Simple Summary
The superfamily Helicoidea is a large and diverse group of Eupulmonata. The superfamily has been the subject of several molecular and phylogenetic studies which greatly improved our knowledge on the evolutionary relationships and historical biogeography of many families. In contrast, the available karyological information on Helicoidea still results in an obscure general picture, lacking a homogeneous methodological approach and a consistent taxonomic record. Nevertheless, the available karyological information highlights the occurrence of a significant chromosomal diversity in the superfamily in terms of chromosome number (varying from 2n = 40 to 2n = 62), chromosome morphology and the distribution of different karyological features among different taxonomic groups. Here we performed a molecular and a comparative cytogenetic analysis on of 15 Helicoidea species of three different families. Furthermore, to provide an updated assessment of the chromosomal diversity of the superfamily we reviewed all the available chromosome data. Finally, superimposing all the chromosome data gathered from different sources on the available phylogenetic relationships of the studied taxa, we discuss the overall observed chromosome diversity in Helicoidea and advance a hypothesis on its chromosomal evolution.
Abstract
We performed a molecular and a comparative cytogenetic analysis on different Helicoidea species and a review of all the available chromosome data on the superfamily to provide an updated assessment of its karyological diversity. Standard karyotyping, banding techniques, and Fluorescence in situ hybridization of Nucleolus Organizer Region loci (NOR-FISH) were performed on fifteen species of three families: two Geomitridae, four Hygromiidae and nine Helicidae. The karyotypes of the studied species varied from 2n = 44 to 2n = 60, highlighting a high karyological diversity. NORs were on a single chromosome pair in Cernuella virgata and on multiple pairs in four Helicidae, representing ancestral and derived conditions, respectively. Heterochromatic C-bands were found on pericentromeric regions of few chromosomes, being Q- and 4′,6-diamidino-2-phenylindole (DAPI) negative. NOR-associated heterochromatin was C-banding and chromomycin A3 (CMA3) positive. Considering the available karyological evidence on Helicoidea and superimposing the chromosome data gathered from different sources on available phylogenetic inferences, we describe a karyotype of 2n = 60 with all biarmed elements as the ancestral state in the superfamily. From this condition, an accumulation of chromosome translocations led to karyotypes with a lower chromosome number (2n = 50–44). This process occurred independently in different lineages, while an augment of the chromosome number was detectable in Polygyridae. Chromosome inversions were also relevant chromosome rearrangements in Helicoidea, leading to the formation of telocentric elements in karyotypes with a relatively low chromosome count.
1. Introduction
The land snails of the superfamily Helicoidea include about 5600 species, constituting a large and diverse group of the about 25,000 Eupulmonata so far described [1,2]. The superfamily has an almost worldwide distribution, being absent only in sub-Saharan continental Africa, southern South America, some Pacific islands, and New Zealand [3,4]. The complex classification and taxonomy of the Helicoidea have been revised several times [5,6,7,8,9], and the species of the superfamily are currently distributed in 16 families, 37 subfamilies, and 359 genera [1].
Helicoidea, due to their peculiar natural history and historical biogeography, are interesting models for studies on evolutionary dynamics, and recent molecular works have started to provide more accurate representations of their evolutionary relationships (e.g., [10,11,12,13]). This is particularly true for several families and subfamilies, whose phylogenetic relationships have been described in several focused works (e.g., [10,13,14]), and more in general for the Helicoidea of the Western Palearctic, whose classification and phylogeny have been recently revised [9]. In particular, Razkin et al. [9] proposed an updated classification and phylogenetic relationships of the western Palearctic Helicoidea, confirming the taxonomic validity of many morphologically defined families and re-defining the systematic boundaries of many different groups respecting the monophyly of families, subfamilies, and tribes [9]. In addition, the origin of the whole superfamily Helicoidea was estimated in the early Cretaceous period, while its families were estimated to be from Late-Cretaceous to Paleogene period [9].
In contrast to a progressively clearer phylogenetic scenario emerging from recent molecular studies, the available karyological information on the superfamily is scattered among older and more recent papers, lacking a homogeneous methodological approach and a consistent taxonomic record, and thus resulting in an obscure general picture. Nevertheless, the available karyological information highlights the occurrence of a significant chromosomal diversity in the superfamily in terms of chromosome number (varying from 2n = 40 to 2n = 62), chromosome morphology, and the distribution of different karyological features among different taxonomic groups (see e.g., [15,16]).
Historically, two different reviews have summarized chromosome information on mollusks in general [17] and gastropods [16], helping to elucidate their overall karyological diversity. However, in Patterson [17] there was some missing information concerning previously described karyotypes of camenids and polygirids (see [18,19]), while in Thiriot-Quiévreux [16], which included published karyological information from Patterson [17] in 2002, missing data involve several different evolutionary lineages (see [20,21,22,23,24,25]). The whole picture on the available chromosome diversity of the Helicoidea thus appears still incomplete and future research as well as evolutionary inferences on the overall karyological diversity of the subfamily would benefit from a new, updated assessment of the available data. Indeed, karyological data, especially when linked to molecular inferences, can be valuable tools to individuate plesio- and apomorphic states, identify and characterize different evolutionary lineages, and to assess taxonomic uncertainties (see e.g., [26,27,28]). The usefulness of cytogenetic studies in evolutionary and taxonomic inferences has been recently shown in different mollusk taxa, highlighting the main evolutionary events of their karyotype diversification (e.g., [29,30,31]), but they usually involved low level taxonomic groups or just a handful of related species.
In this study, we performed a molecular and a comparative karyological analysis with standard, Quinacrine (Q-) staining, DAPI- and CMA3 banding, sequential C banding + fluorochromes, and NOR-FISH on land snail species belonging to three different Helicoidea families (Hygromiidae, Geomitridae, and Helicidae). To provide a molecular taxonomic attribution of the studied specimens, we also performed a molecular analysis using a segment of the mitochondrial 16S rRNA, which has been largely used in previous molecular studies on Helicoidea [9,13,14,32,33,34]. Furthermore, to provide an updated assessment of the chromosomal diversity of the superfamily, we reviewed all the available literature from 1946 to 2021 using an updated taxonomy and nomenclature following World Register of Marine Species (WoRMS) [1] and Mollusca Base [2]. Finally, superimposing all the chromosome data gathered from different sources on the available phylogenetic relationships of the studied taxa, we discuss the overall observed chromosome diversity in the superfamily and in different taxonomic groups and advance a hypothesis on their chromosomal evolution.
2. Materials and Methods
2.1. Sampling
We analyzed a total of 29 specimens of 15 Helicoidea species, including two Geomitridae, nine Helicidae, and four Hygromiidae. Samples were first morphologically determined using conchological and anatomical characters following different sources [35,36,37,38,39,40,41], and subsequently analyzed by molecular methods as described below. A complete list of the studied samples, sampling localities, and their relative taxonomic attribution after morphological and molecular analyses is reported in Table 1.

Table 1.
Number and provenance of the examined specimens of Helicoidae. Biological samples (methanol and acetic acid fixed cell suspensions) are deposited in the Molluscan collection of the Department of Biology, University of Naples Federico II. * Present study; Chr. Nr = chromosome number.
