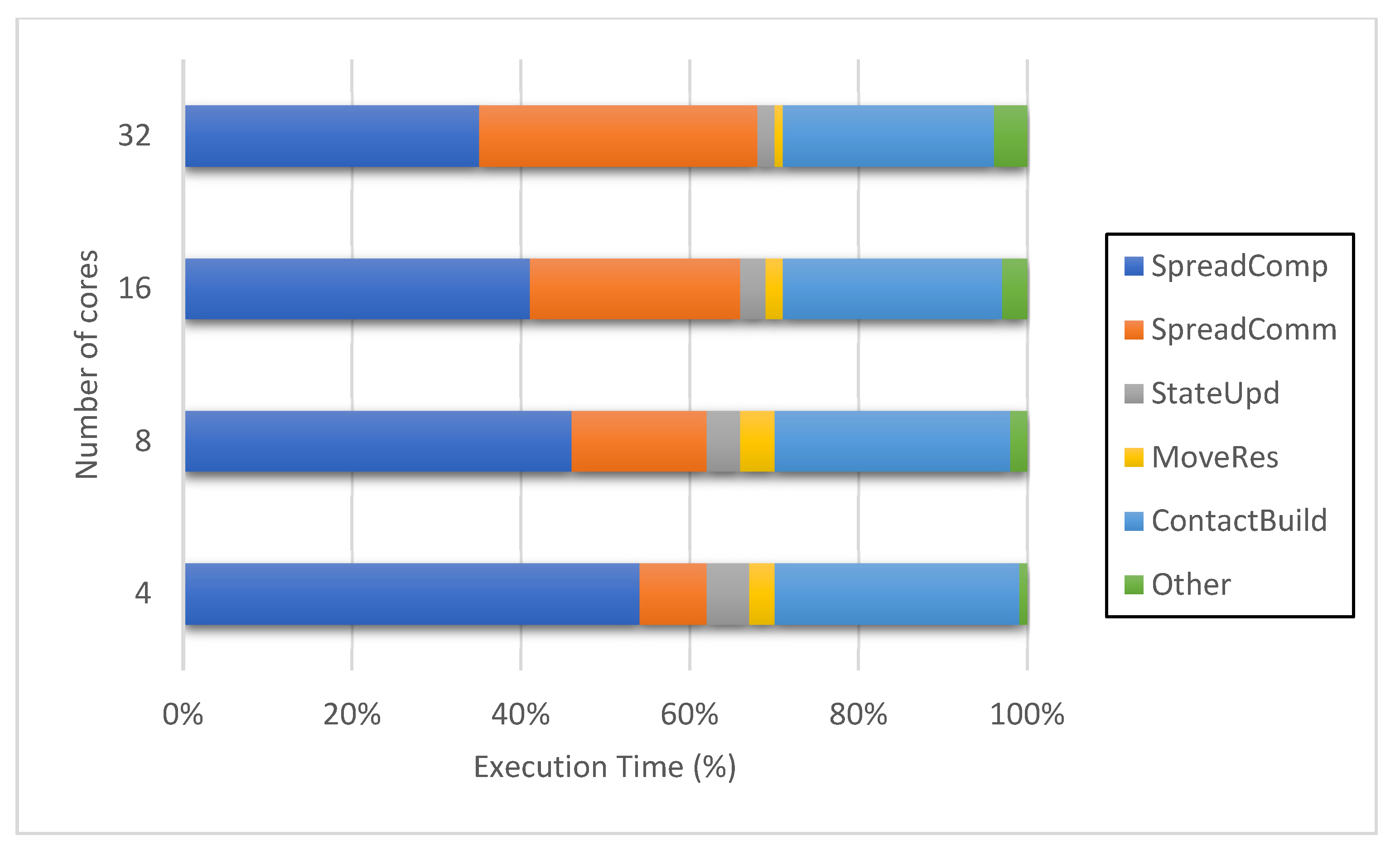
4.4.3. Process Distribution
5. Related Work
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salman, M. The role of veterinary epidemiology in combating infectious animal diseases on a global scale: The impact of training and outreach programs. Prev. Vet. Med. 2009, 92, 284–287. [Google Scholar] [CrossRef]
- Fernández-Carrión, E.; Martínez-Avilés, M.; Ivorra, B.; Martínez-López, B.; Ramos, Á.M.; Sánchez-Vizcaíno, J.M. Motion-based video monitoring for early detection of livestock diseases: The case of African swine fever. PLoS ONE 2017, 12, e0183793. [Google Scholar] [CrossRef]
- Thumbi, S.M.; Njenga, M.K.; Otiang, E.; Otieno, L.; Munyua, P.; Eichler, S.; Widdowson, M.A.; McElwain, T.F.; Palmer, G.H. Mobile phone-based surveillance for animal disease in rural communities: Implications for detection of zoonoses spillover. Philos. Trans. R. Soc. B 2019, 374, 20190020. [Google Scholar] [CrossRef]
- Cook, N.J.; Chabot, B.; Lui, T.; Bench, C.J.; Schaefer, A.L. Infrared thermography detects febrile and behavioural responses to vaccination of weaned piglets. Anim. Int. J. Anim. Biosci. 2015, 9, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Bhatele, A.; Yeom, J.S.; Jain, N.; Kuhlman, C.J.; Livnat, Y.; Bisset, K.R.; Kale, L.V.; Marathe, M.V. Massively Parallel Simulations of Spread of Infectious Diseases over Realistic Social Networks. In Proceedings of the 2017 17th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing (CCGRID), Madrid, Spain, 14–17 May 2017; pp. 689–694. [Google Scholar]
- Rao, D.M.; Chernyakhovsky, A. Parallel simulation of the global epidemiology of Avian Influenza. In Proceedings of the 2008 Winter Simulation Conference, Miami, FL, USA, 7–10 December 2008; pp. 1583–1591. [Google Scholar]
- Holvenstot, P.; Prieto, D.; De Doncker, E. GPGPU parallelization of self-calibrating agent-based influenza outbreak simulation. In Proceedings of the 2014 IEEE High Performance Extreme Computing Conference (HPEC), Waltham, MA, USA, 9–11 September 2014; pp. 1–6. [Google Scholar]
- Eriksson, H.; Timpka, T.; Spreco, A.; Dahlstrom, O.; Stromgren, M.; Holm, E. Dynamic Multicore Processing for Pandemic Influenza Simulation. In Proceedings of the AMIA Annual Symposium Proceedings, Chicago, IL, USA, 12–16 November 2016; pp. 534–540. [Google Scholar]
