Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design and Sampling
2.2. Determination of Feed and Milk Composition and Fatty Acid Profile
2.3. RNA Extraction
2.4. RNA Sequencing
2.5. Differential Expression Analysis
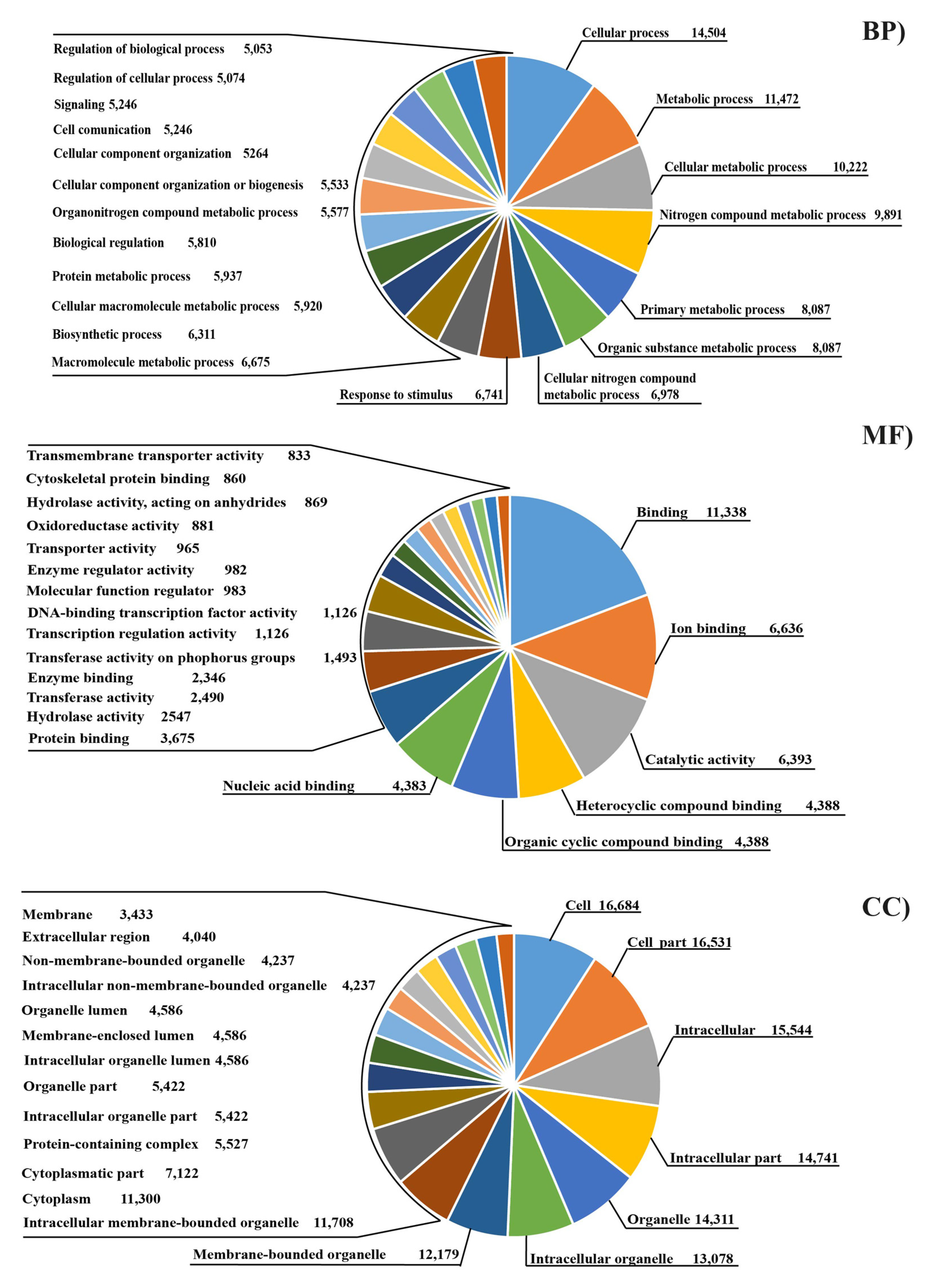
2.6. Transcriptome Functional Analysis
2.7. Statistical Analysis
3. Results
3.1. Effect of Treatment on Milk Composition and Fatty Acid Profile
3.2. cDNA Sequencing and Aligning on the Reference Predicted Transcriptome
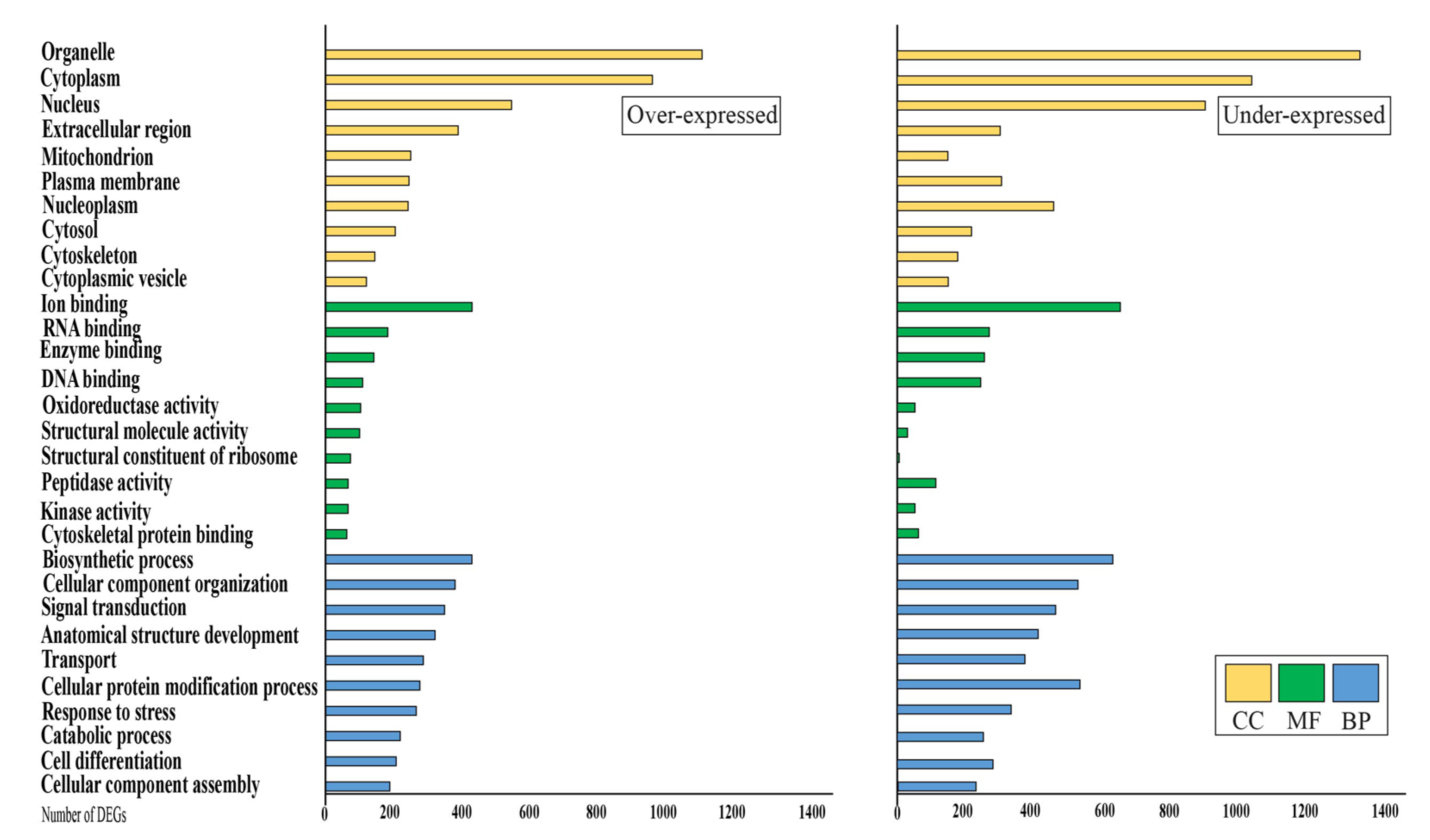
3.3. Analysis of Differential Expressed Genes
3.4. KEGG Analysis
4. Discussion
4.1. Differentially Expressed Genes in Mammary Glands
Effect of Linseed Supplementation on Energy Balance and Protein Pathways
4.2. Milk Production and Composition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conte, G.; Serra, A.; Mele, M. Dairy cow breeding and feeding on the milk fatty acid pattern. In Nutrients in Dairy and Their Implications for Health and Disease; Watson, R.R., Collier, R.J., Preedy, V.R., Eds.; Academic Press: San Diego, CA, USA, 2017; pp. 19–41. [Google Scholar]
- Mele, M. Designing milk fat to improve healthfulness and functional proprieties of dairy products: From feeding strategies to a genetic approach. Ital. J. Anim. Sci. 2009, 8 (Suppl. 2), 365–379. [Google Scholar] [CrossRef]
- Frutos, P.; Toral, P.G.; Hervás, G. Individual variation of the extent of milk fat depression in dairy ewes fed fish oil: Milk fatty acid profile and mRNA abundance of candidate genes involved in mammary lipogenesis. J. Dairy Sci. 2017, 100, 9611–9622. [Google Scholar] [CrossRef] [PubMed]
- Toral, P.G.; Hervás, G.; Belenguer, A.; Carreño, D.; Frutos, P. mRNA abundance of genes involved in mammary lipogenesis during fish oil- or trans -10, cis -12 CLA-induced milk fat depression in dairy ewes. J. Dairy Sci. 2017, 100, 3182–3192. [Google Scholar] [CrossRef] [PubMed]
- Bauman, D.E.; Griinari, J.M. Nutritional regulation of milk fat synthesis. Annu. Rev. Nutr. 2003, 23, 203–227. [Google Scholar] [CrossRef]
- Toral, P.G.; Hervás, G.; Carreño, D.; Belenguer, A.; Frutos, P. Comparison of milk fatty acid responses during fish oil- and trans-10 cis-12 18:2-induced milk fat depression in dairy ewes. Anim. Feed Sci. Technol. 2015, 210, 66–73. [Google Scholar] [CrossRef]
- Bernard, L.; Shingfield, K.G.; Rouel, J.; Ferlay, A.; Chilliard, Y. Effect of plant oils in the diet on performance and milk fatty acid composition in goats fed diets based on grass hay or maize silage. Br. J. Nutr. 2008, 101, 213–224. [Google Scholar] [CrossRef]
- Bell, J.A.; Griinari, J.M.; Kennelly, J.J. Effect of safflower oil, flaxseed oil, monensin, and vitamin E on concentration of conjugated linoleic acid in bovine milk fat. J. Dairy Sci. 2006, 89, 733–748. [Google Scholar] [CrossRef]