For comparative purposes, and to provide an updated evaluation of all the available chromosomal data on Helicoidea, we reviewed all the previously published karyotypes of the superfamily using an updated nomenclature following WoRMS [1] and Mollusca Base [2]. A complete list of all the considered karyotypes, taxonomic attribution, and relative references, including a total of 244 chromosome data for 205 species, 97 genera and 8 families, is provided in Table S1.
2.2. Molecular Analysis
DNA was extracted from foot tissue samples following Sokolov [42]. For molecular analysis, we choose the mitochondrial 16S rRNA as the selected genetic marker considering its wide use in previous molecular studies on Helicoidea (e.g., [9,13,14,32,33,34]) and its adequate taxon sampling available on GenBank. A mitochondrial segment of 16S rRNA of about 600 bp was amplified using the primer pair 16Sa (CGCCTGTTTATCAAAAACAT) and 16Sb (CCGGTCTGAAACTCAGATCAGT) [43]. PCR parameters: initial denaturation at 94 °C for 5 min, 36 cycles at 94 °C for 30 s, 50 °C and 72 °C for 45 s followed by a final step at 72 °C for 7 min. Amplicons were sequenced on an automated sequencer ABI 377 (Applied Biosystems, Foster City, CA, USA) using BigDye Terminator 3.1 (ABI). Chromatograms were checked and edited using Chromas Lite 2.3.1 (Technelysium Pty Ltd, Brisbane, Australia) and BioEdit 7.2.6.1 [44]. All the newly determined sequences were deposited in GenBank (accession numbers: MZ504244-MZ504269).
2.3. Cytogenetic Analysis
Specimens were injected with colchicine (1 mg/mL; 0.1 mL/10 g body weight) and after three hours were killed by immersion in water. Cell suspensions were obtained from gonads as described in [30]. In brief, the gonads were incubated for 30 min in hypotonic solution (KCl 0.075 M and sodium citrate 0.5% 1:1) and fixed for 15 min in methanol-acetic acid, 3:1. Cells were dissociated manually on a steel sieve and 25 μL of chromosome suspension was sprinkled on the slides. Chromosomes were obtained with the air-drying method [45], stained with traditional 5% Giemsa solution at pH 7 and different other staining and banding techniques: Quinacrine (Q) banding according to Schmid [46], CMA3/Methyl green (CMA3/MG) according to Sahar and Latt [47], C-banding according to Sumner [48] but performing the denaturation step with Ba(OH)2 for two min at room temperature, and sequential C-banding + Fluorochromes (CMA3 + DAPI) [49]. NOR-FISH was performed according to [50], using as a probe the 18S rRNA of the Antarctic scallop Adamussium colbecki (Smith, 1902). Ten plates per studied sample were used for karyotype reconstruction and the calculation of relative length (RL) and centromeric index (CI) (Table S2). Chromosome were classified in m = metacentric, sm = submetacentric, st = subtelocentric, and t = telocentric [51].
3. Results
3.1. Molecular Analysis and Taxonomic Attribution
Successful PCR amplifications were obtained for all the examined specimens except for Eobania vermiculata and Campylaea planospira, as their DNA were highly degraded. After searches in Basic Local Alignment Search Tool (BLAST) [52], the newly determined 16S sequences showed an identity versus deposited GenBank sequences ranging from 85.9% to 100%, allowing us to provide the final molecular taxonomic attributions reported in Table 1.
3.2. Karyotype Description
3.2.1. Family Higromiidae
The studied Monacha sp. specimen had a karyotype of 2n = 46 chromosomes gradually decreasing in length. All pairs are metacentric, excluding pairs 2 and 22 that are submetacentric (Figure 1; Table S2).

Figure 1.
Giemsa stained karyotypes of Monacha sp. (Mo.sp.), Trochoidea elegans (Tr.e.), T. pyramidata (Tr.p.), T. trochoides (Tr.t.), Cernuella virgata (Ce.v), Cochlicella acuta (Co.a.).
The three studied Trochoidea species (T. elegans, T. pyramidata, and T. trochoides) all showed a karyotype of 2n = 48, with mostly biarmed chromosomes and the first three pairs distinctively larger than the other pairs. Nevertheless, the three Trochoidea species studied showed a distinct chromosome morphology. In T. elegans, the pairs 1, 8–9, 12, 16–17, 21 are submetacentric, the pairs 11 and 23 are telocentric while all the other pairs are metacentric (Figure 1; Table S2). In Trochoidea pyramidata the pairs 1, 5 and 11 are submetacentric while all the other pairs are metacentric (Figure 1; Table S2). In T. trochoides, the pairs 1, 3, 6, 8, 11 are submetacentric, the pair 24 is telocentric and the remaining pairs are metacentric (Figure 1; Table S2).
3.2.2. Family Geomitridae
Metaphase plates were obtained from specimens of Cernuella virgata, while only haploid plates were obtained from Cochlicella acuta. Cernuella virgata has a karyotype of 2n = 52 with all metacentric chromosome pairs, except for the pair 13 and 15 which are submetacentric (Figure 1, Table S2). Cochlicella acuta showed a karyotype of n = 26 elements; the chromosomes 7–8, 12, 16, 17 and 23 are submetacentric, chromosome 22 is subtelocentric, chromosome 10 is telocentric while all the remaining elements are metacentric (Figure 1, Table S2).
3.2.3. Family Helicidae
The eight studied species of Helicidae showed karyotypes from 2n = 52 to 2n = 60 chromosomes. Variability in the chromosome number was observed both among and within the studied subfamilies and tribes (Table 1; Figure 2 and Figure 3).

Figure 2.
Giemsa-stained karyotypes of Eobania vermiculata (Eo.v.), Otala lactea (Ot.l.), Cantareus apertus (Ca.a.), Erctella mazzullii (Er.m.), Theba pisana from Fusaro lake (Th.p. A), Theba pisana from Messina (Th.p. B).
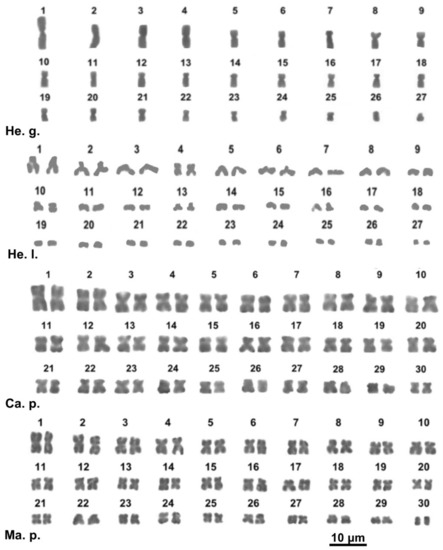
Figure 3.
Giemsa-stained karyotypes of Helix gussoneana (He. g.), H. lucorum. (He. l.), Campylaea planospira (Ca. p.), Marmorana platychela (Ma. p.).
