The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Viral Strains
2.2. Clinical Samples
2.3. DNA Extraction
2.4. Primers and Probes
2.5. Construction of Standard Plasmids
2.6. Optimization of the Reaction Conditions of the Multiplex Real-Time qPCR
2.7. Construction of Standard Curves
2.8. Specificity Analysis of the Multiplex Real-Time qPCR
2.9. Sensitivity Analysis of the Multiplex Real-Time qPCR
2.10. Repeatability Analysis of the Multiplex Real-Time qPCR
2.11. Test of Clinical Samples by the Multiplex Real-Time qPCR
2.12. Statistical Analysis
3. Results
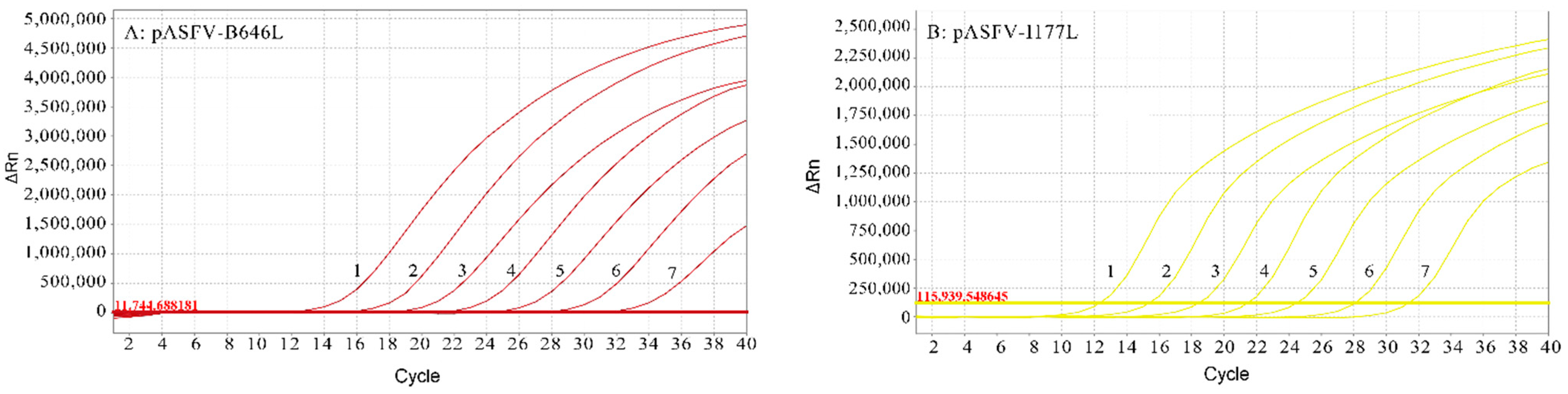
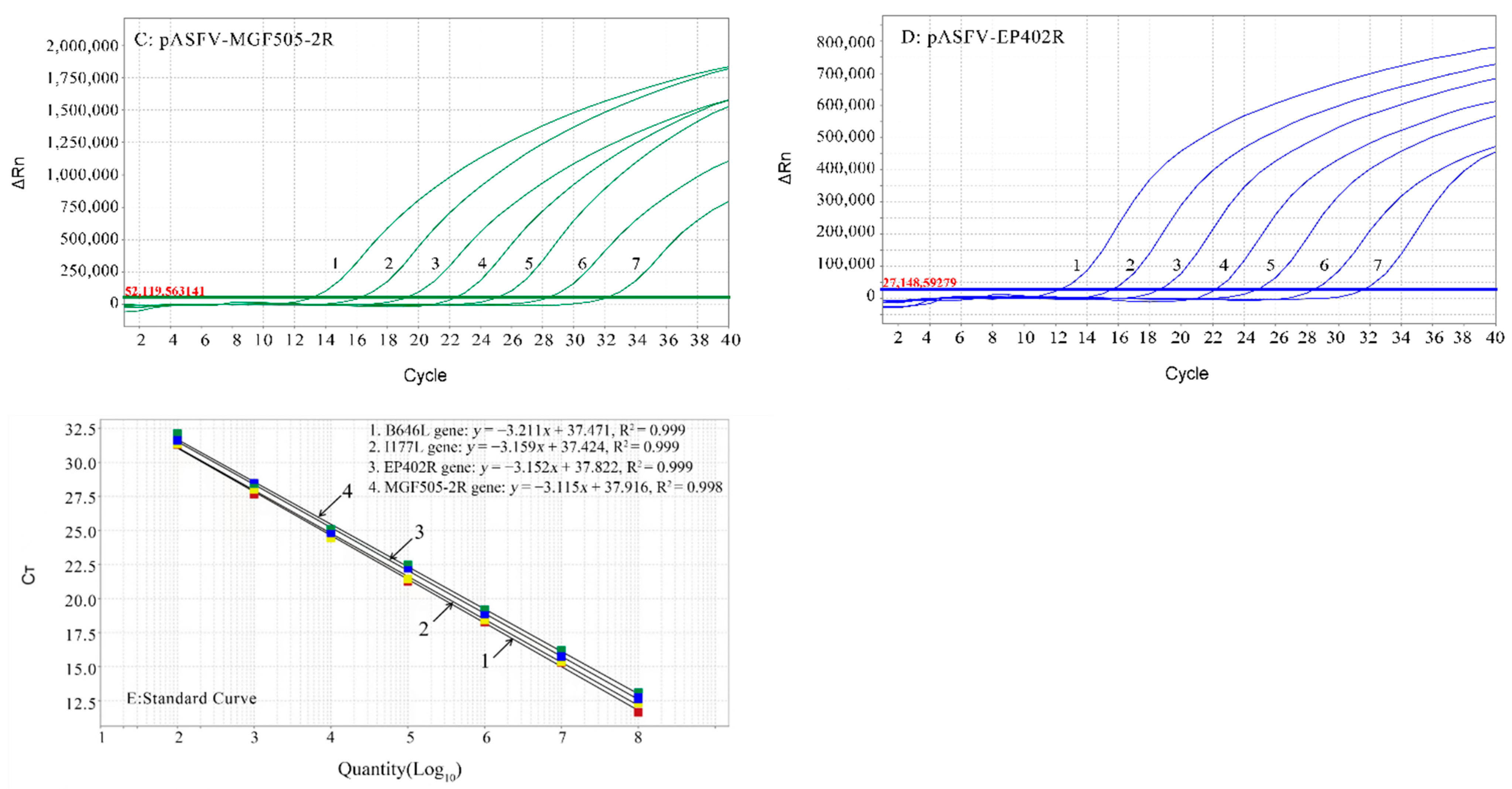
3.1. Construction of Standard Plasmids