- Pachter, L.; Sturmfels, B. (Eds.) Algebraic Statistics for Computational Biology; Cambridge University Press: Cambridge, UK, 2005; 420p. [Google Scholar]
- Beerenwinkel, N.; Sing, T.; Lengauer, T.; Rahnenführer, J.; Roomp, K.; Savenkov, I.; Fischer, R.; Hoffmann, D.; Selbig, J.; Korn, K.; et al. Computational methods for the design of effective therapies against drug resistant HIV strains. Bioinformatics 2005, 21, 3943–3950. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michor, F.; Nowak, M.; Iwasa, Y. Evolution of Resistance to Cancer Therapy. Curr. Pharm. Des. 2006, 12, 261–271. [Google Scholar] [CrossRef] [PubMed]
- Waldrogel, J. The Period in the Volterra–Lotka Predator-Prey Model. SIAM J. Numer. Anal. 1983, 20, 1264–1272. [Google Scholar] [CrossRef]
- Iwasa, Y.; Michor, F.; Nowak, M.A. Evolutionary dynamics of invasion and escape. J. Theor. Biol. 2004, 226, 205–214. [Google Scholar] [CrossRef]
- Watts, D.J.; Muhamad, R.; Medina, D.C.; Dodds, P. Multiscale, resurgent epidemics in a hierarchical metapopulation model. Proc. Natl. Acad. Sci. USA 2005, 102, 11157–11162. [Google Scholar] [CrossRef]
- Hufnagel, L.; Brockmann, D.; Geisel, T. Forecast and control of epidemics in a globalized world. Proc. Natl. Acad. Sci. USA 2004, 101, 15124–15129. [Google Scholar] [CrossRef]
- Diekmann, O. Mathematical Epidemiology of Infectious Diseases: Model Building, Analysis and Interpretation; Mathematical and Computational Biology series; Wiley: Hoboken, NJ, USA, 2000; 303p. [Google Scholar]
- Ezanno, P.; Fourichon, C.; Viet, A.F.; Seegers, H. Sensitivity analysis to identify key-parameters in modelling the spread of bovine viral diarrhea virus in a dairy herd. Prev. Vet. Med. 2007, 80, 49–64. [Google Scholar] [CrossRef]
- Marcé, C.; Ezanno, P.; Seegers, H.; Pfeiffer, D.; Fourichon, C. Within-herd contact structure and transmission of Mycobacterium avium subspecies paratuberculosis in a persistently infected dairy cattle herd. Prev. Vet. Med. 2011, 100, 116–125. [Google Scholar] [CrossRef] [PubMed]
- Ezanno, P.; Fourichon, C.; Seegers, H. Influence of herd structure and type of virus introduction on the spread of bovine viral diarrhoea virus (BVDV) within a dairy herd. Vet. Res. 2008, 39, 1. [Google Scholar] [CrossRef] [PubMed]
- Beaunée, G.; Vergu, E.; Ezanno, P. Modelling of paratuberculosis spread between dairy cattle farms at a regional scale. Vet. Res. 2015, 46, 295. [Google Scholar] [CrossRef] [PubMed]