Cornu apertus and Erctella mazzullii showed a karyotype of 2n = 54 with mostly metacentric chromosomes, excluding pairs 16 and 20 of E. mazzullii and pairs 5, 9, 18, 23 and 25 of C. apertus that are submetacentric (Figure 2, Table S2). The elements of the first pair were about 50% larger than those of pairs 2 and the remaining pairs gradually decreased in length (Figure 2; Table S2). The other two examined Otalini species, Eobania vermiculata and Otala lactea, have a karyotype of 2n = 52 chromosomes. In E. vermiculata the pairs 12 and 22 are submetacentric while all the remaining pairs are metacentric (Figure 2; Table S2). In O. lactea the pairs 8, 9, 12, 17 and 20 are submetacentic, the last pair is telocentric, and all the remaining pairs are metacentric (Figure 2, Table S2). Both species have the chromosomes of the first pair being about 1.8 times larger than the second one, while the remaining pairs gradually decrease in length (Figure 2, Table S2).
The Theba pisana specimens from Messina and Fusaro lake showed a karyotype of 2n = 60 with all metacentric chromosomes, gradually decreasing in length (Figure 2, Table S2).
The studied specimen of Helix gussoneana showed a karyotype of 2n = 54 chromosomes, of which the pairs 7, 12, 14, and 17 are submetacentric, the pairs 13 and 20 are subtelocentric, and all the other pairs are metacentric (Figure 3, Table S2). Helix lucorum only exhibited haploid plates with n = 27, 10 metacentric chromosomes (1, 4, 10, 11, 14, 16, 21, 22 and 26, 27), one submetacentric (pair 2) and 16 telocentric elements (chromosome 3, 5–9, 12, 13, 15, 17–20 and 23–25) (Figure 3, Table S2).
Campylaea planospira (Ariantinae) and Marmorana platychela (Murellinae) show a karyotype of 2n = 60 chromosomes gradually decreasing in length; chromosomes of the two species are mostly metacentric, excluding one pair (27) of C. planospira and two pairs (23 and 27) of M. platychela that are submetacentric (Figure 3, Table S2).
3.3. Chromosome Banding and NOR-FISH
Chromosome banding methods were performed on samples showing an adequate number of metaphase plates, namely Cernuella virgata, Cornu apertus, Otala lactea, Eobania vermiculata, and Theba pisana.
Quinacrine stained uniformly all the chromosomes of all the analyzed taxa (Figure 4 and Figure 5). Staining with CMA3/MG evidenced two loci in Cernuella virgata, differentially highlighted on interstitial regions of two medium-sized chromosomes (Figure 4B). Six loci were CMA3/MG positive on metaphase plates of Cornu apertus and Otala lactea, two on telomeric regions of one of longest pairs (Figure 4B), while the other four loci showed an interstitial position on two pairs of medium-sized chromosomes (Figure 4H). In Eobania vermiculata and Theba pisana, CMA3/MG uniformly stained all chromosome pairs (Figure 5B,G,L).
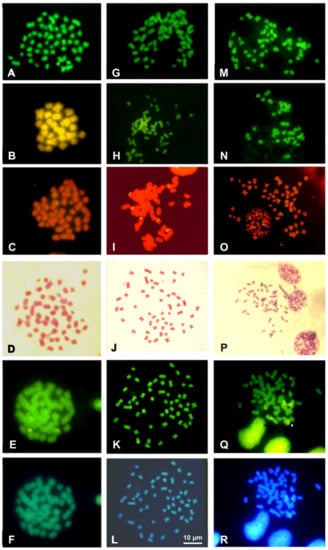
Figure 4.
Metaphase plates of Cernuella virgata (A–F), Cantareus apertus (G–L) and Otala lactea (M–R) stained with Quinacrine (A,G,M), CMA3/MG (B,H,N), NOR-FISH (C,I,O), C-banding Giemsa (D,J,P), sequential C-banding + CMA3 (E,K,Q) + DAPI (F,L,R).
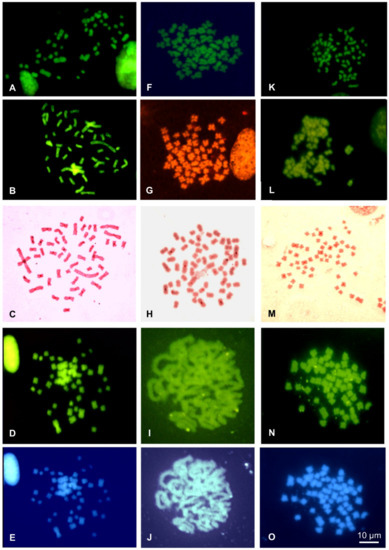
Figure 5.
Metaphase plates of E. vermiculata (A–E), Theba pisana from Naples (F–J) and Theba pisana from Messina (K–O) stained with Quinacrine (A,F,K), CMA3/MG (B,G,L), C-banding Giemsa (C,H,M) and sequential C-banding + CMA3 (D,I,N) + DAPI (E,J,O).
Successful NOR-FISH staining was obtained on metaphase plates of Cernuella virgata, Otala lactea, and Cornu apertus, with hybridization signals distributed on one (C. virgata), three (O. lactea), or four pairs (C. apertus) of medium-sized chromosomes. CMA3/MG staining evidenced positive loci overlapping with those evidenced from NOR-FISH (Figure 4C,I,O). After C-banding, the five considered species showed tiny C-bands on centromeric regions of different chromosome pairs (Figure 4F,J and Figure 5C,H,M). The centromeric C-bands were Q- and DAPI negative (Figure 4E,F,K,L and Figure 5D,E,I,J,N,O), while C-banding and CMA3 performed on metaphase plates of Cernuella virgata, Otala lactea, and Cornu apertus evidenced positive loci overlapping with those highlighted with CMA3/MG staining and NOR-FISH. C-banding and CMA3 evidenced multiple pairs (2–3) showing regions positive to this fluorochromes in Eobania vermiculata and Theba pisana (Figure 4E,K and Figure 5D,I,N). The two examined populations of T. pisana showed very similar patterns of NORs and heterochromatin distribution (Figure 5).
4. Discussion
4.1. Molecular Analysis and Taxonomic Attribution
Searches in GenBank using the newly determined 16S rDNA sequences largely corroborated the preliminary taxonomic attribution of the study samples based on conchological and anatomical characters (Table 1). However, some considerations deserve consideration, such as the nucleotide diversification of the 16S sequences of the examined specimens of T. pyramidata from Capri (Naples, Italy) and the Monacha specimen from Portici (Naples, Italy), compared to the most similar homologous sequences deposited in GenBank. Concerning T. pyramidata, searches in GenBank showed an identity score of 76.3% with a specimen from Djebal Recas, (Tunisia) (AN: KY747545, [53]); 93.5% with a specimen from San Giusto, (Siena, Italy) (AN: AY741444, [54]); 92.5% with a specimen from Siena (Italy) (AN: KU521590, [55]); 93.5% with a specimen from Cala de la Mosca, Alicante (Spain) (AN: KJ458565, [9]) and 88.3% with a specimen from St. Maximin (France) (AN: AY546377, [56]). Interestingly, the 16S rRNA sequences of the two populations from Siena (Italy) show an uncorrected p-distance of 6.5%, highlighting that these populations probably require a taxonomic revision based on a comprehensive taxon sampling of their geographic distribution.