- Mele, M.; Serra, A.; Conte, G.; Pollicardo, A.; Del Viva, M.; Secchiari, P. Whole extruded linseed in the diet of dairy ewes during early lactation: Effect on the fatty acid composition of milk and cheese. Ital. J. Anim. Sci. 2007, 6, 560–562. [Google Scholar] [CrossRef][Green Version]
- Shingfield, K.J.; Bernard, L.; Leroux, C.; Chilliard, Y. Role of trans fatty acids in the nutritional regulation of mammary lipogenesis in ruminants. Animal 2010, 4, 1140–1166. [Google Scholar] [CrossRef] [PubMed]
- Harvatine, K.J.; Perfield, J.W.; Bauman, D.E. Expression of enzymes and key regulators of lipid synthesis is upregulated in adipose tissue during CLA induced milk fat depression in dairy cows. J. Nutr. 2009, 139, 849–854. [Google Scholar] [CrossRef]
- Ibeagha-Awemu, E.M.; Li, R.; Ammah, A.A.; Dudemaine, P.L.; Bissonnette, N.; Benchaar, C.; Zhao, X. Transcriptome adaptation of the bovine mammary gland to diets rich in unsaturated fatty acids shows greater impact of linseed oil over safflower oil on gene expression and metabolic pathways. BMC Genom. 2016, 17, 104. [Google Scholar] [CrossRef]
- Mach, N.; Jacobs, A.A.A.; Kruijt, L.; van Baal, J.; Smits, M.A. Alteration of gene expression in mammary gland tissue of dairy cows in response to dietary unsaturated fatty acids. Animal 2011, 5, 1217–1230. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Toral, P.G.; Gutiérrez-Gil, B.; Hervás, G.; Arranz, J.J.; Frutos, P. Elucidating fish oil-induced milk fat depression in dairy sheep: Milk somatic cell transcriptome analysis. Sci. Rep. 2017, 7, 45905. [Google Scholar] [CrossRef]
- Suárez-Vega, A.; Gutiérrez-Gil, B.; Toral, P.G.; Hervás, G.; Arranz, J.J.; Frutos, P. Conjugated linoleic acid (CLA)-induced milk fat depression: Application of RNA-Seq technology to elucidate mammary gene regulation in dairy ewes. Sci. Rep. 2019, 9, 4473. [Google Scholar] [CrossRef] [PubMed]
- Van Soest, P.J. Collaborative study of acid-detergent fibre and lignin. J. Assoc. Off. Anal. Chem. 1973, 56, 781–784. [Google Scholar]
- AOAC Official Method. Official Methods of Analysis, 16th ed.; AOAC International: Arlington, VA, USA, 1995. [Google Scholar]
- Christie, W.W. Preparation of ester derivatives of fatty acids for chromatographic analysis. In Advances in Lipid Methodology—Two; Christie, W.W., Ed.; Oily Press: Dundee, UK, 1993; pp. 69–111. [Google Scholar]
- Conte, G.; Dimauro, C.; Serra, A.; Macciotta, N.P.P.; Mele, M. A canonical discriminant analysis to study the association between milk fatty acids of ruminal origin and milk fat depression in dairy cows. J. Dairy Sci. 2018, 101, 6497–6510. [Google Scholar] [CrossRef] [PubMed]