3.2. Optimal Reaction Conditions of the Multiplex Real-Time qPCR
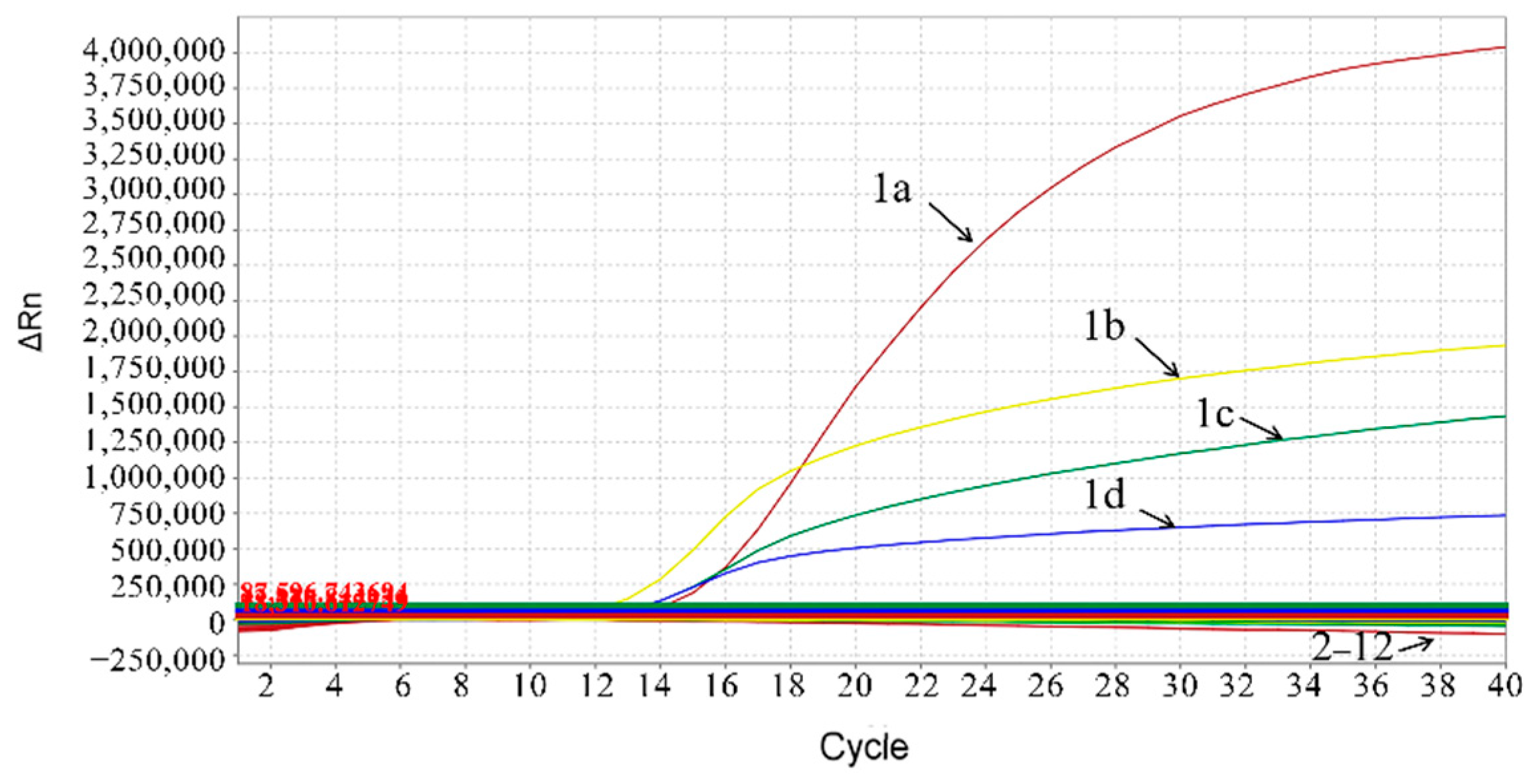
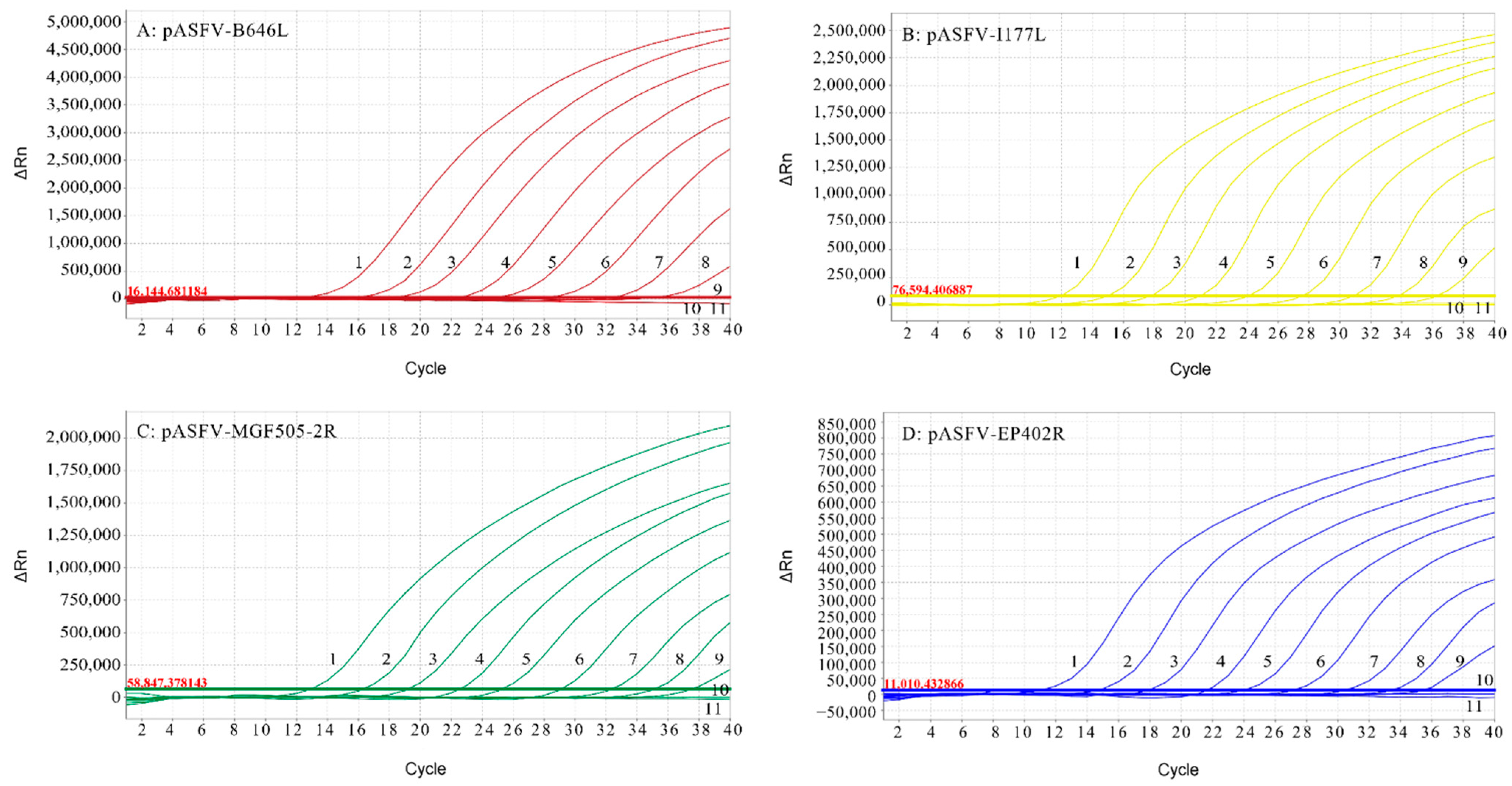
3.3. Standard Curves of the Multiplex Real-Time qPCR
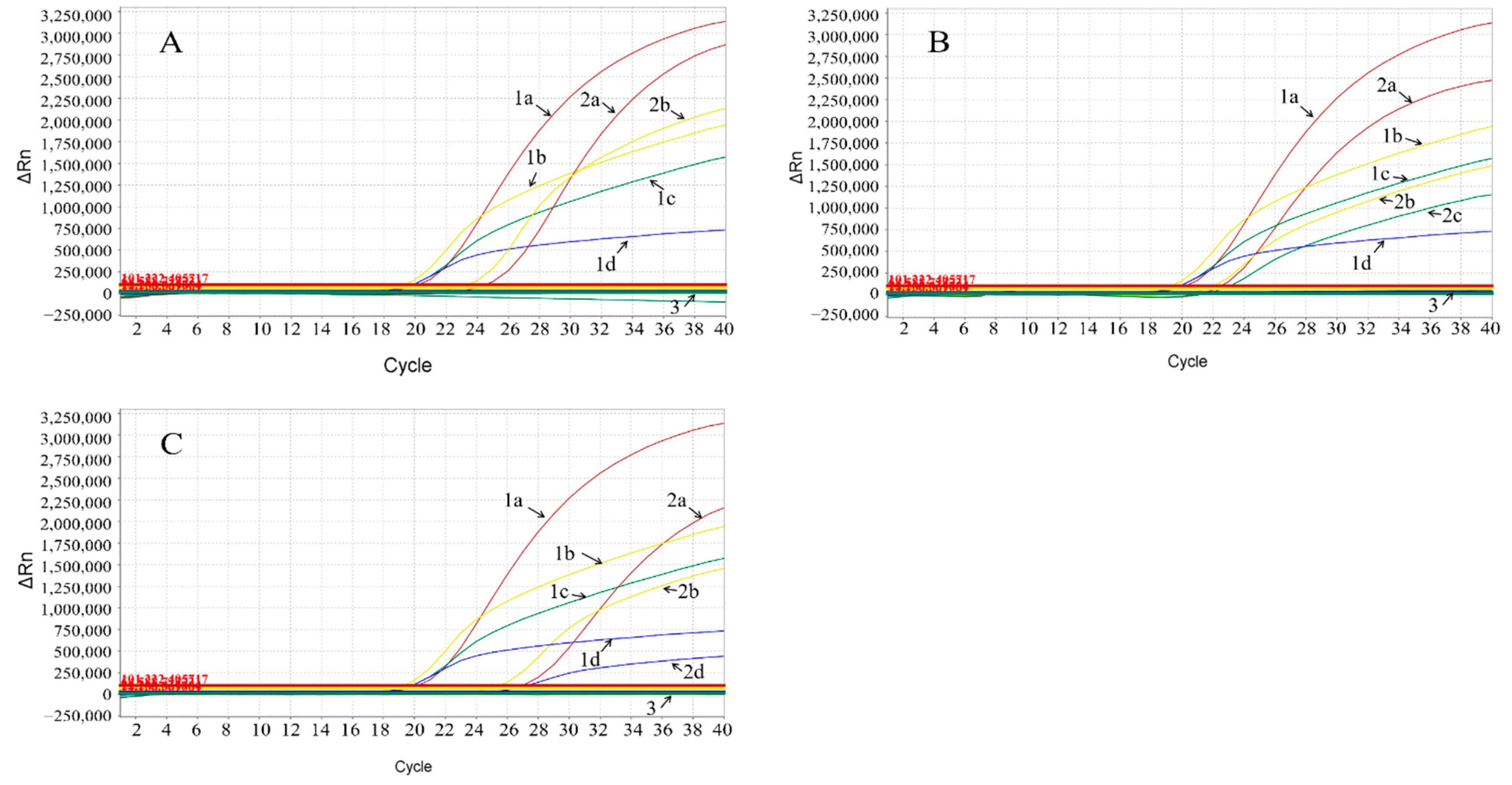
3.4. Specificity of the Multiplex Real-Time qPCR
3.5. Sensitivity of the Multiplex Real-Time qPCR
3.6. Repeatability of the Multiplex Real-Time qPCR
3.7. Evaluation of Clinical Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Galindo, I.; Alonso, C. African swine fever virus: A review. Viruses 2017, 9, 103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauter-Louis, C.; Conraths, F.J.; Probst, C.; Blohm, U.; Schulz, K.; Sehl, J.; Fischer, M.; Forth, J.H.; Zani, L.; Depner, K.; et