Concerning the Monacha specimen from Portici (Naples), it was initially attributed to M. cartusiana based on morphological characters, but the molecular analysis did not support this preliminary determination. In fact, the comparison of homologous 16S sequences deposited in GenBank shows that specimen of the Monacha here studied showed identity scores ranging from 77% to 83.5% with available specimens of M. cartusiana, about 85% with Russian or Lebanese species (M. ciscaucasica, M. roseni, M. nummus) (AN: KX495397, KX495386, KX495427, [57]), and 85.9% with the populations from Siciliy (Italy) of Monacha sp.1 (KX495425, [57]) (see also 16S distance matrix provided in Table S3). Furthermore, available genetic data suggest that M. cartusiana is genetically quite uniform, with populations from Tuscany (AN: AY741416; [54]) and Lombardy (AN: KX495378; [57]) presenting 100% identity in the 16S which, in turn, have 97% identity with Central European populations (e.g., AN: KM247391, MH204083; [58,59]). Considering the above reported molecular evidence, we here consider the specimen from Portici as a new Monacha candidate species, whose taxonomy and phylogenetic relationships have to be better assessed in more focused studies.
4.2. Chromosome Analysis and Karyotype Diversity
In this study, we performed an original molecular and chromosome analysis on different Helicoidea species and a review of all the available karyotype data on the superfamily, providing an updated taxonomic evaluation of the species so far studied (see Table S1).
We here provide for the first time karyological data on seven species of the Helicoidea superfamily: Monaca sp., Trochoidea elegans, Trochoidea pyramidata, Trochoidea trochoides, Campylaea planospira, Helix gussoneana, and Marmorana platychela. We also described the chromosomal formula of Cernuella virgata, Helix lucorum, Otala lactea, Theba pisana and Cochlicella acuta, for which only the chromosome numbers have previously been reported [25,60,61,62], (Figure 1, Figure 2 and Figure 3, Table S1). However, concerning Cochlicella acuta, our results disagree with the chromosome number previously provided by Aparicio [25]. In fact, the specimens examined by us had 2n = 52 chromosomes, while Aparicio [25] found a karyotype of 2n = 46 elements in specimens from Puerto de Vega (Asturias, Spain). The 16S rRNA sequence of the specimens here studied by us shows 99.1% and 97.6% identity with homologous traits of specimens of C. acuta from Siena and Lampedusa (Italy) (AN: AY741442 and AY741443; [54]), respectively, and 95.2% with a specimen from Bakio, Biscay (Spain) (AN: KJ458503; [9]). Unfortunately, no DNA sequences are currently available from the specimens studied in Aparicio [25]. However, considering also their very different karyotype formulae, different Mediterranean populations of C. acuta may belong to independent evolutionary lineages and their taxonomy should be better assessed by further molecular studies. Furthermore, our results confirm the chromosome number and morphology of Cornu apertus, Erctella mazzullii, and Eobania vermiculata already described in [63,64] for Sicilian specimens of these three species.
Overall, the chromosome number of the studied species ranges from 2n = 44 to 2n = 60, highlighting a significant karyological diversity in the study taxa, in line with the range from 2n = 42 to 2n = 62 so far known in Helicoidea [16] (see also Table S1). Concerning the chromosome morphology, most of the studied species have karyotypes typically containing meta- and submetacentric chromosomes, a characteristic which is commonly found in Eupulmonata [16]. However, Helix lucorum shows a karyotype (2n = 54) with 16 telocentric pairs (Figure 3). Although uncommon in Helicoidea, this karyological characteristic is not exclusive of H. lucorum, as karyotypes with a relatively high number of telocentric elements are showed by three Bradybaeninae, namely Acusta ravida (2n = 58), Cathaica fasciola (2n = 60), and Bradybaena similaris (2n = 56), with 7, 22 and 26 telocentric pairs, respectively [16,23,65,66] (see Table S1). Furthermore, in Alopiinae, Medora sp. shows a karyotype (2n = 62) with 11 telocentric pairs [31].
Variations in chromosome number and morphology in the Helicoidea superfamily, and more in general in Eupulmonata, are considered taxonomically relevant and have been highly debated in past studies, with some authors suggesting a progressive reduction of the chromosome number [9,67], while others supporting the opposite hypothesis [16,60]. In this regard, to evaluate the evolutionary trends of karyotype variations in the studied taxa, we superimposed on the evolutionary relationships of the Helicoidea of the Western Palearctic [9,10,14] all the available chromosome data as listed in Table S1, with an updated nomenclature following WoRMS [1] and MolluscaBase [2] (Figure 6).
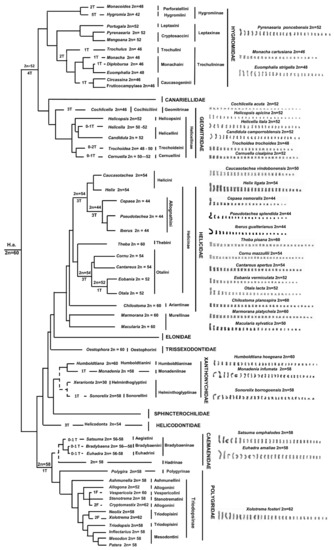
Figure 6.
Phylogenetic tree of Helicoidea (redrawn from [9]) superimposed with our hypothesis on chromosome diversification in Helicoidea. Evolutionary relationships of Hygromiidae, Polygiridae, and Ariantine are from [13,14]. T = translocation; F = Fission. Dashed lines = uncertain relationships. Haploid karyotypes were redrawn from figures presented in the original papers (for References see Table S1; in particular, for Cepaea nemoralis, for which two formulas are given, the presented haploid karyotype is from [24]).
In our hypothesis, accounting for both chromosome number and morphology, we considered a karyotype composed of 2n = 60 as the putative ancestral condition in Helicoidea (Figure 6). This assumption is based on two main considerations: (i) this karyotype is conserved in different families and subfamilies without any noticeable modification; (ii) the most parsimonious hypothesis on chromosomal diversification in the superfamily (with a lower number of chromosome rearrangement per lineage) should account for an overall reduction of the chromosome number from 2n = 60 to 2n = 42. This probably occurred by means of multiple independent, tandem fusions/translocation in different evolutionary lineages. Furthermore, while the putative ancestral karyotype of 2n = 60 shows a conserved morphology in the Helicoidea phylogeny, with mostly metacentric elements gradually decreasing in length (e.g., Marmorana platychela and Theba pisana, present study), distinctively larger pairs are clearly visible in karyotypes with a relatively low chromosome count (2n = 44–42) (e.g., Cepaea or Iberus species, reference in [16], as a clear result of a progressive accumulation of translocations). On the other hand, a general tendency toward an overall decrease in the chromosome number has been hypothesized also in other Gastropoda (e.g., Opisthobranchia and Cephalaspidea [16]), thus possibly representing a significant chromosomal evolutionary trajectory of several groups.