- Beaunée, G.; Vergu, E.; Joly, A.; Ezanno, P. Controlling bovine paratuberculosis at a regional scale: Towards a decision modelling tool. J. Theor. Biol. 2017, 435, 157–183. [Google Scholar] [CrossRef]
- Francis, J.; Klotz, G.; Harvey, N.; Stacey, D. Modeling and support tools for studying disease spread in livestock using networks. In Proceedings of the 2010 5th International Congress on Environmental Modelling and Software, Ottawa, ON, Canada, 5–8 July 2010. [Google Scholar]
- Morris, R.S.; Wilesmith, J.W.; Stern, M.W.; Sanson, R.L.; Stevenson, M.A. Predictive spatial modelling of alternative control strategies for the foot-and- mouth disease epidemic in Great Britain. Acta Hortic. 2001, 566, 337–347. [Google Scholar] [CrossRef]
- Smith, R.L.; Sanderson, M.W.; Renter, D.G.; Larson, R.; White, B. A stochastic risk-analysis model for the spread of bovine viral diarrhea virus after introduction to naïve cow-calf herds. Prev. Vet. Med. 2010, 95, 86–98. [Google Scholar] [CrossRef]
- Halasa, T.; Boklund, A.; Bøtner, A.; Toft, N.; Thulke, H.H. Simulation of Spread of African Swine Fever, Including the Effects of Residues from Dead Animals. Front. Vet. Sci. 2016, 3, 6. [Google Scholar] [CrossRef]
- Barongo, M.B.; Bishop, R.P.; Fèvre, E.M.; Knobel, D.L.; Ssematimba, A. A Mathematical Model that Simulates Control Options for African Swine Fever Virus (ASFV). PLoS ONE 2016, 11, e0158658. [Google Scholar] [CrossRef]
- Parlavantzas, N.; Pham, L.M.; Morin, C.; Arnoux, S.; Beaunée, G.; Qi, L.; Gontier, P.; Ezanno, P. A Service-based Framework for Building and Executing Epidemic Simulation Applications in the Cloud. Concurr. Comput. Pract. Exp. 2020, 32, e5554. [Google Scholar] [CrossRef]
- EFSA (European Food Safety Authority); Zancanaro, G.; Antoniou, S.E.; Bedriova, M.; Boelaert, F.; Gonzales Rojas, J.; Monguidi, M.; Roberts, H.; Saxmose Nielsen, S.; Thulke, H.-H. Scientific report on the SIGMA Animal Disease Data Model: A comprehensive approach for the collection of standardised data on animal diseases. EFSA J. 2019, 17, e05556. [Google Scholar]
- Wooldridge, M. Agent-based software engineering. IEE Proc. Softw. Eng. 1997, 144, 26–37. [Google Scholar] [CrossRef]
- QGIS. Available online: https://www.qgis.org/en/site/about/index.html (accessed on 19 June 2021).