al. African swine fever in wild boar in Europe—A review. Viruses 2021, 13, 1717. [Google Scholar] [CrossRef] [PubMed]
- Cwynar, P.; Stojkov, J.; Wlazlak, K. African swine fever status in Europe. Viruses 2019, 11, 310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Vizcaino, J.M.; Mur, L.; Martinez-Lopez, B. African swine fever: An epidemiological update. Transbound. Emerg. Dis. 2012, 59 (Suppl. S1), 27–35. [Google Scholar] [CrossRef]
- Zhou, X.; Li, N.; Luo, Y.; Liu, Y.; Miao, F.; Chen, T.; Zhang, S.; Cao, P.; Li, X.; Tian, K.; et al. Emergence of African swine fever in China, 2018. Transbound. Emerg. Dis. 2018, 65, 1482–1484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mighell, E.; Ward, M.P. African swine fever spread across Asia, 2018–2019. Transbound. Emerg. Dis. 2021, 68, 2722–2732. [Google Scholar] [CrossRef]
- Gonzales, W.; Moreno, C.; Duran, U.; Henao, N.; Bencosme, M.; Lora, P.; Reyes, R.; Núñez, R.; De Gracia, A.; Perez, A.M. African swine fever in the Dominican Republic. Transbound. Emerg. Dis. 2021, 68, 3018–3019. [Google Scholar] [CrossRef]
- OIE-WAHIS Immediate Notification: African Swine Fever Virus Haiti, Reported on 2021-09-19, Report IDIN_151732. Available online: https://wahis.oie.int/#/report-info?reportId=39928 (accessed on 19 September 2021).