- Giordani, T.; Vangelisti, A.; Conte, G.; Serra, A.; Natali, L.; Ranieri, A.; Mele, M.; Cavallini, A. Transcript profiling in the milk of dairy ewes fed extruded linseed. Genom. Data 2017, 11, 17–19. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smith, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Anders, S.; McCarthy, D.J.; Chen, Y.; Okoniewski, M.O.; Smyth, G.K.; Huber, W.; Robinson, M.D. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat. Protoc. 2013, 8, 1765–1786. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A pratical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization, and analysis in functional genomics research. Bioinformatics 2005, 10, 3674–3676. [Google Scholar] [CrossRef]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999, 27, 29–34. [Google Scholar] [CrossRef]
- Portolano, B.; Spatafora, F.; Bono, G.; Margiotta, S.; Todaro, M.; Oroleva, V.; Leto, G. Application of the Wood model to lactation curves of Comisana sheep. Small Rumin. Res. 1996, 24, 7–13. [Google Scholar] [CrossRef]
- Reynolds, C.K.; Cannon, V.L.; Loerch, S.C. Effects of forage source and supplementation with soybean and marine algal oil on milk fatty acid composition of ewes. Anim. Feed Sci. Technol. 2006, 131, 333–357. [Google Scholar] [CrossRef]
- The Gene Ontology Consotium. Gene Ontology annotations and resources. Nucleic Acids Res. 2013, 41, D530–D535. [Google Scholar] [CrossRef]
- Saez, E.; Rosenfeld, J.; Livolsi, A.; Olson, P.; Lombardo, E.; Nelson, M.; Banayo, E.; Cardiff, R.D.; Izpisua-Belmonte, J.C.; Evans, R.M. PPAR signaling exacerbates mammary gland tumor development. Genes Dev. 2004, 18, 528–540. [Google Scholar] [CrossRef]
- Bionaz, M.; Loor, J.J. Gene networks driving bovine milk fat synthesis during the lactation cycle. BMC Genom. 2008, 9, 366. [Google Scholar] [CrossRef] [PubMed]
- Flores, O.; Lu, H.; Reinberg, D. Factors involved in specific transcription by mammalian RNA polymerase II. Identification and characterization of factor IIH. J. Biol. Chem. 1992, 267, 2786–2793. [Google Scholar] [CrossRef]
- Wang, M.; Zhou, Z.; Khan, M.J.; Gao, J.; Loor, J.J. Clock circadian regulator (CLOCK) gene network expression patterns in bovine adipose, liver, and mammary gland at 3 time points during the transition from pregnancy into lactation. J. Dairy Sci. 2015, 98, 4601–4612. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chen, R.; Kim, H.; Etchegaray, J.P.; Weaver, D.R.; Lee, C. The period of the circadian oscillator is primarily determined by the balance between casein kinase 1 and protein phosphatase 1. Proc. Natl. Acad. Sci. USA 2011, 108, 16451–16456. [Google Scholar] [CrossRef]