- Epi Info. Available online: https://www.cdc.gov/epiinfo/ (accessed on 19 June 2021).
- Keogh, M.; Henry, M. The Implications of Digital Agriculture and Big Data for Australian Agriculture (Research Report, Australian Farm Institute); Australian Farm Institute: Sydney, Australia, 2016. [Google Scholar]
- Bartlett, P.C.; Van Buren, J.W.; Neterer, M.; Zhou, C. Disease surveillance and referral bias in the veterinary medical database. Prev. Vet. Med. 2010, 94, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Mork, M.; Lindberg, A.; Alenius, S.; Vagsholm, I.; Egenvall, A. Comparison between dairy cow disease incidence in data registered by farmers and in data from a disease-recording system based on veterinary reporting. Prev. Vet. Med. 2009, 88, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Shephard, R.W. The Development of a Syndromic Surveillance System for the Extensive Beef Cattle Producing Regions of Australia. Ph.D. Dissertation, University of Sidney, Sidney, Australia, 2006. [Google Scholar]
- Pavlin, J.A.; Mostashari, F.; Kortepeter, M.G.; Hynes, N.A.; Chotani, R.A.; Mikol, Y.B.; Ryan, M.A.K.; Neville, J.S.; Gantz, D.T.; Writer, J.V.; et al. Innovative Surveillance Methods for Rapid Detection of Disease Outbreaks and Bioterrorism: Results of an Interagency Workshop on Health Indicator Surveillance. Am. J. Public Health 2003, 93, 1230–1235. [Google Scholar] [CrossRef] [PubMed]
- Stone, M. The potential for exotic disease syndromic surveillance within veterinary laboratory submissions data. In Proceedings of the Epidemiology and Animal Health Management Branch of the NZVA, Christchurch, New Zealand, 4–7 July 2007; pp. 91–102. [Google Scholar]
- Sintchenko, V.; Gallego, B. Laboratory-guided detection of disease outbreaks: Three generations of surveillance systems. Arch. Pathol. Lab. Med. 2009, 133, 916–925. [Google Scholar] [CrossRef] [PubMed]
- Smith-Akin, K.A.; Bearden, C.F.; Pittenger, S.T.; Bernstam, E.V. Toward a veterinary informatics research agenda: An analysis of the PubMed- indexed literature. Int. J. Med. Inform. 2007, 76, 306–312. [Google Scholar] [CrossRef]
- Tierney, A.; Coyne, K.P.; Dawson, S.; Gaskell, R.M.; Bryan, J.; Newton, R.; Radford, A.D. SAVSNET. 2021. Available online: https://www.liverpool.ac.uk/savsnet/ (accessed on 19 June 2021).
- Moore, G.E.; Ward, M.P.; Dhariwal, J.; Wu, C.C.; Glickman, N.W.; Lewis, H.B.; Glickman, L.T. Development of a national companion animal syndromic surveillance system for bioterrorism. In Proceedings of the GISVET 2004, Guelph, ON, Canada, 23–25 June 2004. [Google Scholar]
- Glickman, L.T.; Moore, G.E.; Glickman, N.W.; Caldanaro, R.J.; Aucoin, D.; Lewis, H.B. Purdue university-Banfield national companion animal surveillance program for emerging and zoonotic diseases. Vector Borne Zoonotic Dis. 2006, 6, 14–23. [Google Scholar] [CrossRef]
- Maciejewski, R.; Tyner, B.; Jang, Y.; Zheng, C.; Nehme, R.; Ebert, D.S.; Cleveland, W.S.; Ouzzani, M.; Grannis, J.; Glickman, L.T. LAHVA: Linked animal-human health visual analytics. In Proceedings of the IEEE Symposium on Visual Analytics Science and Technology, Sacramento, CA, USA, 30 October–1 November 2007; pp. 27–34. [Google Scholar]
- AIMS. Available online: http://aims.fao.org/ (accessed on 19 June 2021).