- Danzetta, M.L.; Marenzoni, M.L.; Iannetti, S.; Tizzani, P.; Calistri, P.; Feliziani, F. African swine fever: Lessons to learn from past eradication experiences: A systematic review. Front. Vet. Sci. 2020, 7, 296. [Google Scholar] [CrossRef]
- Gaudreault, N.N.; Madden, D.W.; Wilson, W.C.; Trujillo, J.D.; Richt, J.A. African swine fever virus: An emerging DNA arbovirus. Front. Vet. Sci. 2020, 7, 215. [Google Scholar] [CrossRef]
- Alonso, C.; Borca, M.; Dixon, L.; Revilla, Y.; Rodriguez, F.; Escribano, J.M.; Ictv Report Consortium. ICTV virus taxonomy profile: Asfarviridae. J. Gen. Virol. 2018, 99, 613–614. [Google Scholar] [CrossRef]
- Wang, G.; Xie, M.; Wu, W.; Chen, Z. Structures and functional diversities of ASFV proteins. Viruses 2021, 13, 2124. [Google Scholar] [CrossRef] [PubMed]
- Achenbach, J.E.; Gallardo, C.; Nieto-Pelegrín, E.; Rivera-Arroyo, B.; Degefa-Negi, T.; Arias, M.; Jenberie, S.; Mulisa, D.D.; Gizaw, D.; Gelaye, E.; et al. Identification of a new genotype of African swine fever virus in domestic pigs from Ethiopia. Transbound. Emerg. Dis. 2017, 64, 1393–1404. [Google Scholar] [CrossRef] [PubMed]
- Giammarioli, M.; Gallardo, C.; Oggiano, A.; Iscaro, C.; Nieto, R.; Pellegrini, C.; Dei Giudici, S.; Arias, M.; De Mia, G.M. Genetic characterization of African swine fever viruses from recent and historical outbreaks in Sardinia (1978–2009). Virus Genes 2011, 42, 377–387. [Google Scholar] [CrossRef] [PubMed]
- Tao, D.; Sun, D.; Liu, Y.; Wei, S.; Yang, Z.; An, T.; Shan, F.; Chen, Z.; Liu, J. One year of African swine fever outbreak in China. Acta Trop. 2020, 211, 105602. [Google Scholar] [CrossRef]
- Njau, E.P.; Domelevo Entfellner, J.B.; Machuka, E.M.; Bochere, E.N.; Cleaveland, S.; Shirima, G.M.; Kusiluka, L.J.; Upton, C.; Bishop, R.P.; Pelle, R.; et al. The first genotype II African swine fever virus isolated in Africa provides insight into the current Eurasian pandemic. Sci. Rep. 2021, 11, 13081. [Google Scholar] [CrossRef]
- Sun, E.; Huang, L.; Zhang, X.; Zhang, J.; Shen, D.; Zhang, Z.; Wang, Z.; Huo, H.; Wang, W.; Huangfu, H.; et al. Genotype I African swine fever viruses emerged in domestic pigs in China and caused chronic infection. Emerg. Microbes Infect. 2021, 10, 2183–2193. [Google Scholar] [CrossRef]
- Teklue, T.; Sun, Y.; Abid, M.; Luo, Y.; Qiu, H.J. Current status and evolving approaches to African swine fever vaccine development. Transbound. Emerg. Dis. 2020, 67, 529–542. [Google Scholar] [CrossRef]
- Wu, K.; Liu, J.; Wang, L.; Fan, S.; Li, Z.; Li, Y.; Yi, L.; Ding, H.; Zhao, M.; Chen, J. Current state of global African swine fever vaccine development under the prevalence and transmission of ASF in China. Vaccines 2020, 8, 531. [Google Scholar] [CrossRef]
- Tran, X.H.; Le, T.T.P.; Nguyen, Q.H.; Do, T.T.; Nguyen, V.D.; Gay, C.G.; Borca, M.V.; Gladue, D.P. African swine fever virus vaccine candidate ASFV-G-DeltaI177L efficiently protects European and native pig breeds against circulating Vietnamese field strain. Transbound. Emerg. Dis. 2021. Early View. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Espinoza, N.; Velazquez-Salinas, L.; Gay, C.G.; Gladue, D.P. ASFV-G-I177L as an effective oral nasal vaccine against the Eurasia strain of Africa swine fever. Viruses 2021, 13, 765. [Google Scholar] [CrossRef]
- Borca, M.V.; Ramirez-Medina, E.; Silva, E.; Vuono, E.; Rai, A.; Pruitt, S.; Holinka, L.G.; Velazquez-Salinas, L.; Zhu, J.; Gladue, D.P. Development of a highly effective African swine fever virus vaccine by deletion of the I177L gene results in sterile immunity against the current epidemic Eurasia strain. J. Virol. 2020, 94, e02017-19. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, V.; Holinka, L.G.; Gladue, D.P.; Sanford, B.; Krug, P.W.; Lu, X.; Arzt, J.; Reese, B.; Carrillo, C.; Risatti, G.R.; et al. African swine fever virus Georgia isolate harboring deletions of MGF360 and MGF505 genes is attenuated in swine and confers protection against challenge with virulent parental virus. J. Virol. 2015, 89, 6048–6056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.; Zhao, D.; He, X.; Liu, R.; Wang, Z.; Zhang, X.; Li, F.; Shan, D.; Chen, H.; Zhang, J.; et al. A seven-gene-deleted African swine fever virus is safe and effective as a live attenuated vaccine in pigs. Sci. China Life Sci. 2020, 63, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo, P.L.; Lacasta, A.; López, E.; Bosch, L.; Collado, J.; Pina-Pedrero, S.; Correa-Fiz, F.; Accensi, F.; Navas, M.J.; Vidal, E.; et al. BA71DeltaCD2: A new recombinant live attenuated African swine fever virus with cross-protective capabilities. J. Virol. 2017, 91, e01058-17. [Google Scholar] [CrossRef] [Green Version]