- Liu, Z.; Huang, M.; Wu, X.; Shi, G.; Xing, L.; Dong, Z.; Qu, Z.; Yan, J.; Yang, L.; Panda, S.; et al. PER1 phosphorylation specifies feeding rhythm in mice. Cell Rep. 2014, 7, 1509–1520. [Google Scholar] [CrossRef]
- Bauman, D.; Griinari, J. Regulation and nutritional manipulation of milk fat: Low-fat milk syndrome. Livest. Prod. Sci. 2001, 70, 15–29. [Google Scholar] [CrossRef]
- Pettersson, A.M.L.; Stenson, B.M.; Lorente-Cebrian, S.; Andersson, D.P.; Mejhert, N.; Kratzel, J.; Astrom, G.; Dahlman, I.; Chibalin, A.V.; Arner, P.; et al. LXR is a nega- tive regulator of glucose uptake in human adipocytes. Diabetologia 2013, 56, 2044–2054. [Google Scholar] [CrossRef] [PubMed]
- Neville, M.C.; Webb, P.; Ramanathan, P.; Mannino, M.P.; Pecorini, C.; Monks, J. The insulin receptor plays an important role in secretory differentiation in the mammary gland. Am. J. Physiol. Endocrinol. Metab. 2013, 305, E1103–E1114. [Google Scholar] [CrossRef]
- Menzies, K.K.; Lefèvre, C.; Macmillan, K.L.; Nicholas, K.R. Insulin regulates milk protein synthesis at multiple levels in the bovine mammary gland. Funct. Integr. Genom. 2009, 9, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.E.; Traugh, J.A. Insulin stimulation of phosphorylation of elongation factor 1 (eEF-1) enhances elongation activity. Eur. J. Biochem. 1998, 251, 201–207. [Google Scholar] [CrossRef]
- Redpath, N.T.; Price, N.T.; Proud, C.G. Cloning and expression of cDNA encoding protein synthesis elongation factor-2 kinase. J. Biol. Chem. 1996, 271, 17547–17554. [Google Scholar] [CrossRef] [PubMed]
- Bionaz, M.; Periasami, K.; Rodriguez-Zas, S.L.; Everts, R.E.; Lewin, H.A.; Hurley, W.L.; Loor, J.J. Old and new stories: Revelation from functional analysisof the bovine mammary transcriptome during the lactation cycle. PLoS ONE 2012, 7, e33268. [Google Scholar] [CrossRef]
- Caroprese, M.; Ciliberti, M.G.; Albenzio, M.; Annicchiarico, G.; Sevi, A. Dietary polyunsaturated fatty acids from flaxseed affect immune responses of dairy sheep around parturition. Vet. Immunol. Immunopathol. 2015, 168, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Lessard, M.; Gagnon, N.; Petit, H.V. Immune Response of Postpartum Dairy Cows Fed Flaxseed. J. Dairy Sci. 2003, 86, 2647–2657. [Google Scholar] [CrossRef]
- Paten, A.M.; Duncan, E.J.; Pain, S.J.; Peterson, S.W.; Kenyon, P.R.; Blair, H.T.; Dearden, K. Functional development of the adult ovine mammary gland—insights from gene expression profiling. BMC Genom. 2015, 16, 748. [Google Scholar] [CrossRef]
- Liu, H.; Weng, W.; Guo, R.; Zhou, J.; Xue, J.; Zhong, S.; Cheng, J.; Zhu, M.X.; Pan, S.J.; Li, Y. Olig2 SUMOylation protects against genotoxic damage response by antagonizing p53 gene targeting. Cell Death Differ. 2020, 27, 3146–3161. [Google Scholar] [CrossRef]