In particular, in Helicoidea, the karyotype of the common ancestor of the clade, including Hygromiidae and Geomitridae (2n = 52), was likely shaped by four chromosome translocations. In the former family, the Leptaxinae inherited this ancestral condition, while in the Hygromiinae, two and five translocations would have produced the karyotype of 2n = 48 in Perforatellini and 2n = 42 in Hygromiini, respectively. In Trochulininae, most species have a karyotype of 2n = 46, which probably originated from the ancestral 2n = 52 by means of three translocations. In Geomitridae, most species of the different subfamilies and tribes show a conserved karyotype of 2n = 52, except for Trochodeini, which shows karyotypes of 2n = 50 (Xerograssa) and 2n = 48 (Trochoidea), which probably originated from one and two translocations, respectively.
The putative primitive Helicoidea karyotype of 2n = 60 is conserved in Trissexodontidae, Xanthonychidae, and some taxa of Helicidae, namely tribes Ariantinae and Thebini, the Murellinae Marmorana platychela and the Helicini Caucasotachea leucoranea. Several species of Otalini and most Helicini have 2n = 52–54, while all Allognathini have 2n = 44, so their karyotype could have been originated from the ancestral 2n = 60 condition by means of a progressive accumulation (three to eight) of translocations. Notably, in Helicidae, the species of several tribes of Helicinae have the chromosomes of pair 1 distinctively larger than the elements of pair 2, (Figure 2, Figure 3 and Figure 6; Table S2), suggesting that the pair 1 was a preferential site for translocations occurred during the transition from 2n = 60 to 2n = 44. This condition is present also in Macularia sylvatica (Murellinae) (see also [16]), suggesting that similar processes occurred independently in different taxonomic groups.
In Helicontidae, the karyotype of 2n = 54 of Helicodonta obvoluta originated from three translocations, while in the clade including Polygeridae and Camaenidae, a single translocation originated the karyotype of 2n = 58 of their common ancestor, which is conserved in most of the 80 Camaenidae and the about 50 Polygeridae species so far analysed. The few exceptions are represented by some Bradybaeninae, whose karyotype of 2n = 56 likely originated by means of one translocation event, while the karyotype of 2n = 60 of Cathaica fasciola probably originated from one fission. Among Polygyridae, the putative ancestral karyotype of the family (2n = 58) is conserved in most studied species, and deviations from this condition concern either a reduction (2n = 52, two Allogona species) or an increase in the chromosome number (2n = 60 in Vespericola columbiana, and 2n = 62 in Cryptomastix germana, Xolotrema fosteri and Triodopsis fraudolenta), involving a progressive accumulation of chromosome translocations and fissions, respectively.
Besides translocation and rare fission events, the available data suggest that also chromosome inversions were relevant to chromosome rearrangements in the karyotype diversification of the Helicoidea. In fact, a progressive accumulation of chromosome inversions explains the differences in the overall karyotype morphology exhibited in different Bradybaeninae genera (e.g., Acusta and Fruticicola, [16]), Trochoidea (2n = 48) and Helix (2n = 54) (present study). In Trochoidea, three and four inversions occurred from the karyotype of T. pyramidata (20 m, 4 sm) to those of T. trochoides (16 m, 7 sm, 1 t) and T. elegans (16 m, 6 sm, 2 t), respectively (Figure 1; Table S1). In Helix, a progressive accumulation of three, six, and sixteen inversions likely occurred from the karyotype of H. straminea (24 m, 2 sm, 1st) [68] to those of H. gussoneana (21 m, 4 sm, 2st), H. pomatia (18 m, 8 sm, 1st), and H. lucorum (10 m, 1 sm, 16 t), respectively (Figure 3; Table S1).
Loci of NORs are generally considered useful taxonomic and phylogenetic markers [69,70,71]. Their localization on a single chromosome pair is considered a primitive character in molluscs, while their occurrence on multiple pairs is regarded as a derived state [16,30,70,72,73,74,75]. Both conditions are present in Helicoidea, but the available data are still scarce to draw phylogenetic considerations. Loci of NORs are on a single pair in two Geometridae species (Helicella virgata, [76]; Cernuella cisalpina, this study) and the polygyrid Xolotrema fosteri [77] and on multiple chromosome pairs in Helicidae (five species of Otalini and Theba pisana; [64], this study) (Table S1). Similarly, studies concerning the location and composition of heterochromatin in Helicoidea concern only one species of Geomitridae and six Helicidae species (see also [63,64]) (Table S1). In these species, tiny heterochromatic C-bands are prevalently localized on centromeric and pericentromeric regions, resulting in Q and DAPI being negative and suggesting a very limited presence of A-T rich clusters [78]. In contrast, C-banding and CMA3 highlighted NOR-associated heterochromatin, which is notoriously rich in G-C [30,79,80,81].
5. Conclusions
We here provide new molecular and cytogenetic data on 15 Helicoidea (Eupulmonata) species and a synthesis on all the available karyological data on the superfamily. The newly generated cytogenetic data include four Hygromiidae, two Geometridae, and nine Helicidae, which show a significant chromosome diversity with karyotypes ranging from 2n = 44 to 2n = 60. Considering the available karyological and phylogenetic data, we hypothesize a karyotype of 2n = 60 with all biarmed elements gradually deceasing in length as the ancestral condition in the superfamily Helicoidea. A reduction of the chromosome number, by means of a progressive accumulation of chromosome translocations, led to the formation of karyotypes with a lower chromosome number (to 2n = 50–44). This process occurred multiple times and independently among different evolutionary lineages, while the opposite process, an augment of the total chromosome count by means of chromosome fissions, is detectable in Polygyridae. Other than translocations and rare fissions, chromosome inversions were relevant to chromosome rearrangements in Helicoidea, leading to the formation of telocentric elements in karyotypes with a relatively low chromosome count.
Supplementary Materials
The following are available online at www.mdpi.com/article/10.3390/ani11092551/s1. Table S1. Available karyological data on Helicoidea; Table S2. Relative length (RL) and Centromeric Index (CI) of the studied taxa. Table S3. Distance Matrix of the Monacha species considered in the study.
Author Contributions
A.P., G.O., N.M. and M.M. conceived the manuscript. A.P., P.C., G.O. and N.M. gathered the studied samples. A.P., G.O., M.M., F.M.G. and O.P. performed the laboratory analyses and evaluated the results obtained. All the authors contributed to the final manuscript version. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Università degli Studi di Napoli Federico II: 000020_Altro2019_Direttore-Assegnazione Ricerca Dipartimentale.
Institutional Review Board Statement
The animals used in our study are non-cephalopod mollusks and as such do not fall under the law in force in Italy for the protection of animals used for scientific purposes (legislative decree N. 26, 4 March 2014) which, in turn, incorporates the European Directive 2010/63 EU L276. Samples from the Cilento, Vallo di Diano and Alburni National Park (PNCVDA) were sampled under authorization n. PNCVDA n. 16341, 19 October 2010.
Data Availability Statement
The data presented in this study are available in the manuscript and in Tables S1–S3. All the newly generated DNA sequences were submitted to GenBank (accession numbers: MZ504244-MZ504269).
Acknowledgments
We are grateful to Gennaro Aprea and Carmen D’Angelo who provided us some of the study specimens and for their help in laboratory preparations. We thank the two anonymous reviewers for their useful comments which improved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WoRMS Editorial Board. World Register of Marine Species. 2021. Available online: http://www.marinespecies.org (accessed on 30 June 2021).