- Lopez, E.; Bosch-Camós, L.; Ramirez-Medina, E.; Vuono, E.; Navas, M.J.; Muñoz, M.; Accensi, F.; Zhang, J.; Alonso, U.; Argilaguet, J.; et al. Deletion mutants of the attenuated recombinant ASF virus, BA71DeltaCD2, show decreased vaccine efficacy. Viruses 2021, 13, 1678. [Google Scholar] [CrossRef] [PubMed]
- Sun, E.; Zhang, Z.; Wang, Z.; He, X.; Zhang, X.; Wang, L.; Wang, W.; Huang, L.; Xi, F.; Huangfu, H.; et al. Emergence and prevalence of naturally occurring lower virulent African swine fever viruses in domestic pigs in China in 2020. Sci. China Life Sci. 2021, 64, 752–765. [Google Scholar] [CrossRef]
- Saijo, M.; Morikawa, S.; Kurane, I. Real-time quantitative polymerase chain reaction for virus infection diagnostics. Expert Opin. Med. Diagn. 2008, 2, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- King, D.P.; Reid, S.M.; Hutchings, G.H.; Grierson, S.S.; Wilkinson, P.J.; Dixon, L.K.; Bastos, A.D.; Drew, T.W. Development of a TaqMan® PCR assay with internal amplification control for the detection of African swine fever virus. J. Virol. Methods 2003, 107, 53–61. [Google Scholar] [CrossRef]
- Wang, A.; Jia, R.; Liu, Y.; Zhou, J.; Qi, Y.; Chen, Y.; Liu, D.; Zhao, J.; Shi, H.; Zhang, J.; et al. Development of a novel quantitative real-time PCR assay with lyophilized powder reagent to detect African swine fever virus in blood samples of domestic pigs in China. Transbound. Emerg. Dis. 2020, 67, 284–297. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, L.; Noll, L.; Stoy, C.; Porter, E.; Fu, J.; Feng, Y.; Peddireddi, L.; Liu, X.; Dodd, K.A.; et al. Development of a real-time PCR assay for detection of African swine fever virus with an endogenous internal control. Transbound. Emerg. Dis. 2020, 67, 2446–2454. [Google Scholar] [CrossRef]
- Trinh, T.B.N.; Truong, T.; Nguyen, V.T.; Vu, X.D.; Dao, L.A.; Nguyen, T.L.; Ambagala, A.; Babiuk, S.; Oh, J.; Song, D.; et al. Development of a novel real-time PCR assay targeting p54 gene for rapid detection of African swine fever virus (ASFV) strains circulating in Vietnam. Vet. Med. Sci. 2021, 7, 2268–2272. [Google Scholar] [CrossRef] [PubMed]
- Velazquez-Salinas, L.; Ramirez-Medina, E.; Rai, A.; Pruitt, S.; Vuono, E.A.; Espinoza, N.; Gladue, D.P.; Borca, M.V. Development real-time PCR assays to genetically differentiate vaccinated pigs from infected pigs with the Eurasian strain of African swine fever virus. Front. Vet. Sci. 2021, 8, 768869. [Google Scholar] [CrossRef] [PubMed]
- Zhan, Y.; Zhang, L.H.; Lin, Y.; Cai, Y.F.; Zou, Y.W.; Hao, Z.Y.; Luo, Z.H.; Wang, N.D.; Deng, Z.B.; Yang, Y.; et al. Development and preliminary testing of a probe-based duplex real-time PCR assay for the detection of African swine fever virus. Mol. Cell Probes 2021, 59, 101764. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Hu, Y.; Liu, P.; Zhu, Z.; Liu, P.; Chen, C.; Wu, X. Development and application of a duplex real-time PCR assay for differentiation of genotypes I and II African swine fever viruses. Transbound. Emerg. Dis. 2022. Early View. [Google Scholar] [CrossRef]
- Guo, Z.; Li, K.; Qiao, S.; Chen, X.X.; Deng, R.; Zhang, G. Development and evaluation of duplex TaqMan real-time PCR assay for detection and differentiation of wide-type and MGF505-2R gene-deleted African swine fever viruses. BMC Vet. Res. 2020, 16, 428. [Google Scholar] [CrossRef]
- Lin, Y.; Cao, C.; Shi, W.; Huang, C.; Zeng, S.; Sun, J.; Wu, J.; Hua, Q. Development of a triplex real-time PCR assay for detection and differentiation of gene-deleted and wild-type African swine fever virus. J. Virol. Methods. 2020, 280, 113875. [Google Scholar] [CrossRef] [PubMed]
- OIE. African swine fever. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals; OIE: Paris, France, 2019; Available online: https://www.oie.int/fileadmin/Home/eng/Health_standards/tahm/3.08.01_ASF.pdf (accessed on 19 September 2021).