- Pal, P.; Hales, K.; Petrik, J.; Hales, D.B. Pro-apoptotic and anti-angiogenic actions of 2-methoxyestradiol and docosahexaenoic acid, the biologically derived active compounds from flaxseed diet, in preventing ovarian cancer. J. Ovarian Res. 2019, 12, 49. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef]
- Zhao, Z.Q.; Wang, L.J.; Sun, X.W.; Zhang, J.J.; Zhao, Y.J.; Na, R.S.; Zhang, J.H. Transcriptome Analysis of the Capra hircus Ovary. PLoS ONE 2015, 10, e0121586. [Google Scholar] [CrossRef] [PubMed]
- Boyer, P.D. The ATP synthase—A splendid molecular machine. Annu. Rev. Biochem. 1997, 66, 717–749. [Google Scholar] [CrossRef]
- Zhang, R.H.; Mustafa, A.F.; Zhao, X. Effects of feeding oilseeds rich in linoleic and linolenic fatty acids to lactating ewes on cheese yield and on fatty acid composition of milk and cheese. Anim. Feed Sci. Tech. 2006, 127, 220–233. [Google Scholar] [CrossRef]
- Gómez Cortés, P.; Bach., A.; Luna, P.; Juárez, M.; de la Fuente, M.A. Effects of extruded linseed supplementation on n-3 fatty acids and conjugated linoleic acid in milk and cheese from ewes. J. Dairy Sci. 2009, 92, 4122–4134. [Google Scholar] [CrossRef]
- Jonson, K.A.; Kincaid, R.L.; Westberg, H.H.; Gaskins, C.T.; Lams, B.K.; Cronrath, J.D. The effect of oilseeds in diets of lactating cows on milk production and methane emissions. J. Dairy Sci. 2002, 85, 1509–1515. [Google Scholar] [CrossRef]
- Kitessa, S.M.; Peake, D.; Bencini, R.; Williams, A.J. Fish oil metabolism in ruminants. Part III: Transfer ofn-3 polyunsaturated fatty acids (PUFA) from tuna oil into sheep’s milk. Anim. Feed Sci. Technol. 2003, 108, 1–14. [Google Scholar] [CrossRef]
- Gonthier, C.; Mustafa, A.F.; Berthiaume, R.; Petit, H.; Ouellet, D. Effects of feeding extruded and micronized flaxseed on blood profile and milk fatty acid composition of dairy cows. J. Dairy Sci. 2005, 88, 748–756. [Google Scholar] [CrossRef]
- Finucane, K.A.; McFadden, T.B.; Bond, J.P.; Kennelly, J.J.; Zhao, F.Q. Onset of lactation in the bovine mammary gland: Gene expression profiling indicates a strong inhibition of gene expression in cell proliferation. Funct. Integr. Genom. 2008, 8, 251–264. [Google Scholar] [CrossRef]
- Faulconnier, Y.; Bernard, L.; Boby, C.; Domagalski, J.; Chilliard, Y.; Leroux, C. Extruded linseed alone or in combination with fish oil modifies mammary gene expression profiles in lactating goats. Animal 2018, 12, 1564–1575. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Beaudoin, F.; Ammah, A.; Bissonnette, N.; Benchaar, C.; Zhao, X.; Lei, C.; Ibeagha-Awemu, E.M. Deep sequencing shows microRNA involvement in bovine mammary gland adaptation to diets supplemented with linseed oil or safflower oil. BMC Genom. 2015, 16, 884. [Google Scholar] [CrossRef]