- AgXML. Available online: https://schemas.liquid-technologies.com/agxml/2.0/ (accessed on 19 June 2021).
- Schmitz, M.; Martini, D.; Kunisch, M.; Mösinger, H.J. agroXML Enabling Standardized, Platform-Independent Internet Data Exchange in Farm Management Information Systems. In Metadata and Semantics; Sicilia, M.A., Lytras, M.D., Eds.; Springer: Boston, MA, USA, 2009. [Google Scholar]
- DCMI. Available online: https://dublincore.org/schemas/xmls/ (accessed on 19 June 2021).
- NetLogo. Available online: https://ccl.northwestern.edu/netlogo/ (accessed on 19 June 2021).
- GAMA. Available online: https://gama-platform.github.io/wiki/Tutorials (accessed on 19 June 2021).
- Harvey, N.; Reeves, A.; Schoenbaum, M.A.; Zagmutt-Vergara, F.J.; Dubé, C.; Hill, A.E.; Corso, B.A.; McNab, W.B.; Cartwright, C.I.; Salman, M.D. The North American Animal Disease Spread Model: A simulation model to assist decision making in evaluating animal disease incursions. Prev. Vet. Med. 2007, 82, 176–197. [Google Scholar] [CrossRef]
- OpenMole. Available online: https://openmole.org/Documentation.html (accessed on 19 June 2021).
- Repast HPC. Available online: https://repast.github.io/repast_hpc.html (accessed on 19 June 2021).
- Wang, X. An SIRS Epidemic Model with Vital Dynamics and a Ratio-Dependent Saturation Incidence Rate. Discret. Dyn. Nat. Soc. 2015, 2015, 720682. [Google Scholar] [CrossRef]
- Guinat, C.; Gubbins, S.; Vergne, T.; Gonzales, J.L.; Dixon, L.; Pfeiffer, D.U. Experimental pig-to-pig transmission dynamics for African swine fever virus, Georgia 2007/1 strain. Epidemiol. Infect. 2016, 144, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Epidemic Simulation Front End Source Code. Available online: https://github.com/fimocode/GamaNew/tree/gama (accessed on 19 June 2021).
- Pham, M.L.; Parlavantzas, N.; Morin, C.; Arnoux, S.; Qi, L.; Gontier, P.; Ezanno, P. DiFFuSE, a distributed framework for cloud-based epidemic simulations: A case study in modelling the spread of bovine viral diarrhea virus. In Proceedings of the 2017 IEEE International Conference on Cloud Computing Technology and Science (CloudCom), Hong Kong, China, 11–14 December 2017. [Google Scholar]
- Disease Data Converter. Available online: https://github.com/fimocode/diseasedataconverter (accessed on 19 June 2021).
- Amazon EC2 On-Demand Pricing. Available online: https://aws.amazon.com/ec2/pricing/on-demand/ (accessed on 19 June 2021).
- Cost of Server Ownership: On-Premise vs. IaaS. Available online: https://www.sherweb.com/blog/cloud-server/total-cost-of-ownership-of-servers-iaas-vs-on-premise/ (accessed on 28 June 2021).