- Wang, T.; Luo, R.; Sun, Y.; Qiu, H.J. Current efforts towards safe and effective live attenuated vaccines against African swine fever: Challenges and prospects. Infect. Dis. Poverty 2021, 10, 137. [Google Scholar] [CrossRef]
- Liu, L.; Wang, X.; Mao, R.; Zhou, Y.; Yin, J.; Sun, Y.; Yin, X. Research progress on live attenuated vaccine against African swine fever virus. Microb. Pathog. 2021, 158, 105024. [Google Scholar] [CrossRef]
- Rathakrishnan, A.; Connell, S.; Petrovan, V.; Moffat, K.; Goatley, L.C.; Jabbar, T.; Sánchez-Cordón, P.J.; Reis, A.L.; Dixon, L.K. Differential effect of deleting members of African swine fever virus multigene families 360 and 505 from the genotype II Georgia 2007/1 isolate on virus replication, virulence and induction of protection. J. Virol. 2022, 96, e0189921. [Google Scholar] [CrossRef]
- Hawkins, S.F.C.; Guest, P.C. Multiplex analyses using real-time quantitative PCR. Methods Mol. Biol. 2017, 1546, 125–133. [Google Scholar]
- Hoffmann, B.; Beer, M.; Reid, S.M.; Mertens, P.; Oura, C.A.; van Rijn, P.A.; Slomka, M.J.; Banks, J.; Brown, I.H.; Alexander, D.J.; et al. A review of RT-PCR technologies used in veterinary virology and disease control: Sensitive and specific diagnosis of five livestock diseases notifiable to the World Organization for Animal Health. Vet. Microbiol. 2009, 139, 1–23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, M.; Wang, H.H.; Hua, Y.; Qin, F.; Yang, J. African swine fever in China: Impacts, responses, and policy implications. Food Policy 2021, 192, 102065. [Google Scholar] [CrossRef]
- Mazur-Panasiuk, N.; Żmudzki, J.; Woźniakowski, G. African swine fever virus—Persistence in different environmental conditions and the possibility of its indirect transmission. J. Vet. Res. 2019, 63, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Target Gene | Name | Sequences (5′→3′) | Amplicon (bp) |
|---|---|---|---|
| B646L | P72-F | GGCGTATAAAAAGTCCAGGAAATTC | 79 |
| P72-R | TTCGGCGAGCGCTTTATC | ||
| P72-P | FAM-TCACCAAATCCTTTTGCGATGCAAGCT-BHQ1 | ||
| MGF505-2R | 505-2R-F | AGTCATGCACGGCATATACAA | 153 |
| 505-2R-R | GGTTTAAACCGTGCCACATCC | ||
| 505-2R-P | VIC-ACGCGGCCACCCAATTCAGAGAC-BHQ1 | ||
| EP402R | EP402R-F | TACTACATGCGTCCCTCAACAC | 182 |
| EP402R-R | AATGGCGGGATATTGGGTAGT | ||
| EP402R-P | CY5-ACCGTGTCCTCCACCCAAACCAT-BHQ2 | ||
| I177L | I177L-F | GGCATAATTATCAAATGCGAAGGG | 122 |
| I177L-R | TGGAAAGTTAATGATCAGGGCTT | ||
| I177L-P | Texas Red-AATCCTAGCTTGCCGGTAATGGCT-BHQ2 |
| Multiplex Real-Time qPCR | OIE-Recommended Real-Time qPCR | ||||
|---|---|---|---|---|---|
| Reagent | Volume (μL) | Final Concentration (nM) | Reagent | Volume (μL) | Final Concentration (nM) |
| Premix Ex Taq (2×) | 10 | 1× | Premix Ex Taq (2×) | 12.5 | 1× |
| P72-F (20 μM) | 0.2 | 200 | P72-P1 (50 μM) | 1 | 2000 |
| P72-R (20 μM) | 0.2 | 200 | P72-P2 (50 μM) | 1 | 2000 |
| P72-P (20 μM) | 0.2 | 200 | P72-P (5 μM) | 1 | 200 |
| I177L-F (20 μM) | 0.1 | 100 | / | / | / |