- Merida, I.; Avila-Flores, A.; Merino, E. Diacylglycerol kinases: At the hub of cell signalling. Biochem. J. 2008, 409, 1–18. [Google Scholar] [CrossRef]
- Harvatine, K.J.; Bauman, D.E. SREBP1 and thyroid hormone responsive spot 14 (S14) are involved in the regulation of bovine mammary lipid synthesis during diet-induced milk fat depression and treatment with CLA. J. Nutr. 2006, 136, 2468–2474. [Google Scholar] [CrossRef]
- Palmquist, D.L.; Lock, A.L.; Shingfield, K.J.; Bauman, D.E. Biosynthesis of conjugated linoleic acid in ruminants and humans. Adv. Food Nutr. Res. 2005, 50, 179–217. [Google Scholar]
- Mamedova, L.K.; Robbins, K.; Johnson, B.J.; Bradford, B.J. Tissue expression of angiopoietin-like protein 4 in cattle. J. Anim. Sci. 2010, 88, 124–130. [Google Scholar] [CrossRef]
- Koltes, D.A.; Spurlock, D.M. Adipose tissue angiopoietin-like protein 4 messenger RNA changes with altered energy balance in lactating Holstein cows. Domest. Anim. Endocrin. 2012, 43, 307–316. [Google Scholar] [CrossRef]
- Köster, A.; Chao, Y.B.; Mosior, M.; Ford, A.; Gonzalez-DeWhitt, P.A.; Hale, J.E.; Li, D.; Qiu, Y.; Fraser, C.C.; Yang, D.D.; et al. Transgenic Angiopoietin-Like (Angptl)4 overexpression and targeted disruption of angptl4 and angptl3: Regulation of triglyceride metabolism. Endocrinology 2005, 146, 4943–4950. [Google Scholar] [CrossRef] [PubMed]
- Bursten, S.L.; Harris, W.E.; Bomsztyk, K.; Lovett, D. Interleukin-1 rapidly stimulates lysophosphatidate acyltransferase and phosphatidate phosphohydrolase activities in human mesangial cells. J. Biol. Chem. 1991, 266, 20732–20743. [Google Scholar] [CrossRef]

| C 1 | L 2 | SE 3 | p-Value | ||
|---|---|---|---|---|---|
| n = 8 | n = 8 | ||||
| Milk production | g/d | 1148.36 | 798.55 | 68.91 | <0.001 |
| Fat | g/100 g of milk | 7.35 | 7.85 | 0.26 | 0.211 |
| Lactose | g/100 g of milk | 4.34 | 4.40 | 0.06 | 0.424 |
| Proteins | g/100 g of milk | 6.51 | 6.49 | 0.11 | 0.942 |
| Fat secretion | mg | 83.99 | 62.70 | 5.85 | 0.004 |
| Lactose secretion | mg | 50.51 | 35.39 | 3.35 | 0.003 |
| Protein secretion | mg | 73.88 | 51.22 | 4.07 | <0.001 |
| C4:0 | g/100 g of total lipids | 3.11 | 2.62 | 0.07 | <0.001 |
| C6:0 | g/100 g of total lipids | 2.52 | 1.45 | 0.06 | <0.001 |
| C8:0 | g/100 g of total lipids | 2.41 | 1.16 | 0.08 | <0.001 |
| C10:0 | g/100 g of total lipids | 7.43 | 3.23 | 0.27 | <0.001 |
| C10:1c9 | g/100 g of total lipids | 0.29 | 0.11 | 0.01 | <0.001 |
| C11:0 | g/100 g of total lipids | 0.07 | 0.03 | 0.01 | <0.001 |
| C12:0 | g/100 g of total lipids | 4.28 | 1.96 | 0.16 | <0.001 |
| C13:0iso | g/100 g of total lipids | 0.03 | 0.01 | 0.00 | <0.001 |