- Perumalla, K.S.; Seal, S.K. Reversible parallel discrete-event execution of large-scale epidemic outbreak models. In Proceedings of the 2010 IEEE Workshop on Principles of Advanced and Distributed Simulation (PADS’10), Atlanta, GA, USA, 17–19 May 2010. [Google Scholar]
- Bisset, K.R.; Chen, J.; Deodhar, S.; Feng, X.; Ma, Y.; Marathe, M.V. Indemics: An interactive high-performance computing framework for data-intensive epidemic modeling. ACM Trans. Model. Comput. Simul. 2014, 24, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Zou, P.; Lü, Y.-S.; Wu, L.-D.; Chen, L.-L.; Yao, Y.-P. Epidemic simulation of a large-scale social contact network on GPU clusters. SIMULATION 2013, 89, 1154–1172. [Google Scholar] [CrossRef]
- Sukcharoen, P.; Pumma, S.; Mongkolsermporn, O.; Achalakul, T.; Li, X. Design and analysis of a cloud-based epidemic simulation framework. In Proceedings of the 2012 9th International Conference on Electrical Engineering/Electronics, Computer, Telecommunications and Information Technology, Phetchaburi, Thailand, 16–18 May 2012. [Google Scholar]
- Price, R.C.; Pettey, W.; Freeman, T.; Keahey, K.; Leecaster, M.; Samore, M.; Tobias, J.; Facelli, J.C. SaTScan on a Cloud: On-Demand Large Scale Spatial Analysis of Epidemics. Online J. Public Health Inform. 2010, 2. [Google Scholar] [CrossRef]
- Haris, M.; Manzoor, M.S. Spatiotemporal study of dengue virus infection via cloud based framework. Int. J. Adv. Res. Comput. Sci. Softw. Eng. 2016, 6, 155–161. [Google Scholar]
| Farm Category | Small | Medium | Large |
|---|---|---|---|
| Small | Frequently (0.06/0.29) | Barely (-/0.29) | Barely (-/-) |
| Medium | Frequently (0.06/0.29) | Rarely (0.06/0.26) | Barely (-/3.2) |
| Large | Barely (-/-) | Rarely (0.06/0.26) | Barely (-/3.2) |
| Environment | I | II | III | IV |
|---|---|---|---|---|
| On-Premises | Cloud | Hybrid | ||
| Platform | Acer Nitro 5 | Dell PowerEdge R740XD | VM | II + III |
| CPU (core) | 4 | 4/80 | 4 | 4 |
| Memory (GB) | 8 | 8/256 | 8 | 8 |
| Cost (USD/hour) | 0.15 | 0.67 | 0.17 | 0.42 |
| Performance (P) | 6 | 72 | 48 | 56 |
| Cost-effective Performance (Pc) | 40 | 108 | 282 | 133 |
| Compute Nodes | Distribution Policy | 20% | 40% | 80% |
|---|---|---|---|---|
| 1 | Round-robin | 7.32% | 8.22% | 7.62% |
| 1 | Job-based | 12.90% | 10.08% | 9.00% |
| 2 | Round-robin | 1.32% | 4.68% | 3.66% |
| 2 | Job-based | 4.38% | 7.68% | 6.78% |
| 3 | Round-robin | 3.54% | 3.78% | 3.66% |
| 3 | Job-based | 8.04% | 6.24% | 5.40% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pham, L.M.; Parlavantzas, N.; Le, H.-H.; Bui, Q.H. Towards a Framework for High-Performance Simulation of Livestock Disease Outbreak: A Case Study of Spread of African Swine Fever in Vietnam. Animals 2021, 11, 2743. https://doi.org/10.3390/ani11092743
Pham LM, Parlavantzas N, Le H-H, Bui QH. Towards a Framework for High-Performance Simulation of Livestock Disease Outbreak: A Case Study of Spread of African Swine Fever in Vietnam. Animals. 2021; 11(9):2743. https://doi.org/10.3390/ani11092743
Chicago/Turabian StylePham, Linh Manh, Nikos Parlavantzas, Huy-Ham Le, and Quang Hung Bui. 2021. "Towards a Framework for High-Performance Simulation of Livestock Disease Outbreak: A Case Study of Spread of African Swine Fever in Vietnam" Animals 11, no. 9: 2743. https://doi.org/10.3390/ani11092743
APA StylePham, L. M., Parlavantzas, N., Le, H.-H., & Bui, Q. H. (2021). Towards a Framework for High-Performance Simulation of Livestock Disease Outbreak: A Case Study of Spread of African Swine Fever in Vietnam. Animals, 11(9), 2743. https://doi.org/10.3390/ani11092743