- Mollusca Base. 2021. Available online: http://www.molluscabase.org (accessed on 30 June 2021).
- Scott, B. Phylogenetic relationships of the Camaenidae (Pulmonata, Stylommatophora, Helicoidea). J. Molluscan Stud. 1996, 62, 65–74. [Google Scholar] [CrossRef]
- Scott, B. Biogeography of the Helicoidea (Mollusca, Gastropoda, Pulmonata), land snails with a Pangean distribution. J. Biogeogr. 1997, 24, 399–407. [Google Scholar] [CrossRef]
- Schileyko, A.A. Treatise on recent terrestrial pulmonate molluscs. Part 12. Bradybaenidae, Monadeniidae, Xanthonychidae, Epiphragmophoridae, Helminthoglyptidae, Elonidae, Humboldtianidae, Sphincterochilidae, Cochlicellidae. Ruthenica 2004, 2, 1627–1763. [Google Scholar]
- Schileyko, A.A. Treatise on recent terrestrial pulmonate molluscs. Part 13. Helicidae, Pleurodontidae, Polygyridae, Ammonitellidae, Oreohelicidae, Thysanophoridae. Ruthenica 2006, 2, 1765–1906. [Google Scholar]
- Schileyko, A.A. Treatise on recent terrestrial pulmonate molluscs. Part 14. Helicodontidae, Ciliellidae, Hygromiidae. Ruthenica 2006, 2, 1907–2047. [Google Scholar]
- Bouchet, P.; Rocroi, J.P. Classification and nomenclature of gastropod families. Malacologia 2005, 47, 1–397. [Google Scholar]
- Razkin, O.; Gómez-Moliner, B.J.; Prieto, C.E.; Martínez-Ortí, A.; Arrébola, J.R.; Muñoz, B.; Chueca, L.J.; Madeira, M.J. Molecular phylogeny of the western Palaearctic Helicoidea (Gastropoda, Stylommatophora). Mol. Phylogenet. Evol. 2015, 83, 99–117. [Google Scholar] [CrossRef]
- Perez, K.E.; Defreitas, N.; Slapcinsky, J.; Minton, R.L.; Anderson, F.E.; Pearcet, A. Molecular phylogeny, evolution of shell shape, and DNA barcoding in Polygyridae (Gastropoda, Pulmonata), an endemic North American clade of land snails. Am. Malacol. Bull. 2014, 32, 1–31. [Google Scholar] [CrossRef]
- Köhler, F.; Criscione, F. A molecular phylogeny of camaenid land snails from north-western Australia unravels widespread homoplasy in morphological characters (Gastropoda, Helicoidea). Mol. Phylogenet. Evol. 2015, 83, 44–55. [Google Scholar] [CrossRef]
- Wade, C.M.; Mordan, P.B.; Naggs, F. Evolutionary relationships among the Pulmonate land snails and slugs (Pulmonata, Stylommatophora). Biol. J. Linn. Soc. 2016, 87, 593–610. [Google Scholar] [CrossRef]
- Caro, A.; Neiber, M.T.; Gomez-Moliner, B.J.; Madeira, M.J. Molecular phylogeny and biogeography of the land snail subfamily Leptaxinae (Gastropoda, Hygromiidae). Mol. Phylogenet. Evol. 2019, 139, 106570. [Google Scholar] [CrossRef]
- Groenenberg, D.S.J.; Subai, P.; Gittenberger, E. Systematics of Ariantinae (Gastropoda, Pulmonata, Helicidae), a new approach to an old problem. Contrib. Zool. 2016, 85, 37–65. [Google Scholar] [CrossRef]
- Tatewaki, R.; Kitada, J. Karyological studies of five species of land snails (Helicoidea, Mollusca). Genetica 2004, 74, 73–80. [Google Scholar] [CrossRef]
- Thiriot-Quiévreux, C. Advances in chromosomal studies of gastropod molluscs. J. Molluscan Stud. 2003, 69, 187–201. [Google Scholar] [CrossRef]
- Patterson, C.M. Chromosomes of molluscs. In Proceedings of the 2nd Symposium of Mollusca, Cochin, India, 12–16 January 1968; Series 3, Part 2. Marine Biological Association of India: Ernakulam, India, 1968; pp. 635–686. [Google Scholar]
- Husted, L.; Burch, P.R. The Chromosomes of Polygyrid Snails. Am. Nat. 1946, 80, 410–429. Available online: http://www.jstor.org/stable/2458185 (accessed on 18 August 2021). [CrossRef] [PubMed]
- Laws, H.M. Chromosomes of snails introduced into South Australia and the Northern Territory. Rec. Aust. Mus. 1965, 15, 79–87. [Google Scholar]
- Bantock, C.R. Localisation of chiasmata in Cepaea nemoralis L. Heredity 1972, 29, 213–221. [Google Scholar] [CrossRef][Green Version]
- Laws, H.M. The chromosomes of some Australian camaenid land snails. Cytologia 1973, 38, 229–235. [Google Scholar] [CrossRef][Green Version]
- Stern, E.M.; Metcalf, A. L Chromosome Numbers in Ashmunella (Gastropoda Eupulmonata Polygyridae). Veliger 1974, 17, 19–22. [Google Scholar]
- Babrakzai, N. Chromosomes of Helminthoglyptidae (Pulmonata: Gastropoda). Ph.D. Thesis, University of Arizona, Tucson, AZ, USA, 1975. [Google Scholar]
- Page, C. The Karyotype of the Land Snail Cepaea nemoralis (L.). Heredity 1978, 41, 321–325. [Google Scholar] [CrossRef]
- Aparicio, M.T. Cytotaxonomic studies of the family Helicidae (Gastropoda, Pulmonata). Genét. Ibér. 1981, 33, 211–224. [Google Scholar]
- Mezzasalma, M.; Guarino, F.M.; Aprea, G.; Petraccioli, A.; Crottini, A.; Odierna, G. Karyological evidence for diversification of Italian slow worm populations. Comp. Cytogenet. 2013, 7, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Mezzasalma, M.; Andreone, F.; Branch, W.R.; Glaw, F.; Guarino, F.M.; Nagy, Z.T.; Odierna, G.; Aprea, G. Chromosome evolution in pseudoxyrhophiine snakes from Madagascar, a wide range of karyotypic variability. Biol. J. Linn. Soc. 2014, 112, 450–460. [Google Scholar] [CrossRef]
- Mezzasalma, M.; Andreone, F.; Aprea, G.; Glaw, F.; Odierna, G.; Guarino, F.M. Molecular phylogeny, biogeography and chromosome evolution of Malagasy dwarf geckos of the genus Lygodactylus (Squamata, Gekkonidae). Zool. Scr. 2017, 46, 42–54. [Google Scholar] [CrossRef]
- Odierna, G.; Aprea, G.; Barucca, M.; Biscotti, M.A.; Canapa, A.; Capriglione, T.; Olmo, E. Karyology of the Antarctic chiton Nuttalochiton mirandus (Thiele, 1906) (Mollusca, Polyplacophora) with some considerations on chromosome evolution in chitons. Chromosome Res. 2008, 16, 899–906. [Google Scholar] [CrossRef]
- Petraccioli, A.; Maio, N.; Odierna, G. Chromosomes of Lepidochitona caprearum (Scacchi, 1836) (Polyplacophora, Acanthochitonina, Tonicellidae) provide insights into Acanthochitonina karyological evolution. Comp. Cytogenet. 2012, 6, 397–407. [Google Scholar] [CrossRef]