| C13:0ante | g/100 g of total lipids | 0.01 | 0.01 | 0.00 | 0.202 |
| C12:1c11 | g/100 g of total lipids | 0.06 | 0.02 | 0.00 | <0.001 |
| C13:0 | g/100 g of total lipids | 0.06 | 0.04 | 0.00 | <0.001 |
| C14:0iso | g/100 g of total lipids | 0.12 | 0.07 | 0.00 | <0.001 |
| C14:0 | g/100 g of total lipids | 8.63 | 5.89 | 0.24 | <0.001 |
| C15:0iso | g/100 g of total lipids | 0.27 | 0.14 | 0.01 | <0.001 |
| C15:0ante | g/100 g of total lipids | 0.44 | 0.27 | 0.01 | <0.001 |
| C14:1c9 | g/100 g of total lipids | 0.17 | 0.10 | 0.01 | <0.001 |
| C15:0 | g/100 g of total lipids | 0.94 | 0.71 | 0.02 | <0.001 |
| C16:0iso | g/100 g of total lipids | 0.27 | 0.14 | 0.01 | <0.001 |
| C16:0 | g/100 g of total lipids | 19.93 | 15.62 | 0.54 | <0.001 |
| C16:1t6/7 | g/100 g of total lipids | 0.02 | 0.06 | 0.00 | <0.001 |
| C16:1t9 | g/100 g of total lipids | 0.04 | 0.32 | 0.01 | <0.001 |
| C17:0iso | g/100 g of total lipids | 0.37 | 0.26 | 0.01 | <0.001 |
| C16:1c7 | g/100 g of total lipids | 0.23 | 0.26 | 0.01 | 0.035 |
| C16:1c9 | g/100 g of total lipids | 0.74 | 0.52 | 0.03 | <0.001 |
| C17:0ante | g/100 g of total lipids | 0.42 | 0.22 | 0.01 | <0.001 |
| C16:1c13 | g/100 g of total lipids | 0.13 | 0.04 | 0.01 | <0.001 |
| C17:0 | g/100 g of total lipids | 0.59 | 0.43 | 0.02 | <0.001 |
| C17:1c9 | g/100 g of total lipids | 0.24 | 0.12 | 0.01 | <0.001 |
| C18:0 | g/100 g of total lipids | 5.20 | 8.87 | 0.35 | <0.001 |
| C18:1t6-8 | g/100 g of total lipids | 0.17 | 0.96 | 0.03 | <0.001 |
| C18:1t9 | g/100 g of total lipids | 0.15 | 0.72 | 0.02 | <0.001 |
| C18:1t10 | g/100 g of total lipids | 0.21 | 0.81 | 0.03 | <0.001 |
| C18:1t11 | g/100 g of total lipids | 0.67 | 4.94 | 0.17 | <0.001 |
| C18:1t12 | g/100 g of total lipids | 0.24 | 0.83 | 0.02 | <0.001 |
| C18:1c9 | g/100 g of total lipids | 15.39 | 21.81 | 0.55 | <0.001 |
| C18:1t15 | g/100 g of total lipids | 0.15 | 0.46 | 0.02 | <0.001 |
| C18:1c11 | g/100 g of total lipids | 0.34 | 0.45 | 0.01 | <0.001 |
| C18:1c12 | g/100 g of total lipids | 0.23 | 0.29 | 0.01 | <0.001 |
| C18:1t16 | g/100 g of total lipids | 0.19 | 0.51 | 0.02 | <0.001 |
| C18:1c14 | g/100 g of total lipids | 0.04 | 0.09 | 0.00 | <0.001 |
| C18:2n6 | g/100 g of total lipids | 3.03 | 2.06 | 0.11 | <0.001 |
| C18:3n3 | g/100 g of total lipids | 1.41 | 2.18 | 0.10 | <0.001 |
| C18:2c9t11 | g/100 g of total lipids | 0.47 | 2.38 | 0.08 | <0.001 |
| C22:0 | g/100 g of total lipids | 0.16 | 0.15 | 0.01 | 0.196 |
| C20:3n6 | g/100 g of total lipids | 0.03 | 0.01 | 0.00 | <0.001 |
| C20:3n3 | g/100 g of total lipids | 0.03 | 0.02 | 0.00 | 0.043 |
| C20:4n6 | g/100 g of total lipids | 0.22 | 0.10 | 0.01 | <0.001 |