| I177L-R (20 μM) | 0.1 | 100 | / | / | / |
| I177L-P (20 μM) | 0.1 | 100 | / | / | / |
| EP402R-F (20 μM) | 0.2 | 200 | / | / | / |
| EP402R-R (20 μM) | 0.2 | 200 | / | / | / |
| EP402R-P (20 μM) | 0.2 | 200 | / | / | / |
| MGF505-2R-F (20 μM) | 0.2 | 200 | / | / | / |
| MGF505-2R-R (20 μM) | 0.2 | 200 | / | / | / |
| MGF505-2R-P (20 μM) | 0.3 | 300 | / | / | / |
| Template | 2 | / | Template | 2.5 | / |
| Distilled water | Up to 20 | / | Distilled water | Up to 25 | / |
| Plasmid | Concentration (Copies/μL) | Ct Values of Intra-Assay for Repeatability | Ct Values of Inter-Assay for Reproducibility | ||||
|---|---|---|---|---|---|---|---|
| SD | CV (%) | SD | CV (%) | ||||
| pASFV-B646L | 3.21 × 108 | 12.60 | 0.10 | 0.80 | 12.69 | 0.13 | 1.01 |
| 3.21 × 106 | 18.45 | 0.16 | 0.88 | 18.45 | 0.11 | 0.62 | |
| 3.21 × 104 | 24.53 | 0.18 | 0.71 | 24.75 | 0.30 | 1.20 | |
| pASFV-I177L | 3.21 × 108 | 12.41 | 0.07 | 0.59 | 12.45 | 0.10 | 0.84 |
| 3.21 × 106 | 18.42 | 0.17 | 0.92 | 18.37 | 0.13 | 0.69 | |
| 3.21 × 104 | 24.44 | 0.23 | 0.92 | 24.37 | 0.20 | 0.83 | |
| pASFV-MGF505-2R | 3.21 × 108 | 12.57 | 0.09 | 0.62 | 12.59 | 0.17 | 1.36 |
| 3.21 × 106 | 18.70 | 0.17 | 0.89 | 18.75 | 0.21 | 1.10 | |
| 3.21 × 104 | 24.81 | 0.11 | 0.43 | 24.79 | 0.16 | 0.63 | |
| pASFV-EP402R | 3.21 × 108 | 13.18 | 0.07 | 0.53 | 13.29 | 0.13 | 0.99 |
| 3.21 × 106 | 19.05 | 0.09 | 0.46 | 19.03 | 0.30 | 1.56 | |
| 3.21 × 104 | 25.13 | 0.07 | 0.26 | 25.19 | 0.13 | 0.53 | |
| Source | Tissue | Environmental Swab | Whole Blood | Nasopharyngeal Swab | Total |
|---|---|---|---|---|---|
| Pig farm | / | 2/107 (1.87%) c | 11/1134 (0.97%) | 3/37 (8.11%) | 15/1278 (1.25%) d |
| Slaughterhouse | 58/480 (12.08%) b | 214/752 (28.46%) a | / | / | 330/1908 (17.30%) b |
| Farmer’s market | 14/175 (8.00%) b | 53/451 (11.75%) b | / | / | 67/626 (10.70%) c |
| Harmless disposal site | 105/365 (28.77%) a | 16/62 (25.81%) a | / | / | 121/427 (28.34%) a |
| Total | 177/1020 (17.35%) B | 285/1372 (20.77%) A | 11/1134 (0.97%) D | 3/37 (8.11%) C | 534/4239 (12.60%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, K.; Shi, K.; Zhou, Q.; Xiong, C.; Mo, S.; Zhou, H.; Long, F.; Wei, H.; Hu, L.; Mo, M. The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals 2022, 12, 1754. https://doi.org/10.3390/ani12141754
Zhao K, Shi K, Zhou Q, Xiong C, Mo S, Zhou H, Long F, Wei H, Hu L, Mo M. The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals. 2022; 12(14):1754. https://doi.org/10.3390/ani12141754
Chicago/Turabian StyleZhao, Kang, Kaichuang Shi, Qingan Zhou, Chenyong Xiong, Shenglan Mo, Hongjin Zhou, Feng Long, Haina Wei, Liping Hu, and Meilan Mo. 2022. "The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus" Animals 12, no. 14: 1754. https://doi.org/10.3390/ani12141754
APA StyleZhao, K., Shi, K., Zhou, Q., Xiong, C., Mo, S., Zhou, H., Long, F., Wei, H., Hu, L., & Mo, M. (2022). The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals, 12(14), 1754. https://doi.org/10.3390/ani12141754