- Petraccioli, A.; Capriglione, T.; Colomba, M.; Crovato, P.; Odierna, G.; Sparacio, I.; Maio, N. Comparative cytogenetic study in four alopiinae door snails (Gastropoda, Clausiliidae). Malacologia 2015, 58, 225–232. [Google Scholar] [CrossRef]
- Fiorentino, V.; Salomone, N.; Manganelli, G.; Giusti, F. Historical biogeography of Tyrrhenian land snails, the Marmorana-Tyrrheniberus radiation (Eupulmonata, Helicidae). Mol. Phylogenet. Evol. 2010, 55, 26–37. [Google Scholar] [CrossRef]
- Fiorentino, V.; Manganelli, G.; Giusti, F.; Ketmaier, V. Recent expansion and relic survival, phylogeography of the land snail genus Helix (Mollusca, Gastropoda) from south to north Europe. Mol. Phylogenet. Evol. 2016, 98, 358–372. [Google Scholar] [CrossRef] [PubMed]
- Dayrat, B.; Conrad, M.; Balayan, S.; White, T.R.; Albrecht, C.; Golding, R.; Gomes, S.R.; Harasewych, M.G.; Martins, A.M.D.F. Phylogenetic relationships and evolution of pulmonate gastropods (Mollusca), new insights from increased taxon sampling. Mol. Phylogenet. Evol. 2011, 59, 425–437. [Google Scholar] [CrossRef]
- Giusti, F. Notulae malacologicae XVI. I molluschi terrestri e di acqua dolce viventi sul massiccio dei Monti Reatini (Appennino Centrale). Lav. Soc. Ital. Biogeog. 1971, 2, 423–576. [Google Scholar] [CrossRef][Green Version]
- Giusti, F.; Castagnolo, L. I molluschi terrestri delle dune italiane, brevi cenni di ecologia, elenco delle specie e chiavi per il loro riconoscimento. In Quaderni Sulla “Struttura delle Zoocenosi Terrestri”, 3. Ambienti Mediterranei, 1. Le Coste Sabbiose; Pubblicazioni AQ/1/174; C.N.R.: Roma, Italy, 1982; pp. 51–102. [Google Scholar]
- Giusti, F.; Castagnolo, L.; Manganelli, G. La fauna malacologica delle faggete Italiane, brevi cenni di ecologia, elenco delle specie e chiavi per il riconoscimento dei generi e delle entità più comuni. Boll. Malacol. 1985, 21, 69–144. [Google Scholar]
- Kerney, M.P.; Cameron, R.A.D. Guide des Escargots et limaces d’Europe Identification et Biologie de Plus de 300 Espèces; Delachaux et Niestlé: Paris, France, 1999. [Google Scholar]
- Gavetti, E.; Birindelli, S.; Bodon, M. Molluschi terrestri e d’acqua dolce della valle di Susa. Monogr. Mus. Reg. Sci. Nat. Torino 2008, 44, 1–273. [Google Scholar]
- Welter-Schultes, F.W. European Non-Marine Molluscs, a Guide for Species Identification; Planet Poster Editions: Göttingen, Germany, 2012. [Google Scholar]
- Maio, N.; Petraccioli, A.; Crovato, P.; Niero, I.; Pignataro, C.; Odierna, G. Guida naturalistica di campo ai Molluschi terrestri dei Monti Alburni. Biologia, ecologia, distribuzione e conservazione. PNCVDA Quad. Della Biodivers. 2016, 4, 1–354. [Google Scholar]
- Sokolov, E.P. An improved method for DNA isolation from mucopolysaccharide-rich molluscan tissues. J. Molluscan Stud. 2000, 66, 573–575. [Google Scholar] [CrossRef]
- Palumbi, S.R.; Martin, A.; Romano, S.; Mcmillan, W.O.; Stice, L.; Grabowski, G. The Simple Fool’s Guide to PCR; Version 2.0; University of Hawaii: Honolulu, HI, USA, 1991. [Google Scholar]
- Hall, T.A. BioEdit, a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Sidhom, M.; Said, K.; Chatti, N.; Guarino, F.M.; Odierna, G.; Petraccioli, A.; Picariello, O.; Mezzasalma, M. Karyological characterization of the common chameleon (Chamaeleo chamaeleon) provides insights on the evolution and diversification of sex chromosomes in Chamaeleonidae. Zoology 2020, 141, 125738. [Google Scholar] [CrossRef]
- Schmid, M. Chromosome banding in Amphibia. Constitutive heterochromatin and nucleolus organizer regions in Bufo and Hyla. Chromosoma 1978, 66, 361–388. [Google Scholar] [CrossRef]
- Sahar, E.; Latt, S.A. Energy transfer and binding competition between dyes used to enhance staining differentiation in metaphase chromosomes. Chromosoma 1980, 79, 1–28. [Google Scholar] [CrossRef]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Odierna, G.; Aprea, G.; Capriglione, T. Chromosomal and molecular analysis of some repeated families in Discoglossus Otth, 1837 (Anura, Discoglossidae), taxonomic and phylogenetic implications. Ital. J. Zool. 1999, 66, 275–283. [Google Scholar] [CrossRef][Green Version]
- Mezzasalma, M.; Visone, V.; Petraccioli, A.; Odierna, G.; Capriglione, T.; Guarino, F.M. Non-random accumulation of LINE1-like sequences on differentiated snake W chromosomes. J. Zool. 2016, 300, 67–75. [Google Scholar] [CrossRef]
- Levan, A.R.; Fredga, K.; Sandberg, A.A. Nomenclature for centromeric position on chromosomes. Hereditas 1964, 52, 201–220. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Ezzine, I.K.; Dimassi, N.; Pfarrer, B.; Said, K.; Neubert, E. New records of the endemic Sicilian land snail species Marmorana (Murella) muralis (O. F. Muller, 1774) from the north of Tunisia. (Eupulmonata, Gastropoda). Zookeys 2018, 775, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Manganelli, G.; Salomone, N.; Giusti, F. A molecular approach to the phylogenetic relationships of the western Palaearctic Helicoidea (Gastropoda, Stylommatophora). Biol. J. Linn. Soc. 2005, 85, 501–512. [Google Scholar] [CrossRef]
- Boeckers, A.; Greve, C.; Hutterer, R.; Misof, B.; Haase, M. Testing heterogeneous base composition as potential cause for conflicting phylogenetic signal between mitochondrial and nuclear DNA in the land snail genus Theba Risso 1826 (Gastropoda, Stylommatophora, Helicoidea). Org. Divers. Evol. 2016, 16, 835–846. [Google Scholar] [CrossRef]
- Steinke, D.; Albrecht, C.; Pfenninger, M. Molecular phylogeny and character evolution in the Western Palaearctic Helicidae s.l. (Gastropoda, Stylommatophora). Mol. Phylogenet. Evol. 2004, 32, 724–734. [Google Scholar] [CrossRef]