| C23:0 | g/100 g of total lipids | 0.12 | 0.09 | 0.01 | 0.011 |
| C20:5n3 | g/100 g of total lipids | 0.09 | 0.06 | 0.00 | <0.001 |
| C24:0 | g/100 g of total lipids | 0.08 | 0.07 | 0.00 | 0.832 |
| C22:3n3 | g/100 g of total lipids | 0.02 | 0.01 | 0.00 | 0.003 |
| C22:5n3 | g/100 g of total lipids | 0.19 | 0.09 | 0.00 | <0.001 |
| C22:6n3 | g/100 g of total lipids | 0.07 | 0.04 | 0.00 | <0.001 |
| SFA | g/100 g of total lipids | 57.45 | 43.43 | 1.02 | <0.001 |
| MUFA | g/100 g of total lipids | 19.67 | 33.43 | 0.74 | <0.001 |
| PUFA | g/100 g of total lipids | 5.60 | 6.97 | 0.23 | <0.001 |
| PUFAn6 | g/100 g of total lipids | 3.28 | 2.12 | 0.11 | <0.001 |
| PUFAn3 | g/100 g of total lipids | 1.84 | 2.45 | 0.10 | <0.001 |
| BCFA | g/100 g of total lipids | 1.92 | 1.12 | 0.04 | <0.001 |
| OCFA | g/100 g of total lipids | 3.19 | 2.08 | 0.07 | <0.001 |
| n6/n3 | 1.87 | 0.87 | 0.07 | <0.001 | |
| DI10 4 | 0.04 | 0.03 | 0.00 | <0.001 | |
| DI14 4 | 0.02 | 0.02 | 0.00 | 0.016 | |
| DI18 4 | 0.75 | 0.71 | 0.01 | 0.013 | |
| DI/RA 4 | 0.43 | 0.33 | 0.01 | <0.001 |
| Library | Number of Raw Reads | Number of Trimmed Reads | Number of Mapped Reads on Ovis aries Reference Transcriptome | Percentage of Mapped Reads on Ovis aries Reference Transcriptome |
|---|---|---|---|---|
| C1 | 28,098,370 | 25,682,138 | 24,120,413 | 93.92% |
| C2 | 26,880,020 | 23,336,744 | 20,150,560 | 86.35% |
| C3 | 48,397,516 | 39,530,734 | 37,211,397 | 94.13% |
| C4 | 25,537,932 | 20,359,560 | 16,085,295 | 79.01% |
| L1 | 45,305,912 | 23,821,170 | 20,585,520 | 86.42% |
| L2 | 34,764,246 | 11,941,984 | 10,888,233 | 91.18% |
| L3 | 26,732,292 | 24,451,846 | 22,165,383 | 90.65% |
| L4 | 46,172,488 | 18,620,748 | 16,039,894 | 86.14% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Conte, G.; Giordani, T.; Vangelisti, A.; Serra, A.; Pauselli, M.; Cavallini, A.; Mele, M. Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed. Animals 2021, 11, 2707. https://doi.org/10.3390/ani11092707
Conte G, Giordani T, Vangelisti A, Serra A, Pauselli M, Cavallini A, Mele M. Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed. Animals. 2021; 11(9):2707. https://doi.org/10.3390/ani11092707
Chicago/Turabian StyleConte, Giuseppe, Tommaso Giordani, Alberto Vangelisti, Andrea Serra, Mariano Pauselli, Andrea Cavallini, and Marcello Mele. 2021. "Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed" Animals 11, no. 9: 2707. https://doi.org/10.3390/ani11092707
APA StyleConte, G., Giordani, T., Vangelisti, A., Serra, A., Pauselli, M., Cavallini, A., & Mele, M. (2021). Transcriptome Adaptation of the Ovine Mammary Gland to Dietary Supplementation of Extruded Linseed. Animals, 11(9), 2707. https://doi.org/10.3390/ani11092707