- Neiber, M.T.; Hausdorf, B. Phylogeography of the land snail genus Circassina (Gastropoda, Hygromiidae) implies multiple Pleistocene refugia in the western Caucasus region. Mol. Phylogenet. Evol. 2015, 93, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Pienkowska, J.R.; Manganelli, G.; Giusti, F.; Lesicki, A. Monacha claustralis (Rossmässler, 1834) new to Polish and Czech malacofauna (Gastropoda, Eupulmonata, Hygromiidae). J. Conchol. 2015, 42, 79–93. [Google Scholar]
- Pienkowska, J.R.; Prockow, M.; Gorka, M.; Lesicki, A. Distribution of Monacha claustralis (Rossmässler, 1834) and M. cartusiana (O. F. Mueller, 1774) (Eupulmonata, Hygromiidae) in central European and Balkan countries, new data. Folia Malacol. 2018, 26, 103–120. [Google Scholar] [CrossRef][Green Version]
- Patterson, C.M.; Burch, J.B. Chromosomes of Pulmonated Molluscs. In Pulmonates; Systematics, Evolution and Ecology; Fretter, V., Peake., J., Eds.; Academic Press: New York, NY, USA, 1978; Volume 2A, pp. 171–217. [Google Scholar]
- Aparicio, M.T. The chromosomes of eight species of the subfamily Helicinae (Gastropoda, Pulmonata, Helicidae) from Spain. Malacol. Rev. 1983, 16, 71–78. [Google Scholar]
- Bakhtadze, N.G.; Chakvetadze, N.L.; Mumladze, L.J.; Bakhtadze, G.I.; Tskhadaia, E.A. Karyological data of terrestrial mollusks (Mollusca: Gastropoda: Pulmonata) of Georgia. Proc. Inst. Zool. 2014, 25, 23–27. [Google Scholar] [CrossRef]
- Vitturi, R.; Rasotto, M.B.; Farinella-Ferruzza, N. The chromosomes of 16 molluscan species. Boll. Zool. 1982, 49, 61–71. [Google Scholar] [CrossRef]
- Vitturi, R.; Libertini, A.; Sineo, L.; Sparacio, I.; Lannino, A.; Gregorini, A.; Colomba, M. Cytogenetics of the land snails Cantareus aspersus and C. mazzullii (Mollusca, Gastropoda, Eupulmonata). Micron 2005, 36, 351–357. [Google Scholar] [CrossRef]
- Sun, T. Chromosomal studies in three land snails. Sinozoologia 1995, 12, 154–162. [Google Scholar]
- Babrakzai, N.; Samsam, S.; Miller, W.B. Cytological Relationships of Helminthoglyptidae and Bradybaenidae (Mollusca, Pulmonata). In Proceedings of the Twenty-Fourth Annual Meeting of the Arizona-Nevada Academy of Science and the Fifty-Sixth Annual Meeting Southwestern and Rocky Mountain Division American Association for the Advancement of Science, Las Vegas, NV, USA, 9–12 April 1980; p. 31. [Google Scholar]
- Rainer, M. Chromosomenuntersuchungen an Gastropoden (Stylommatophora). Malacologia 1967, 5, 341–373. [Google Scholar]
- Petraccioli, A.; Crovato, P.; De Vico, G.; Odierna, G.; Picariello, O.L.A.; Tardy, E.; Viglietti, S.; Guarino, F.M.; Maio, N. Helix straminea Briganti, 1825 in Italy (Gastropoda, Pulmonata), taxonomic history, morphology, biology, distribution and phylogeny. Eur. Zool. J. 2021, 88, 390–416. [Google Scholar] [CrossRef]
- Odierna, G.; Olmo, E.; Cobror, O. Taxonomic implications of NOR-localization in lacertid lizards. Amphib. Reptil. 1987, 87, 373–382. [Google Scholar]
- Carvalho, K.A.; Garcia, P.C.A.; Recco-Pimentel, S.M. NOR dispersion, telomeric sequence detection in centromeric regions and meiotic multivalent configurations in species of the Aplastodiscus albofrenatus group (Anura, Hylidae). Cytogenet. Genome Res. 2009, 126, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Britton-Davidian, J.; Cazaux, B.; Catalan, J. Chromosomal dynamics of nucleolar organizer regions (NORs) in the house mouse, micro-evolutionary insights. Heredity 2012, 108, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Thiriot-Quiévreux, C. Review of the literature on bivalve cytogenetics in the last ten years. Cah. Biol. Mar. C 2002, 43, 17–26. [Google Scholar]
- Wang, Y.; Guo, X. Chromosomal rearrangement in Pectinidae revealed by rRNA loci and implications for bivalve evolution. Biol. Bull. 2004, 207, 247–256. [Google Scholar] [CrossRef]
- Colomba, M.S.; Gregorini, A.; Liberto, F.; Reitano, A.; Giglio, S.; Sparacio, I. Monographic revision of the endemic Helix mazzullii De Cristofori & Jan, 1832 complex from Sicily and re-introduction of the genus Erctella Monterosato, 1894 (Pulmonata, Stylommatophora, Helicidae). Zootaxa 2011, 3134, 1–42. [Google Scholar] [CrossRef]
- Colomba, M.S.; Vitturi, R.; Rampin, M.; Lannino, A.; Taravella, A.; Libertini, A. Comparative cytogenetic analysis of three Stylommatophoran slugs (Mollusca, Pulmonata). Malacologia 2009, 51, 173–179. [Google Scholar] [CrossRef]
- Vitturi, R.; Colombera, D.; Catalano, E.; Amico, F.P. Spermatocyte chromosome analysis of Helicella virgata (Eupulmonata, Helicidae). J. Hered. 1991, 82, 339–343. [Google Scholar] [CrossRef]
- Diopotex-Chong, M.E.; Babrazkai, N. Preliminary cytological studies in Triodopsis fosteri (Baker, 1920) Mollusca-Pulmonata Polygiridae. An. Inst. Cienc. Mar Limnol. 1993, 19, 13–18. [Google Scholar]
- Schweizer, D. Counterstain-enhanced chromosome banding. Hum. Genet. 1981, 57, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Odierna, G.; Aprea, G.; Barucca, M.; Canapa, A.; Capriglione, T.; Olmo, E. Karyology of the Antarctic scallop Adamussium colbecki, with some comments on the karyological evolution of pectinids. Genetica 2006, 127, 341–349. [Google Scholar] [CrossRef] [PubMed]
- Odierna, G.; Aprea, G.; Barucca, M.; Canapa, A.; Capriglione, T.; Olmo, E. The karyology of the Antarctic whelk, Neobuccinum eatoni (Mollusca, Neogastropoda). Ital. J. Zool. 2006, 73, 303–308. [Google Scholar] [CrossRef]
- Petraccioli, A.; Guarino, F.M.; Maio, N.; Odierna, G. Molecular cytogenetic study of three common Mediterranean limpets, Patella caerulea, P. rustica and P. ulyssiponensis (Archaeogastropoda, Mollusca). Genetica 2010, 138, 219–225. [Google Scholar] [CrossRef] [PubMed][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).